Abstract
Many networks of interest in the sciences, including social networks, computer networks, and metabolic and regulatory networks, are found to divide naturally into communities or modules. The problem of detecting and characterizing this community structure is one of the outstanding issues in the study of networked systems. One highly effective approach is the optimization of the quality function known as “modularity” over the possible divisions of a network. Here I show that the modularity can be expressed in terms of the eigenvectors of a characteristic matrix for the network, which I call the modularity matrix, and that this expression leads to a spectral algorithm for community detection that returns results of demonstrably higher quality than competing methods in shorter running times. I illustrate the method with applications to several published network data sets.
Keywords: clustering, partitioning, modules, metabolic network, social network
Many systems of scientific interest can be represented as networks, sets of nodes or vertices joined in pairs by lines or edges. Examples include the internet and the worldwide web, metabolic networks, food webs, neural networks, communication and distribution networks, and social networks. The study of networked systems has a history stretching back several centuries, but it has experienced a particular surge of interest in the last decade, especially in the mathematical sciences, partly as a result of the increasing availability of accurate large-scale data describing the topology of networks in the real world. Statistical analyses of these data have revealed some unexpected structural features, such as high network transitivity (1), power-law degree distributions (2), and the existence of repeated local motifs (3); see refs. 4–6 for reviews.
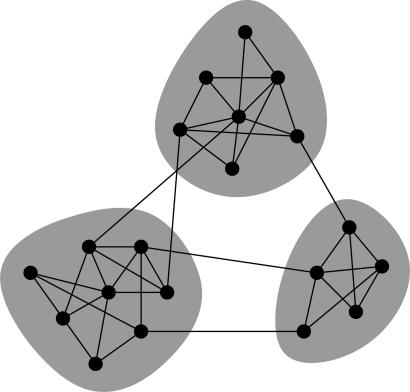
One issue that has received a considerable amount of attention is the detection and characterization of community structure in networks (7, 8), meaning the appearance of densely connected groups of vertices, with only sparser connections between groups (Fig. 1). The ability to detect such groups could be of significant practical importance. For instance, groups within the worldwide web might correspond to sets of web pages on related topics (9); groups within social networks might correspond to social units or communities (10). Merely the finding that a network contains tightly knit groups at all can convey useful information: if a metabolic network were divided into such groups, for instance, it could provide evidence for a modular view of the network's dynamics, with different groups of nodes performing different functions with some degree of independence (11, 12).
Fig. 1.
The vertices in many networks fall naturally into groups or communities, sets of vertices (shaded) within which there are many edges, with only a smaller number of edges between vertices of different groups.
Past work on methods for discovering groups in networks divides into two principal lines of research, both with long histories. The first, which goes by the name of graph partitioning, has been pursued particularly in computer science and related fields, with applications in parallel computing and integrated circuit design, among other areas (13, 14). The second, identified by names such as block modeling, hierarchical clustering, or community structure detection, has been pursued by sociologists and more recently by physicists, biologists, and applied mathematicians, with applications especially to social and biological networks (7, 15, 16).
It is tempting to suggest that these two lines of research are really addressing the same question, albeit by somewhat different means. There are, however, important differences between the goals of the two camps that make quite different technical approaches desirable. A typical problem in graph partitioning is the division of a set of tasks between the processors of a parallel computer so as to minimize the necessary amount of interprocessor communication. In such an application the number of processors is usually known in advance and at least an approximate figure for the number of tasks that each processor can handle. Thus we know the number and size of the groups into which the network is to be split. Also, the goal is usually to find the best division of the network regardless of whether a good division even exists; there is little point in an algorithm or method that fails to divide the network in some cases.
Community structure detection, by contrast, is perhaps best thought of as a data analysis technique used to shed light on the structure of large-scale network data sets, such as social networks, internet and web data, or biochemical networks. Community structure methods normally assume that the network of interest divides naturally into subgroups and the experimenter's job is to find those groups. The number and size of the groups are thus determined by the network itself and not by the experimenter. Moreover, community structure methods may explicitly admit the possibility that no good division of the network exists, an outcome that is itself considered to be of interest for the light it sheds on the topology of the network.
This article focuses on community structure detection in network data sets representing real-world systems of interest. However, both the similarities and differences between community structure methods and graph partitioning will motivate many of the developments that follow.
The Method of Optimal Modularity
Suppose then that we are given, or discover, the structure of some network and that we want to determine whether there exists any natural division of its vertices into nonoverlapping groups or communities, where these communities may be of any size.
Let us approach this question in stages and focus initially on the problem of whether any good division of the network exists into just two communities. Perhaps the most obvious way to tackle this problem is to look for divisions of the vertices into two groups so as to minimize the number of edges running between the groups. This “minimum cut” approach is the approach most often adopted in the graph-partitioning literature. However, as discussed above, the community structure problem differs crucially from graph partitioning in that the sizes of the communities are not normally known in advance. If community sizes are unconstrained then we are, for instance, at liberty to select the trivial division of the network that puts all of the vertices in one of our two groups and none in the other, which guarantees we will have zero intergroup edges. This division is, in a sense, optimal, but clearly it does not tell us anything of any worth. We can, if we want, artificially forbid this solution, but then a division that puts just one vertex in one group and the rest in the other will often be optimal, and so forth.
The problem is that simply counting edges is not a good way to quantify the intuitive concept of community structure. A good division of a network into communities is not merely one in which there are few edges between communities; it is one in which there are fewer than expected edges between communities. If the number of edges between two groups is only what one would expect on the basis of random chance, then few thoughtful observers would claim this constitutes evidence of meaningful community structure. On the other hand, if the number of edges between groups is significantly less than we expect by chance, or equivalent if the number within groups is significantly more, then it is reasonable to conclude that something interesting is going on.
This idea, that true community structure in a network corresponds to a statistically surprising arrangement of edges, can be quantified by using the measure known as modularity (17). The modularity is, up to a multiplicative constant, the number of edges falling within groups minus the expected number in an equivalent network with edges placed at random. (A precise mathematical formulation is given below.)
The modularity can be either positive or negative, with positive values indicating the possible presence of community structure. Thus, one can search for community structure precisely by looking for the divisions of a network that have positive, and preferably large, values of the modularity (18).
The evidence so far suggests that this approach, of looking for divisions with high modularity, is a very effective way to tackle the problem. For instance, Guimerà and Amaral (12) and later Danon et al. (8) optimized modularity over possible partitions of computer-generated test networks by using simulated annealing. In direct comparisons using standard measures, Danon et al. found that this method outperformed all other methods for community detection of which they were aware, in most cases by an impressive margin. On the basis of such results we consider maximization of the modularity to be perhaps the definitive current method of community detection, being at the same time based on sensible statistical principles and highly effective in practice. Unfortunately, optimization by simulated annealing is not a workable approach for the large network problems facing today's scientists, because it demands too much computational effort. A number of alternative heuristic methods have been investigated, such as greedy algorithms (18) and extremal optimization (19). Here we take a different approach based on a reformulation of the modularity in terms of the spectral properties of the network of interest.
Suppose our network contains n vertices. For a particular division of the network into two groups let si = 1 if vertex i belongs to group 1 and si = −1 if it belongs to group 2. And let the number of edges between vertices i and j be Aij, which will normally be 0 or 1, although larger values are possible in networks where multiple edges are allowed. (The quantities Aij are the elements of the so-called adjacency matrix.) At the same time, the expected number of edges between vertices i and j if edges are placed at random is kikj/2m, where ki and kj are the degrees of the vertices and m = ½Σi ki is the total number of edges in the network. Thus the modularity Q is given by the sum of Aij − kikj/2m over all pairs of vertices i, j that fall in the same group.
Observing that the quantity ½(sisj + 1) is 1 if i and j are in the same group and 0 otherwise, we can then express the modularity as
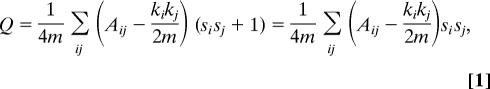 |
where the second equality follows from the observation that 2m = Σi ki = Σij Aij. The leading factor of 1/4m is merely conventional: it is included for compatibility with the previous definition of modularity (17).
Eq. 1 can conveniently be written in matrix form as
where s is the column vector whose elements are the si and we have defined a real symmetric matrix B with elements
which we call the modularity matrix. Much of our attention in this article will be devoted to the properties of this matrix. For the moment, note that the elements of each of its rows and columns sum to zero, so that it always has an eigenvector (1,1,1, …) with eigenvalue zero. This observation is reminiscent of the matrix known as the graph Laplacian (20), which is the basis for one of the best-known methods of graph partitioning, spectral partitioning (21, 22), and has the same property. And indeed, the methods presented here have many similarities to spectral partitioning, although there are some crucial differences as well, as we will see.
Given Eq. 2, we proceed by writing s as a linear combination of the normalized eigenvectors ui of B so that s = Σi=1n aiui with ai = uiT·s. Then we find
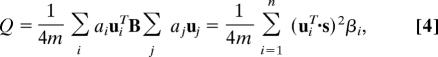 |
where βi is the eigenvalue of B corresponding to eigenvector ui.
Let us assume that the eigenvalues are labeled in decreasing order, β1 ≥ β2 ≥ … ≥ βn. We want to maximize the modularity by choosing an appropriate division of the network, or equivalently by choosing the value of the index vector s. This means choosing s so as to concentrate as much weight as possible in terms of the sum in Eq. 4 involving the largest (most positive) eigenvalues. If there were no other constraints on our choice of s (apart from normalization), this would be an easy task: we would simply chose s proportional to the eigenvector u1. This places all of the weight in the term involving the largest eigenvalue β1, the other terms being automatically zero, because the eigenvectors are orthogonal.
Unfortunately, there is another constraint on the problem imposed by the restriction of the elements of s to the values ±1, which means s cannot normally be chosen parallel to u1. Let us do our best, however, and make it as close to parallel as possible, which is equivalent to maximizing the dot product u1T·s. It is straightforward to see that the maximum is achieved by setting si = +1 if the corresponding element of u1 is positive and si = −1 otherwise. In other words, all vertices whose corresponding elements are positive go in one group and all of the rest in the other. This then gives us the algorithm for dividing the network: we compute the leading eigenvector of the modularity matrix and divide the vertices into two groups according to the signs of the elements in this vector.
We immediately notice some satisfying features of this method. First, as has been made clear, it works even though the sizes of the communities are not specified. Unlike conventional partitioning methods that minimize the number of between-group edges, there is no need to constrain the group sizes or artificially forbid the trivial solution with all vertices in a single group. There is an eigenvector (1,1,1, …) corresponding to such a trivial solution, but its eigenvalue is zero. All other eigenvectors are orthogonal to this one and hence must possess both positive and negative elements. Thus, as long as there is any positive eigenvalue this method will not put all vertices in the same group.
It is, however, possible for there to be no positive eigenvalues of the modularity matrix. In this case the leading eigenvector is the vector (1,1,1, …) corresponding to all vertices in a single group together. But this is precisely the correct result: the algorithm is in this case telling us that there is no division of the network that results in positive modularity, as can immediately be seen from Eq. 4, because all terms in the sum will be zero or negative. The modularity of the undivided network is zero, which is the best that can be achieved. This is an important feature of the algorithm. The algorithm has the ability not only to divide networks effectively, but also to refuse to divide them when no good division exists. The networks in this latter case will be called indivisible. That is, a network is indivisible if the modularity matrix has no positive eigenvalues. This idea will play a crucial role in later developments.
The algorithm as described makes use only of the signs of the elements of the leading eigenvector, but the magnitudes convey information, too. Vertices corresponding to elements of large magnitude make large contributions to the modularity, Eq. 4, and conversely for small ones. Alternatively, if we take the optimal division of a network into two groups and move a vertex from one group to the other, the vector element for that vertex gives an indication of how much the modularity will decrease: vertices corresponding to elements of large magnitude cannot be moved without incurring a large modularity penalty, whereas those corresponding to smaller elements can be moved at relatively little cost. Thus, the elements of the leading eigenvector measure how firmly each vertex belongs to its assigned community, those with large vector elements being strong central members of their communities, whereas those with smaller elements are more ambivalent.
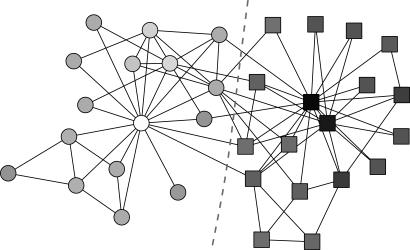
As an example of the operation of this algorithm, Fig. 2 shows the result of its application to a famous network from the social science literature, which has become something of a standard test for community detection algorithms. The network is the “karate club” network of Zachary (23), which shows the pattern of friendships between the members of a karate club at an American university in the 1970s. This example is of particular interest because, shortly after the observation and construction of the network, the club in question split in two as a result of an internal dispute. Applying our eigenvector-based algorithm to the network, we find the division indicated by the dotted line in Fig. 2, which coincides exactly with the known division of the club in real life, indicated by the shapes of the vertices.
Fig. 2.
Application of the eigenvector-based method to the karate club network of ref. 23. Shapes of vertices indicate the membership of the corresponding individuals in the two known factions of the network, and the dotted line indicates the split found by the algorithm, which matches the factions exactly. The shades of the vertices indicate the strength of their membership, as measured by the value of the corresponding elements of the eigenvector.
The vertices in Fig. 2 are shaded according to the values of the elements in the leading eigenvector of the modularity matrix, and these values seem also to accord well with known social structure within the club. In particular, the three vertices with the heaviest weights, either positive or negative (black and white vertices in Fig. 2), correspond to the known ringleaders of the two factions.
Dividing Networks into More than Two Communities
In the preceding section a simple matrix-based method for finding a good division of a network into two parts is described. Many networks, however, contain more than two communities, so we would like to extend the method to find good divisions of networks into larger numbers of parts. The standard approach to this problem, and the one adopted here, is repeated division into two: we use the algorithm of the previous section first to divide the network into two parts, then divide those parts, and so forth.
In doing this it is crucial to note that it is not correct, after first dividing a network in two, to simply delete the edges falling between the two parts and then apply the algorithm again to each subgraph. This is because the degrees appearing in the definition, Eq. 1, of the modularity will change if edges are deleted, and any subsequent maximization of modularity would thus maximize the wrong quantity. Instead, the correct approach is to write the additional contribution ΔQ to the modularity upon further dividing a group g of size ng in two as
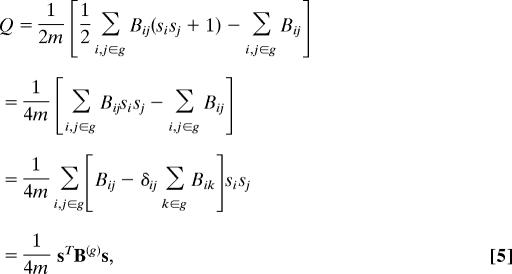 |
where δij is the Kronecker δ-symbol, we have made use of si2 = 1, and B(g) is the ng × ng matrix with elements indexed by the labels i,j of vertices within group g and having values
Because Eq. 5 has the same form as Eq. 2 we can now apply the spectral approach to this generalized modularity matrix, just as before, to maximize ΔQ. Note that the rows and columns of B(g) still sum to zero and that ΔQ is correctly zero if group g is undivided. Note also that for a complete network Eq. 6 reduces to the previous definition of the modularity matrix, Eq. 3, because Σk Bik is zero in that case.
In repeatedly subdividing the network, an important question we need to address is at what point to halt the subdivision process. A nice feature of this method is that it provides a clear answer to this question: if there exists no division of a subgraph that will increase the modularity of the network, or equivalently that gives a positive value for ΔQ, then there is nothing to be gained by dividing the subgraph and it should be left alone; it is indivisible in the sense of the previous section. This happens when there are no positive eigenvalues to the matrix B(g), and thus the leading eigenvalue provides a simple check for the termination of the subdivision process: if the leading eigenvalue is zero, which is the smallest value it can take, then the subgraph is indivisible.
Note, however, that although the absence of positive eigenvalues is a sufficient condition for indivisibility, it is not a necessary one. In particular, if there are only small positive eigenvalues and large negative ones, the terms in Eq. 4 for negative βi may outweigh those for positive. It is straightforward to guard against this possibility, however; we simply calculate the modularity contribution ΔQ for each proposed split directly and confirm that it is greater than zero.
Thus the algorithm is as follows. We construct the modularity matrix, Eq. 3, for the network and find its leading (most positive) eigenvalue and the corresponding eigenvector. We divide the network into two parts according to the signs of the elements of this vector, and then repeat the process for each of the parts, using the generalized modularity matrix, Eq. 6. If at any stage we find that a proposed split makes a zero or negative contribution to the total modularity, we leave the corresponding subgraph undivided. When the entire network has been decomposed into indivisible subgraphs in this way, the algorithm ends.
One immediate corollary of this approach is that all “communities” in the network are, by definition, indivisible subgraphs. A number of authors have in the past proposed formal definitions of what a community is (9, 16, 24). The present method provides an alternative first-principles definition of a community as an indivisible subgraph.
Further Techniques for Modularity Maximization
In this section I describe briefly another method for dividing networks in two by modularity optimization, which is entirely different from the spectral method. Although not of special interest on its own, this second method is, as will be shown shortly, very effective when combined with the spectral method.
Suppose we are given some initial division of our vertices into two groups. We then find among the vertices the one that, when moved to the other group, will give the biggest increase in the modularity of the complete network, or the smallest decrease if no increase is possible. We make such moves repeatedly, with the constraint that each vertex is moved only once. When all n vertices have been moved, we search the set of intermediate states occupied by the network during the operation of the algorithm to find the state that has the greatest modularity. Starting again from this state, we repeat the entire process iteratively until no further improvement in the modularity results. Those familiar with the literature on graph partitioning may find this algorithm reminiscent of the Kernighan–Lin algorithm (25), and indeed the Kernighan–Lin algorithm provided the inspiration for the method.
Despite its simplicity, we find that this method works moderately well. It is not competitive with the best previous methods, but it gives respectable modularity values in the trial applications we have made. However, the method really comes into its own when it is used in combination with the spectral method introduced earlier. It is a common approach in standard graph partitioning problems to use spectral partitioning based on the graph Laplacian to give an initial broad division of a network into two parts, and then refine that division by using the Kernighan–Lin algorithm. For community structure problems we find that the equivalent joint strategy works very well. The spectral approach based on the leading eigenvector of the modularity matrix gives an excellent guide to the general form that the communities should take and this general form can then be fine-tuned by the vertex moving method to reach the best possible modularity value. The whole procedure is repeated to subdivide the network until every remaining subgraph is indivisible, and no further improvement in the modularity is possible. (We note in passing that in principle the fine-tuning method could also be used to refine results from other modularity maximization algorithms, such as the extremal optimization algorithm of ref. 19.)
Typically, the fine-tuning stages of the algorithm add only a few percent to the final value of the modularity. For the karate club network of Fig. 2, for instance, the spectral method on its own finds a division of the network with modularity Q = 0.393, which improves to Q = 0.419 upon fine-tuning. Nonetheless, an improvement of this magnitude is enough, as we will see, to make the difference between a method that is merely good and one that is exceptional.
Example Applications
In practice, the algorithm developed here gives excellent results. For a quantitative comparison between this algorithm and others we follow Duch and Arenas (19) and compare values of the modularity for a variety of networks drawn from the literature. Results are shown in Table 1 for six different networks, the exact same six used by Duch and Arenas. We compare modularity figures against three previously published algorithms: the betweenness-based algorithm of Girvan and Newman (10), which is widely used and has been incorporated into some of the more popular network analysis programs; the fast algorithm of Clauset et al. (26), which optimizes modularity by using a greedy algorithm; and the extremal optimization algorithm of Duch and Arenas (19), which is arguably the best previously existing method, by standard measures (8), if one discounts methods impractical for large networks, such as exhaustive enumeration of all partitions or simulated annealing. Table 1 reveals some interesting patterns. The algorithm clearly outperforms the methods of Girvan and Newman and Clauset et al. for all of the networks in the task of optimizing the modularity. The extremal optimization method, on the other hand, is more competitive. For the smaller networks, up to ≈1,000 vertices, there is essentially no difference in performance between the method of this article and extremal optimization; the modularity values for the divisions found by the two algorithms differ by no more than a few parts in 1,000 for any given network. For the larger networks, however, the spectral algorithm does better than extremal optimization, and furthermore the gap widens as network size increases, to a maximum modularity difference of ≈6% for the largest network studied. For the very large networks that have been of particular interest in the last few years, therefore, it appears that the spectral method for detecting community structure may be the most effective of the methods considered here.
Table 1.
Comparison of modularities for the network divisions found by the algorithm described here and three other previously published methods as described in the text, for six networks of varying sizes
| Network | Size n | Modularity Q |
|||
|---|---|---|---|---|---|
| GN | CNM | DA | This article | ||
| Karate | 34 | 0.401 | 0.381 | 0.419 | 0.419 |
| Jazz musicians | 198 | 0.405 | 0.439 | 0.445 | 0.442 |
| Metabolic | 453 | 0.403 | 0.402 | 0.434 | 0.435 |
| 1,133 | 0.532 | 0.494 | 0.574 | 0.572 | |
| Key signing | 10,680 | 0.816 | 0.733 | 0.846 | 0.855 |
| Physicists | 27,519 | — | 0.668 | 0.679 | 0.723 |
The networks are, in order, the karate club network of Zachary (23), the network of collaborations between early jazz musicians of Gleiser and Danon (27), a metabolic network for the nematode Caenorhabditis elegans (28), a network of e-mail contacts at a university (29), a trust network of mutual signing of cryptography keys (30), and a coauthorship network of scientists working in condensed matter physics (31). No modularity figure is given for the last network with the GN algorithm because the slow O(n3) operation of the algorithm prevents its application to such large systems. GN, Glrvan and Newman (10); CNM, Clauset et al. (26); DA, Duch and Arenas (19).
The modularity values given in Table 1 provide a useful quantitative measure of the success of the algorithm when applied to real-world problems. It is worthwhile, however, also to confirm that it returns sensible divisions of networks in practice. I have given one example demonstrating such a division in Fig. 2. I have also checked the method against many of the example networks used in previous studies (10, 17). Here I give two more examples, both involving network representations of American politics.
The first example is a network of books about politics, compiled by V. Krebs (personal communication). In this network the vertices represent 105 recent books on American politics bought from the on-line bookseller Amazon.com, and edges join pairs of books that are frequently purchased by the same buyer. Books were divided (by me) according to their stated or apparent political alignment, liberal or conservative, except for a small number of books that were explicitly bipartisan or centrist, or had no clear affiliation.
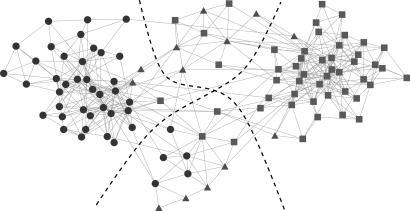
Fig. 3 shows the result of feeding this network through the algorithm. The algorithm finds four communities of vertices, denoted by the dotted lines in Fig. 3, with a modularity of 0.526. As can be seen, one of these communities consists almost entirely of liberal books and one almost entirely of conservative books. Most of the centrist books fall in the two remaining communities. Thus the books appear to form communities of copurchasing that align closely with political views, a finding that encourages us to believe that the algorithm is capable of extracting meaningful results from raw network data. It is particularly interesting to note that the centrist books belong to their own communities and are not, in most cases, merely lumped in with the liberals or conservatives; this finding may indicate that political moderates form their own purchasing community.
Fig. 3.
Krebs' network of books on American politics. Vertices represent books and edges join books frequently purchased by the same readers. Dashed lines divide the four communities found by the algorithm, and shapes represent the political alignment of the books (circles are liberal, squares are conservative, and triangles are centrist or unaligned).
For the second example, we consider a network of political commentary web sites, also called “weblogs” or “blogs,” compiled from online directories by Adamic and Glance,† who also assigned a political alignment, conservative or liberal, to each blog based on content. The 1,225 vertices in the network studied here correspond to the 1,225 blogs in the largest component of Adamic and Glance's network, and undirected edges connect vertices if either of the corresponding blogs contained a hyperlink to the other on its front page. On feeding this network through the algorithm we discover that the network divides cleanly into conservative and liberal communities and, remarkably, the optimal modularity of Q = 0.426 is found for a division into just two communities. One community has 638 vertices of which 620 (97%) represent conservative blogs. The other has 587 vertices of which 548 (93%) represent liberal blogs. The algorithm found no division of either of these two groups that gives any positive contribution to the modularity; as near as the algorithm is able to tell, these groups are “indivisible” in the sense defined here. This behavior is unique in my experience among networks of this size and is perhaps a testament not only to the widely noted polarization of the current political landscape in the United States but also to the strong cohesion of the two factions.
Implementation
This algorithm is fast as well as accurate. The most time-consuming part of the algorithm is the evaluation of the leading eigenvector of the modularity matrix. The fastest method for finding this eigenvector is the simple power method, the repeated multiplication of the matrix into a trial vector. Although it appears at first glance that matrix multiplications will be slow, taking O(n2) operations each because the modularity matrix is dense, we can in fact perform them much faster by exploiting the particular structure of the matrix. Writing B = A − kkT/2m, where A is the adjacency matrix and k is the vector whose elements are the degrees of the vertices, the product of B and an arbitrary vector x can be written
The first term is a standard sparse matrix multiplication taking time O(m + n). The inner product kT·x takes time O(n) to evaluate and hence the second term can be evaluated in total time O(n) also. Thus the complete multiplication can be performed in O(m + n) time. Typically O(n) such multiplications are needed to converge to the leading eigenvector, for a running time of O[(m + n)n] overall. Often we are concerned with sparse graphs with m ∝ n, in which case the running time becomes O(n2). It is a simple matter to extend this procedure to find the leading eigenvector of the generalized modularity matrix, Eq. 6, also.
Although I will not go through the details here, it is straightforward to show that the fine-tuning stage of the algorithm can also be completed in O[(m + n)n] time, so that the combined running time for a single split of a graph or subgraph scales as O[(m + n)n], or O(n2) on a sparse graph.
We then repeat the division into two parts until the network is reduced to its component indivisible subgraphs. The running time of the entire process depends on the depth of the tree or “dendrogram” formed by these repeated divisions (10, 16). In the worst case the dendrogram has depth linear in n, but only a small fraction of possible dendrograms realize this worst case. A more realistic figure for running time is given by the average depth of the dendrogram, which goes as log n, giving an average running time for the whole algorithm of O(n2log n) in the sparse case. This is considerably better than the O(n3) running time of the betweenness algorithm (10), and slightly better than the O(n2log2 n) of the extremal optimization algorithm (19). It is not as good as the O(nlog2 n) running time for the greedy algorithm of ref. 26, but the results are of far better quality than those for the greedy algorithm. In practice, running times are reasonable for networks up to ≈100,000 vertices with current computers. For the largest of the networks studied here, the collaboration network, which has ≈27,000 vertices, the algorithm takes ≈20 min to run on a standard personal computer (circa 2006).
Conclusions
In this article we have examined the problem of detecting community structure in networks, which is framed as an optimization task in which one searches for the maximal value of the quantity known as modularity over possible divisions of a network. We have shown that this problem can be rewritten in terms of the eigenvalues and eigenvectors of a matrix called the modularity matrix, and by exploiting this transformation created a computer algorithm for community detection that demonstrably outperforms the best previous general-purpose algorithms in terms of both quality of results and speed of execution. The algorithm has been applied to a variety of real-world network data sets, including social and biological examples, the results showing it to give both intuitively reasonable divisions of networks and quantitatively better divisions as measured by the modularity.
Acknowledgments
I thank Lada Adamic, Alex Arenas, and Valdis Krebs for graciously providing network data. This work was funded in part by National Science Foundation Grant DMS-0405348 and the James S. McDonnell Foundation.
Note.
After this article was submitted, I was informed of a recent conference presentation by White and Smyth (32) in which a result similar to Eq. 2 was derived and used as the basis for a modularity maximization algorithm quite different from the one presented here. I thank Christian Pich for bringing this presentation to my attention.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Adamic, L. A. & Glance, N., WWW-2005 Workshop on the Weblogging Ecosystem, May 10–14, 2005, Chiba, Japan.
References
- 1.Watts D. J., Strogatz S. H. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 2.Barabási A.-L., Albert R. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 3.Milo R., Shen-Orr S., Itzkovitz S., Kashtan N., Chklovskii D., Alon U. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 4.Albert R., Barabási A.-L. Rev. Mod. Phys. 2002;74:47–97. [Google Scholar]
- 5.Dorogovtsev S. N., Mendes J. F. F. Adv. Phys. 2002;51:1079–1187. [Google Scholar]
- 6.Newman M. E. J. SIAM Rev. 2003;45:167–256. [Google Scholar]
- 7.Newman M. E. J. Eur. Phys. J. B. 2004;38:321–330. [Google Scholar]
- 8.Danon L., Duch J., Diaz-Guilera A., Arenas A. J. Stat. Mech. 2005:P09008. [Google Scholar]
- 9.Flake G. W., Lawrence S. R., Giles C. L., Coetzee F. M. IEEE Computer. 2002;35:66–71. [Google Scholar]
- 10.Girvan M., Newman M. E. J. Proc. Natl. Acad. Sci. USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holme P., Huss M., Jeong H. Bioinformatics. 2003;19:532–538. doi: 10.1093/bioinformatics/btg033. [DOI] [PubMed] [Google Scholar]
- 12.Guimerà R., Amaral L. A. N. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsner U. Graph Partitioning: A Survey. Chemnitz, Germany: Technische Universität Chemnitz; 1997. Technical Report 97-27. [Google Scholar]
- 14.Fjällström P.-O. Linköping Electronic Articles in Computer and Information Science. 1998;Vol. 3 Available at www.ep.liu.se/ea/cis/1998/006. Accessed May 10, 2006.
- 15.White H. C., Boorman S. A., Breiger R. L. Am. J. Sociol. 1976;81:730–779. [Google Scholar]
- 16.Wasserman S., Faust K. Social Network Analysis. Cambridge, U.K.: Cambridge Univ. Press; 1994. [Google Scholar]
- 17.Newman M. E. J., Girvan M. Phys. Rev. E. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- 18.Newman M. E. J. Phys. Rev. E. 2004;69:066133. doi: 10.1103/PhysRevE.69.066133. [DOI] [PubMed] [Google Scholar]
- 19.Duch J., Arenas A. Phys. Rev. E. 2005;72:027104. doi: 10.1103/PhysRevE.72.027104. [DOI] [PubMed] [Google Scholar]
- 20.Chung F. R. K. Spectral Graph Theory, CBMS Regional Conference Series in Mathematics; Providence, RI: Am. Math. Soc.; 1997. no. 92. [Google Scholar]
- 21.Fiedler M. Czech. Math. J. 1973;23:298–305. [Google Scholar]
- 22.Pothen A., Simon H., Liou K.-P. SIAM J. Matrix Anal. Appl. 1990;11:430–452. [Google Scholar]
- 23.Zachary W. W. J. Anthropol. Res. 1977;33:452–473. [Google Scholar]
- 24.Radicchi F., Castellano C., Cecconi F., Loreto V., Parisi D. Proc. Natl. Acad. Sci. USA. 2004;101:2658–2663. doi: 10.1073/pnas.0400054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kernighan B. W., Lin S. Bell System Tech. J. 1970;49:291–307. [Google Scholar]
- 26.Clauset A., Newman M. E. J., Moore C. Phys. Rev. E. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 27.Gleiser P., Danon L. Adv. Complex Systems. 2003;6:565–573. [Google Scholar]
- 28.Jeong H., Tombor B., Albert R., Oltvai Z. N., Barabási A.-L. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 29.Guimerà R., Danon L., Díaz-Guilera A., Giralt F., Arenas A. Phys. Rev. E. 2003;68:065103. doi: 10.1103/PhysRevE.68.065103. [DOI] [PubMed] [Google Scholar]
- 30.Guardiola X., Guimerà R., Arenas A., Diaz-Guilera A., Streib D., Amaral L. A. N. e-Print Archive. 2002 http://xxx.lanl.gov/abs/cond-mat/0206240.
- 31.Newman M. E. J. Proc. Natl. Acad. Sci. USA. 2001;98:404–409. doi: 10.1073/pnas.021544898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White S., Smyth P. In: Kargupta H., Srivastava J., Kamath C., Goodman A., editors. Proceedings of the 5th SIAM International Conference on Data Mining; Philadelphia: Society for Industrial and Applied Mathematics; 2005. pp. 274–285. [Google Scholar]