Abstract
There is growing evidence that dissolved phosphorus can regulate planktonic production in the oceans’ subtropical gyres, yet there is little quantitative information about the biochemical fate of phosphorus in planktonic communities. We observed in the North Pacific Subtropical Gyre (NPSG) that the synthesis of membrane lipids accounted for 18–28% of the phosphate (PO43−) taken up by the total planktonic community. Paradoxically, Prochlorococcus, the cyanobacterium that dominates NPSG phytoplankton, primarily synthesizes sulfoquinovosyldiacylglycerol (SQDG), a lipid that contains sulfur and sugar instead of phosphate. In axenic cultures of Prochlorococcus, it was observed that <1% of the total PO43− uptake was incorporated into membrane lipids. Liquid chromatography/mass spectrometry of planktonic lipids in the NPSG confirmed that SQDG was the dominant membrane lipid. Furthermore, the analyses of SQDG synthesis genes from the Sargasso Sea environmental genome showed that the use of sulfolipids in subtropical gyres was confined primarily to picocyanobacteria; no sequences related to known heterotrophic bacterial SQDG lineages were found. This biochemical adaptation by Prochlorococcus must be a significant benefit to these organisms, which compete against phospholipid-rich heterotrophic bacteria for PO43−. Thus, evolution of this “sulfur-for-phosphorus” strategy set the stage for the success of picocyanobacteria in oligotrophic environments and may have been a major event in Earth’s early history when the relative availability of sulfate and PO43− were significantly different from today’s ocean.
Keywords: bacteria, cyanobacteria, genomics, lipids, marine phosphorus cycle
The oceans’ subtropical gyres, once likened to marine deserts, are now recognized to play an important role in the global carbon cycle (1, 2). Collectively, these regions are responsible for as much as half of the total organic carbon export to the deep sea (3). Yet the relative importance of nitrogen, phosphorus, or trace metal availability in controlling the production of organic matter within each gyre is poorly constrained (1, 4). Although phosphorus must regulate global production on geological timescales (5), subtropical gyres are highly dynamic and may experience periods when phosphorus is either replete or scarce relative to other essential nutrients (2).
The climatic and oceanographic regime of the North Pacific has undergone dramatic changes since the 1970s, which have led to numerous responses in the chemical cycling and ecology of the North Pacific subtropical gyre (NPSG), the world’s largest biome (2). The most notable of these changes include enhanced photosynthesis (6) driven by nitrogen fixation (1), increased inventories of organic carbon (7), and decreased availability of dissolved phosphorus (1, 6). It has been hypothesized that these biological and chemical changes were accompanied by a “domain shift” in the composition of the phytoplankton community, during which algae of the domain Eukarya were supplanted by photosynthetic organisms of the domain Bacteria, primarily the unicellular cyanobacterium Prochlorococcus (6). Prochlorococcus is characterized by its extremely small physical size (0.6 μm), relatively small genome (<2.5 Mbp), and physiologically diverse genotypes present throughout the euphotic zone in subtropical and tropical latitudes (8, 9). It also apparently is able to outcompete eukaryotic phytoplankton for dissolved nutrients in the NPSG (10) and other oligotrophic gyres where phosphorus availability may be an important control on various aspects of planktonic growth and activity (11, 12). In the NPSG, Prochlorococcus are often so dominant that they are, in effect, the only phytoplankton cells present (10). Indeed, culture studies with Prochlorococcus have indicated that these organisms contain very little phosphorus per cell (13, 14); phosphorus:carbon ratios may approach 1:500, much lower than the canonical Redfield ratio of 1:106 for mean plankton (13).
Very little is known of the biochemical partitioning of phosphorus by living plankton. Nearly all of the phosphorus found in suspended marine particulate matter is present as phosphoesters (15, 16), but these molecular structures are characteristic of all three major classes of phosphorus-containing biochemicals: DNA, RNA, and phospholipids. Calculations based on the genome size of the MED4 strain of Prochlorococcus suggest that as much as half of the cellular phosphorus is contained in DNA (13), which would leave the rest as RNA, phospholipids, and other biochemicals (e.g., ATP, phosphorylated proteins, etc.). We sought to elucidate the biochemical adaptations that allow Prochlorococcus to maintain low cellular phosphorus content and dominate potentially phosphorus-limiting gyre environments. Therefore, we investigated the cellular reservoirs of membrane phospholipids from the total planktonic community in the NPSG as well as laboratory cultures of Prochlorococcus and other plankton.
Results and Discussion
Incorporation of Phosphorus into Membrane Lipids by Subtropical Gyre Plankton.
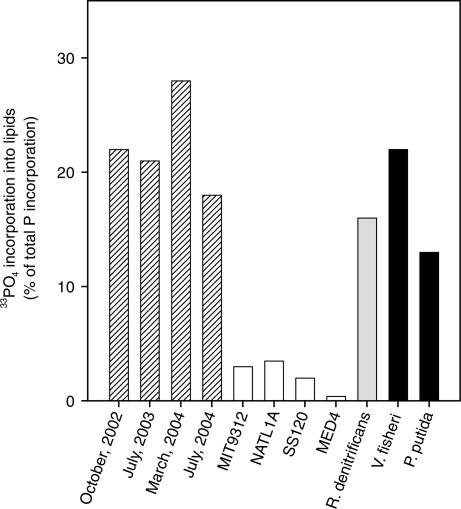
We traced the incorporation of33PO43− into phospholipids of the total planktonic community during four cruises to station ALOHA (22.75oN, 158oW) in the NPSG. Although ortho-phosphate is by no means the only source of dissolved phosphorus to plankton at this location, it is the most biologically available chemical form and may account for as much as half of the total uptake from dissolved phosphorus sources (10). We found that 18–28% of the 33PO43− taken up into the total planktonic community during a 24-h incubation was incorporated into phospholipids (Fig. 1). This observation was supported by measurements of lipid-phosphorus concentrations (3.1 ± 2.5 nM phosphorus in total seawater), which composed approximately one-fourth of the total suspended particulate phosphorus (13.6 ± 2.2 nM phosphorus; data provided by K. Björkman and D. Karl, University of Hawai’i, Manoa). This proportion of phospholipids is higher than that reported in one study of coastal waters (3–14%) (17) and may reflect the higher relative abundance of bacterial cells in open ocean vs. coastal waters. Our data from the NPSG show that the total planktonic community, which in addition to Prochlorococcus is composed almost exclusively of heterotrophic bacteria (10), devotes a significant fraction of its phosphorus uptake to cell membrane synthesis.
Fig. 1.
The fraction of total 33PO43− uptake that was recovered as phospholipids in incubations of NPSG total plankton (striped), Prochlorococcus cultures (white), AP bacteria cultures (gray), and heterotrophic bacteria cultures (black).
We repeated 33PO43− uptake incubations with laboratory cultures of Prochlorococcus and found that in contrast to the total planktonic community of the NPSG, <4% of the 33PO43− taken up by these organisms was incorporated into lipids (Fig. 1). Of the four Prochlorococcus strains we tested, only the MED4 cultures were axenic (i.e., completely pure; devoid of contaminating heterotrophic bacteria), and we observed that <1% of 33PO43− taken up was incorporated into lipids by this strain. Although the concentration of PO43− in the culture media was 3 orders of magnitude higher than that typically observed in the NPSG (50 μM vs. 50 nM), it was the same as that used in other studies that showed a low cell quota for phosphorus in Prochlorococcus (13, 14). Thus, it was highly unlikely that luxury uptake of phosphorus occurred in these incubations, which could have skewed the relative incorporation of 33PO43− into phospholipids. Furthermore, because PO43− was the only source of dissolved phosphorus in the culture media, we make the relatively safe assumption that 33PO43− representatively traced total phosphorus incorporation. The observations of MED4 are consistent with a measurement reported >20 years ago for Synechococcus WH 7803 (18), another marine unicellular picocyanobacterium representing a sister genus to Prochlorococcus (19). Other strains we tested, MIT9312, NATL1A, and SS120, although not axenic, still showed only 2–4% incorporation of 33PO43− into lipids. Thus, the values for 33PO43− uptake presented for these latter three strains represent upper limits on the amount of phosphorus dedicated to lipid synthesis by Prochlorococcus. This observation that much more 33PO43− was incorporated into lipids by the total planktonic community than could be accounted for by Prochlorococcus is consistent with published data showing that these organisms have a much higher C:P ratio than Redfield plankton (13, 14). It also suggests that heterotrophic bacteria, which are relatively phosphorus-rich (i.e., lower C:P) (20), are responsible for most of the phospholipid synthesis in the NPSG.
Molecular Distribution of Membrane Lipids in Gyre Plankton.
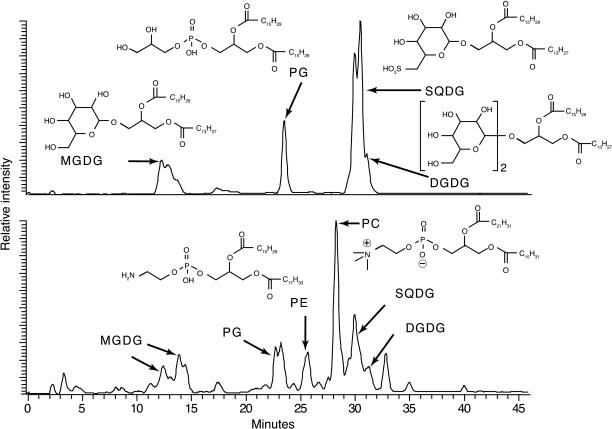
We screened the structural diversity of polar membrane lipids extracted from the Prochlorococcus cultures using HPLC/electrospray-ionization ion-trap mass spectrometry (HPLC/ESI-IT-MSn) (21). We found that the vast majority of membrane lipids in Prochlorococcus were either sulfolipids or glycolipids, compounds that contain either sulfate/sugar- or sugar-based polar head groups instead of PO43−-based polar head groups (Fig. 2 and Table 1). Specifically, the sulfolipid sulfoquinovosyldiacylglycerol (SQDG) and the glycolipids monogalactosyldiacylglycerol and digalactosyldiacylglycerol composed 94 ± 5% of the total mass of lipids extracted from marine picocyanobacteria (i.e., Prochlorococcus and Synechococcus). The primary remaining cellular lipid was the phospholipid phosphatidylglycerol (PG), but in some of the nonaxenic cultures phosphatidylethanolamine (PE) and phosphatidylcholine (PC) also were observed.
Fig. 2.
HPLC/ESI-IT-MSn base peak chromatograms of membrane lipids for Prochlorococcus 9312 (Upper) and the total planktonic community from the NPSG (Lower). The vertical axis approximates relative abundance, although the MS is more sensitive to PG than SQDG, which exaggerates the relative size of the PG peaks. Fractions of each membrane lipid are listed in Table 1. Broad or doublet peaks are due to variations in fatty acid chain length. Generic formulas are used to represent common fatty acid tail groups of the intact lipid molecules. Abbreviations for chemical names are given in the text.
Table 1.
Molecular distribution of membrane lipids in plankton cultures and total planktonic community of the NPSG
| Culture or sample | Axenic | MGDG | DGDG | SQDG | PG | PE | PC | SQDG/PG |
|---|---|---|---|---|---|---|---|---|
| Prochlorococcus | ||||||||
| MED4 | Yes | 22 | 10 | 66 | 2 | – | – | 33.5 |
| NATL1A | No | 25 | 10 | 50 | 9 | 5 | tr | NA |
| SS120 | No | 17 | 7 | 61 | 8 | 5 | 1 | NA |
| MIT9312 | No | 31 | 53 | 7 | 9 | tr | – | NA |
| Synechococcus | ||||||||
| WH8103 | Yes | 7 | 37 | 52 | 3 | – | – | 17.3 |
| WH8113 | Yes | 5 | 26 | 56 | 8 | – | – | 7.0 |
| WH8020 | Yes | 11 | 31 | 47 | 7 | – | – | 6.7 |
| WH8018 | Yes | 4 | 48 | 45 | 3 | – | – | 15.0 |
| WH8101 | Yes | 18 | 41 | 38 | 3 | – | – | 12.7 |
| WH8102 | No | 14 | 25 | 50 | 8 | 2 | – | NA |
| Other cyanobacteria | ||||||||
| Synechocystis sp. PCC6803 | Yes | 14 | 56 | 19 | 10 | – | – | 1.9 |
| Synechocystis sp. PCC6803* | Yes | 59 | 17 | 16 | 8 | – | – | 2.0 |
| Gloeobacter violaceus | Yes | 5 | 78 | – | 17 | – | – | NA |
| Trichodesmium erythraeum | Yes | 45 | 16 | 20 | 18 | – | – | 1.1 |
| Heterotrophic bacteria | ||||||||
| Vibrio fischeri | Yes | – | – | – | 10 | 76 | – | NA |
| Roseobacter denitrificans | Yes | – | – | – | 44 | – | 39 | NA |
| Pseudomonas putida | Yes | – | – | – | 13 | 65 | – | NA |
| NPSG plankton | ||||||||
| October, 2002 | NA | ND | ND | ND | ND | ND | ND | ND |
| July, 2003 | NA | ND | ND | ND | ND | ND | ND | ND |
| March, 2004 | NA | 16 | 7 | 25 | 21 | 9 | 22 | 1.2 |
| July, 2004 | NA | 15 | 8 | 24 | 24 | 10 | 19 | 1.0 |
Values are percent of total polar membrane lipids as determined by HPLC/ESI-IT-MSn. –, none detected; tr, trace (<0.5%); ND, not determined; NA, not applicable; MGDG, monogalactosyldiacylglycerol; DGDG, digalactosyldiacylglycerol.
*Ref. 22.
A few freshwater cyanobacteria have been investigated previously, and these also contained SQDG, monogalactosyldiacylglycerol, and digalactosyldiacylglycerol, whereas PG was the only phospholipid (22, 23). However, none of these strains showed nearly as strong a preference for SQDG over PG as seen in marine Prochlorococcus and Synechococcus, and it is uncertain whether the difference between these previous studies and the observations presented here are an artifact of the lower SO42− concentrations that are used in “freshwater” culture media vs. “seawater” media. These previous studies used TLC for the analysis of intact membrane lipids, yet HPLC/ESI-IT-MSn analysis of SQDG and PG in the cyanobacteria Synechocystissp. PCC6803 (Table 1) agreed almost exactly with the data generated by other investigators using TLC (22).
SQDG and PG are the only membrane lipids in cyanobacteria with a net negative charge and are likely to serve a similar purpose in the cell (23). Benning (23) hypothesized that SQDG may replace PG in cyanobacteria when ambient phosphate is scarce. If this “sulfolipid–phospholipid substitution hypothesis” is true, then we might expect that the SQDG/PG ratio of Prochlorococcus cells to be even higher in the NPSG than the 37 ± 7 that we observe in cultures because the ratio of dissolved SO42− to PO43− is nearly 3 orders of magnitude higher in the NPSG than in the culture media.
We know of no reports of PE or PC in cultured cyanobacteria (22), and, thus, it is likely that these molecules we observed in the nonaxenic cultures originated from heterotrophic bacterial contamination. We examined cultures of the marine heterotrophic bacteria Vibrio fischeri and Pseudomonas putida and observed that PG, PE, and PC were all abundant, whereas no sulfolipids or glycolipids were detected (Table 1). This result is consistent with the substantive body of literature showing that marine heterotrophic bacteria have polar membrane lipids composed almost exclusively of phospholipids (e.g., ref. 24).
Despite the dominance of Prochlorococcus in the NPSG, on average 52% of lipids extracted from the total planktonic community in the NPSG were phospholipids (Fig. 2 and Table 1). This observation is consistent with cell counts reported for this environment indicating that heterotrophic bacteria compose at least half of the total planktonic biomass (10). The abundant phospholipids in the NPSG samples, PG, PE, and PC, are all common in heterotrophic bacterial cells (24). The SQDG/PG ratio of whole plankton from the NPSG was ≈1, and, given the SQDG/PG ratio of our cultures of Prochlorococcus, it is likely that >90% of the PG in the NPSG was derived from organisms other than Prochlorococcus.
We used preparative HPLC to preclude the possibility that all of the 33PO43− that we traced into planktonic lipids was simply due to the rapid synthesis of PG by cyanobacteria. Our data showed that more than half of the uptake of 33PO43− could be accounted for by the synthesis of PE and PC alone (see Fig. 4, which is published as supporting information on the PNAS web site); again, these two compounds were not present in our axenic Prochlorococcus and are not found in cyanobacteria. Thus, organisms of noncyanobacterial origin contributed substantially to the incorporation of 33PO43− into lipids in the NPSG.
We have observed that the nitrogen-fixing cyanobacterium Trichodesmium erythreaum consistently contains more PG than Prochlorococcus (Table 1), but in the NPSG Trichodesmium are only observed during ephemeral blooms and were not present during our cruises. Eukaryotic algae, although rich in chloroplast-derived glycolipids, also contain a variety of phospholipids (predominantly PC). However, these organisms are unlikely to be the primary source of the phospholipids because they compose only a small fraction of the total planktonic biomass in the near-surface waters of the NPSG (11), and our mass spectra show that the phospholipids from the NPSG did not contain an abundance of the polyunsaturated fatty acids that are characteristic of eukaryotic algae (25). In summary, our findings support the conclusion that most of the phospholipids that were present in the NPSG originated from heterotrophic bacteria and that these organisms were responsible for most of the uptake of 33PO43− into phospholipids.
Potential Planktonic Sources of Sulfolipids in Gyres.
Aside from cyanobacteria and the chloroplast membranes of photosynthetic eukaryotes, SQDG has only been confirmed in bacteria from the α- and γ-proteobacterial lineage (23), although some Archaea possess homologs of sqdB, a gene involved in sulfolipid biosynthesis (23, 26). Among the proteobacteria, SQDG has been reported in some caulobacteria (27) and in the anoxygenic photosynthetic (AP) bacteria. AP bacteria are heterotrophs that use light to subsidize energy requirements and appear to compose a small but ubiquitous fraction of bacterial cells in oligotrophic marine ecosystems (28, 29). However, the presence of SQDG is not universal in AP bacteria (23), and it was not present in cultures of Roseobacter denitrificans that we analyzed (Table 1). Instead, R. denitrificans allocated 16 ± 6% of PO43− uptake to the synthesis of phospholipids (Fig. 1). This allocation of PO43− to lipids is more similar to that of heterotrophic bacteria we investigated (17 ± 9%) than to that of axenic Prochlorococcus (0.4 ± 0.3%). Indeed, it is likely that an even greater fraction of PO43− uptake is used for phospholipid synthesis by heterotrophic bacteria under NPSG conditions than under our culturing conditions because rapidly growing heterotrophic bacterial cells synthesize greater amounts of phosphorus-rich RNA than slowly growing cells (30, 31). Our heterotrophic bacterial cultures grew 2 orders of magnitude faster (≈20 d−1) than is typical of oligotrophic environments (0.05–0.3 d−1) (32).
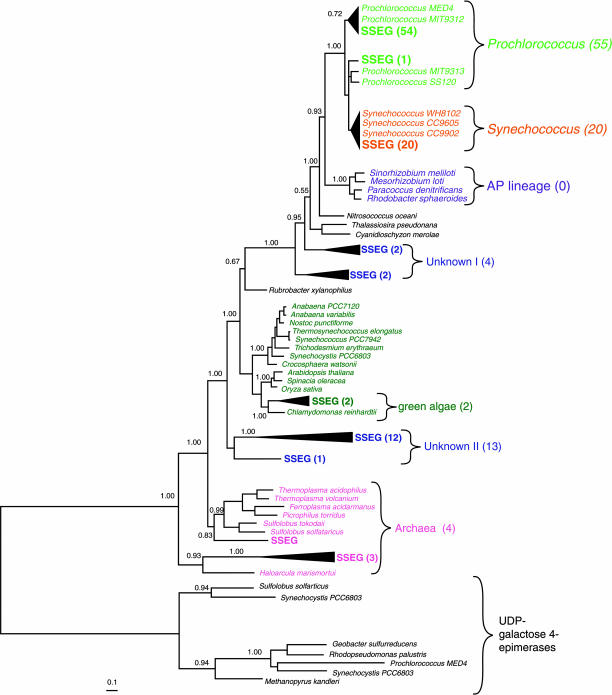
To evaluate the potential contribution of heterotrophic bacteria to SQDG pools in the oligotrophic ocean, we examined sequence reads from a shotgun genomic DNA library collected from the Sargasso Sea (33) for the presence and phylogenetic affinity of sqdB. The numerically dominant fraction of sqdB genes (77%) were picocyanobacterial in origin (Fig. 3), whereas none were found to be closely related to the AP bacterial lineage. A scenario where AP bacteria contribute significantly to SQDG synthesis without contributing a single copy of the sqdB gene to the environmental genome is unlikely. So although AP bacteria are present in both the NPSG (34) and the Sargasso Sea (M. Koblíz̆ek, personal communication), these organisms, similar to R. denitrificans, apparently do not carry the capability to synthesize SQDG. Thus, the environmental genome strongly supports our other data suggesting that the majority of the SQDG we observe in the NPSG is derived from cyanobacteria, rather than from AP or heterotrophic bacteria.
Fig. 3.
Tree of SqdB sequences from the Sargasso Sea environmental genome (denoted SSEG) (33) and representative phytoplankton and heterotrophic bacteria. UDP-galactose 4-epimerases are used as outgroups. The quantities of closely related SSEG sequences are indicated in parentheses. (Scale bar indicates substitutions site−1.)
Significance of Sulfolipids in Cyanobacteria.
The biochemical requirement of heterotrophic bacteria to use environmentally scarce phosphorus atoms to synthesize phospholipids, vs. abundant sulfur and carbon atoms to synthesize sulfolipids, could put heterotrophic bacteria at a disadvantage against Prochlorococcus in the nutrient-depleted waters of the subtropical gyres. Because phospholipids in heterotrophic bacteria are such a large biochemical sink for phosphorus, these organisms, which are responsible for respiration, could be more “limited” by phosphorus availability than those responsible for photosynthesis (7, 35). This result would be at odds with work suggesting that respiration outweighs photosynthesis in the open ocean (36) but could explain, at least to some degree, the conflicting trends of increasing inventories of organic carbon (7) and decreasing inventories of nutrient phosphorus in the NPSG (1, 6). Indeed, Prochlorococcus is abundant in other oligotrophic environments where PO43− is an order of magnitude less abundant than in NPSG (11). Yet it appears that not all marine cyanobacteria are equal in their ability to substitute SQDG for PG. For example, our lipid extracts from marine Synechococcus, which were grown at phosphate concentrations similar to Prochlorococcus cultures, show significantly lower SQDG/PG (12 ± 5 vs. 37 ± 7), which is consistent with observations that Prochlorococcus are more tolerant of low-phosphorus conditions than Synechococcus (11, 12).
Evolution of the ability of photosynthetic cells to substitute sulfate (SO42−) for PO43− in lipids may have led to major changes in the cycling of carbon, phosphorus, and sulfur in the ocean. There is much debate about how photosynthetic bacteria diversified into their current major lineages (37, 38). Although it is likely that the biochemical pathways for SQDG synthesis evolved only once in Earth’s history (23), it is not clear in which bacterial lineage this innovation occurred; the observation that the marine picocyanobacterial sqdB lineage is more similar to that of the AP bacteria than to other cyanobacteria such as T. erythraeum further complicates this issue (Fig. 3). In any case, the absence of both SQDG (Table 1) and the sqdB gene in the basally branching cyanobacterial genome Gloeobacter violaceus, and the presence of the lipid and the gene in at least four of the seven major lineages of cyanobacteria, suggests that the capability for SQDG synthesis either evolved or was acquired early in the cyanobacterial radiation. If our results from AP bacteria and other strictly heterotrophic bacteria that lack SQDG (Fig. 1) are representative of photosynthetic bacteria that existed before the evolution of SQDG, then the switch from PO43− to SO42− for lipid synthesis would have reduced cellular phosphorus requirements by 15–25%. In an ocean limited by PO43−, this reduction also would have led to a concomitant 15–25% increase in organic carbon production, because the PO43− that normally would have been used for phospholipid synthesis would be available to fuel further production.
During the Archean, cyanobacteria and other photosynthetic bacteria evolved in an ocean that probably contained much lower concentrations of PO43− and SO42− than the modern ocean (5, 39, 40); SO42− concentrations were apparently low enough to limit bacterial uptake in some regions of the ocean (40). But by the mid-Proterozoic, SO42− inventories increased substantially (40), and we hypothesize that the elevation of the ratio of dissolved SO42− to PO43− could have provided the environmental impetus for the evolution of SQDG. Depending on the exact timing of SQDG evolution, the rate at which SQDG-containing cyanobacteria radiated throughout the oceans, and the extent of phosphorus limitation, the upshift in organic carbon production allowed by the use of SQDG in cyanobacteria could have contributed to the observed major shifts in carbon cycling in the Proterozoic. Furthermore, the potential for SO42− to act as a substitute for PO43− calls into question the often assumed constancy of phosphorus (vs. nitrogen) limitation of photosynthetic carbon production over geological timescales.
In phosphorus-limited region of the modern ocean, it is quite likely that both picocyanobacteria and heterotrophic bacteria are under intense pressure to reduce their cellular phosphorus demands. Indeed, it has been observed that respiration by heterotrophic bacteria may currently be limited by available PO43− in the Sargasso Sea (35). Thus, the distinctive ability of picocyanobacteria to minimize their phosphorus requirements through the synthesis of SQDG could be the basic biochemical adaptation that has allowed these organisms to dominate photosynthetic carbon production in phosphorus-limited marine environments.
Methods
Seawater was collected from a depth of 5 m in the NPSG at station ALOHA, aboard research cruises supported by the Hawaiian Ocean Time-series (HOT) program. Incubations were conducted in acid-cleaned polycarbonate bottles, spiked with tracer-level quantities (≈25 pM) of carrier-free 33PO43−, and incubated in on deck flowing seawater incubators under ambient light conditions for 24 h. Aboard the ship, the incorporation of 33PO43− into suspended particulate matter was determined by the ratio of 33P radioactivity in filtered (0.2 μm pore size) and unfiltered aliquots of seawater. For lipid analysis, suspended particulate matter from the incubations was collected by filtration on precombusted alumina membranes (0.2 μm), which were immediately placed in liquid nitrogen and transported to the laboratory on dry ice. In the laboratory, lipids were extracted from both suspended particles and cell cultures using a modified Bligh–Dyer method. Phospholipid concentrations in suspended particles were determined from an aliquot of the extraction by analysis of soluble reactive phosphorus after persulfate oxidation (41).
For the Prochlorococcus culture experiments, cells were grown in artificial seawater supplemented with 800 μM NH4Cl, 50 μM NaH2PO4, and trace metal mix described in refs. 9 and 13. Exponential phase cultures were maintained at constant growth rates (ranging from 0.35 to 0.5 day−1 depending on the strain) at 22–24°C under constant light provided by cool-white fluorescent lamps at irradiances of 30 μM·quanta·m−2·s−1. Cultures of heterotrophic bacteria were grown in Marine Broth at 30°C and harvested in late exponential phase. Prochlorococcus and heterotrophic bacteria cultures were spiked with 33PO43−, incubated for 2–24 h, and analyzed for 33P uptake into lipids as was done with NPSG whole plankton.
Intact polar lipids were analyzed as described in ref. 21 from duplicate cultures (not 33PO43−-labeled). Briefly, a LiChrospher Diol column was used with a linear gradient of normal phase eluents. MSn experiments were performed by using a LCQ Deca XP ion trap mass spectrometer (ThermoFinnigan, San Jose, CA) with an electrospray (ESI) interface. The mass spectrometer was configured such that the base peak from each full scan (500–2000 Da) was fragmented up to MS3. Separate runs for positive and negative ion modes yielded complementary structural information. Authentic standards were used for quantification and to aid structural identification.
The entire data set of individual sequence reads from the Sargasso Sea environmental genome (SSEG) sequencing project (33) was downloaded from GenBank and searched by using tblastn with the SqdB amino acid sequences of Prochlorococcus MED4, Synechococcus WH8102, Trichodesmium erythraeum, Rubrobacter xylanophilus, Sinorhizobium meliloti, Rhodobacter sphaeroides, Nitrosococcus oceani, and Sulfolobus tokodaii. All reads with a score >100 from each search were retained for further analyses. This analysis resulted in 106 sequence reads representing 99 independent clones. The raw trace data for each read was trimmed so that no more than 5 of 25 consecutive bases on either the 5′ or 3′ end had a confidence score of <20. The resulting sequences were aligned with full-length SqdB sequences from cultured organisms. Phylogenetic trees were inferred with mrbayes (http://mrbayes.csit.fsu.edu). Samples used for the assembly of the SSEG were collected by Venter et al. (33) from waters with below-detectable levels of PO43− (data from Bermuda Atlantic Time-Series Study; http://bats.bbsr.edu).
Supplementary Material
Acknowledgments
We thank K. Björkman, T. Gregory, D. Karl, and the HOT program for particulate phosphorus data, logistical support, and access to cruises; M. Hullar and J. Smoot for important advice/encouragement concerning the 33Phospholipid measurements; L. Moore (University of Southern Maine, Portland, ME) for the strains of Prochlorococcus; E. Webb (Woods Hole Oceanographic Institution) for strains of Synechococcus and Trichodesmium; K.-U. Hinrichs for generous access to the HPLC/ESI-IT-MS in the early stages of this study; and T. Lyons, J. Waterbury, and J. Hayes for insightful comments on a draft of this paper. B.A.S.V.M. was supported by Department of Energy Grant DE-FG02-98ER62535 (to A.H.D.), the Postdoctoral Program at Woods Hole Oceanographic Institution (WHOI) with funding provided by the Watson Chair to D. Repeta, and the Gryce B. Kerr and the Penzance Endowed fund in Support of Assistant Scientists at WHOI. G.R. was supported by National Science Foundation (NSF) Grant OCE 0453029. H.F.F. was supported by postdoctoral funding from the National Aeronautics and Space Administration Astrobiology Institute University of Rhode Island/WHOI Team. C.T.E. was supported by NSF Grant OCE 09984163 (to R. Keil).
Abbreviations
- NPSG
North Pacific subtropical gyre
- SQDG
sulfoquinovosyldiacylglycerol
- PG
phosphatidylglycerol
- PE
phosphatidylethanolamine
- PC
phosphatidylcholine
- AP
anoxygenic photosynthetic
- ESI
electrospray ionization
- IT
ion-trap.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Karl D., Letelier R., Tupas L., Dore J., Christian J., Hebel D. Nature. 1997;388:533–538. [Google Scholar]
- 2.Karl D. M. Ecosystems. 1999;2:181–214. [Google Scholar]
- 3.Emerson S., Quay P., Karl D., Winn C., Tupas L., Landry M. Nature. 1997;389:951–954. [Google Scholar]
- 4.Wu J., Sunda W., Boyle E. A., Karl D. M. Science. 2000;289:759–762. doi: 10.1126/science.289.5480.759. [DOI] [PubMed] [Google Scholar]
- 5.Bjerrum C. J., Canfield D. E. Nature. 2002;417:159–162. doi: 10.1038/417159a. [DOI] [PubMed] [Google Scholar]
- 6.Karl D. M., Bidigare R. R., Lelelier R. M. Deep-Sea Res. II. 2001;48:1449–1470. [Google Scholar]
- 7.Church M. J., Ducklow H. D., Karl D. M. Limnol. Oceanogr. 2002;47:1–10. [Google Scholar]
- 8.Rocap G., Larimer F. W., Lamerdin J., Malfatti S., Chain P., Ahlgren N. A., Arellano A. R., Coleman M., Hauser L., Hess W. R., et al. Nature. 2003;424:1042–1046. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 9.Moore L. R., Post A. F., Rocap G., Chisholm S. W. Limnol. Oceanogr. 2002;47:989–996. [Google Scholar]
- 10.Björkman K., Thomson-Bulldis A. L., Karl D. M. Aquat. Microb. Ecol. 2000;22:185–198. [Google Scholar]
- 11.Moutin T., Thingstad T. F., Wambeke V., Marie D., Slawyk G., Raimbault P., Claustre H. Limnol. Oceanogr. 2002;47:1562–1567. [Google Scholar]
- 12.Fuller N. J., West N. J., Marie D., Yallop M., Rivlin T., Post A. F., Scanlan D. J. Limnol. Oceanogr. 2005;50:363–375. [Google Scholar]
- 13.Bertilsson S., Berglund O., Karl D. M., Chisholm S. W. Limnol. Oceanogr. 2003;48:1721–1731. [Google Scholar]
- 14.Heldal M., Scanlan D. J., Norland S., Thingstad F., Mann N. H. Limnol. Oceanogr. 2003;48:1732–1743. [Google Scholar]
- 15.Kolowith L. C., Ingall E. D., Benner R. Limnol. Oceanogr. 2001;46:309–320. [Google Scholar]
- 16.Paytan A., Cade-Menum B. J., McLaughlin K., Faul K. L. Mar. Chem. 2003;82:55–70. [Google Scholar]
- 17.Suzumura M., Ingall E. D. Mar. Chem. 2001;75:141–149. [Google Scholar]
- 18.Cuhel R. L., Waterbury J. B. Limnol. Oceanogr. 1984;29:370–374. [Google Scholar]
- 19.Rocap G., Distel D. L., Waterbury J. B., Chisholm S. W. Appl. Environ. Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gundersen K., Heldal M., Norland S., Purdie D. A., Knap A. H. Limnol. Oceanogr. 2002;47:1525–1530. [Google Scholar]
- 21.Sturt H. F., Summons R. E., Smith K., Elvert M., Hinrichs K.-U. Rapid Commun. Mass Spectrom. 2004;18:617–628. doi: 10.1002/rcm.1378. [DOI] [PubMed] [Google Scholar]
- 22.Wada H., Murata N. In: Lipids in Photosynthesis: Structure, Function and Genetics. Siegenthaler P.-A., Murata N., editors. Amsterdam: Kluwer Academic; 1998. pp. 65–81. [Google Scholar]
- 23.Benning C. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:53–75. doi: 10.1146/annurev.arplant.49.1.53. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson S. G. In: Microbial Lipids. Ratledge C., Wilkinston S. G., editors. Vol. 1. London: Academic Limited; 1988. pp. 299–488. [Google Scholar]
- 25.Wakeham S. G., Lee C., Hedges J., Hernes P. J., Peterson M. L. Geochim. Cosmochim. Acta. 1997;61:5363–5369. [Google Scholar]
- 26.Sato N. J. Plant Res. 2004;117:495–505. doi: 10.1007/s10265-004-0183-1. [DOI] [PubMed] [Google Scholar]
- 27.Abraham W.-R., Strömpl C., Vancanneyt M., Bennasar A., Swings J., Lünsdorf H., Smit J., Moore E. R. B. Int. J. Syst. Evol. Microbiol. 2004;54:1227–1234. doi: 10.1099/ijs.0.02943-0. [DOI] [PubMed] [Google Scholar]
- 28.Schwalbach M. S., Fuhrman J. A. Limnol. Oceanogr. 2005;50:620–628. [Google Scholar]
- 29.Van Mooy B. A. S., Devol A. H., Keil R. G. Limnol. Oceanogr. 2004;49:1056–1062. [Google Scholar]
- 30.Kerkhof L., Kemp P. FEMS Microbiol. Ecol. 1999;30:253–260. doi: 10.1111/j.1574-6941.1999.tb00653.x. [DOI] [PubMed] [Google Scholar]
- 31.Kemp P. F., Lee S., LaRoche J. Appl. Environ. Microbiol. 1993;59:2594–2601. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ducklow H. In: Microbial Ecology of the Ocean. Kirchman D. L., editor. New York: Wiley–Liss; 2000. pp. 85–120. [Google Scholar]
- 33.Venter J. C., Remington K., Heidelberg J. F., Halpern A. L., Rusch D., Eisen J. A., Wu D., Paulsen I., Nelson K. E., Nelson W., et al. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 34.Cottrell M. T., Mannino A., Kirchman D. L. Appl. Environ. Microbiol. 2006;72:557–564. doi: 10.1128/AEM.72.1.557-564.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obernosterer I., Kawasaki N., Benner R. Aquat. Microbial Ecol. 2003;32:229–237. [Google Scholar]
- 36.del Giorgio P. A., Duarte C. M. Nature. 2002;420:379–384. doi: 10.1038/nature01165. [DOI] [PubMed] [Google Scholar]
- 37.Xiong J., Fischer W. M., Inoue K., Nakahara M., Bauer C. E. Science. 2000;289:1724–1730. doi: 10.1126/science.289.5485.1724. [DOI] [PubMed] [Google Scholar]
- 38.Woese C. R. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kah L. C., Lyons T. W., Frank T. D. Nature. 2004;431:831–838. doi: 10.1038/nature02974. [DOI] [PubMed] [Google Scholar]
- 40.Habicht K. S., Gade M., Thamdrup B., Berg P., Canfield D. E. Science. 2002;298:2372–2374. doi: 10.1126/science.1078265. [DOI] [PubMed] [Google Scholar]
- 41.White D. C., Davis W. M., Nickels J. S., King J. D., Bobbie R. J. Oecologia. 1979;40:51–62. doi: 10.1007/BF00388810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.