Abstract
Site-specific incorporation of unnatural amino acids (UAAs) into proteins is a valuable tool for studying structure–function relationships, incorporating biophysical probes, and elucidating protein–protein interactions. In higher eukaryotic cells, the methodology is currently limited to incorporation of a single UAA in response to a stop codon, which is known as nonsense suppression. Frameshift suppression is a unique methodology for incorporating UAAs in response to quadruplet codons, but currently, it is mostly limited to in vitro protein translation systems. Here, we evaluate the viability of frameshift suppression in Xenopus oocytes. We demonstrate UAA incorporation by using yeast phenylalanine frameshift suppressor (YFFS) tRNAs that recognize two different quadruplet codons (CGGG and GGGU) in vivo. Suppression efficiency of the YFFS tRNAs increases nonlinearly with the amount of injected tRNA, suggesting a significant competition with endogenous, triplet-recognizing tRNA. Both frameshift suppressor tRNAs are less efficient than the amber suppressor tRNA THG73 (Tetrahymena thermophila G73), which has been used extensively for UAA incorporation in Xenopus oocytes. However, the two YFFS tRNAs are more “orthogonal” to the Xenopus system than THG73, and they offer a viable replacement when suppressing at promiscuous sites. To illustrate the potential of combining nonsense and frameshift suppression, we have site-specifically incorporated two and three UAAs simultaneously into a neuroreceptor expressed in vivo.
Keywords: nicotinic receptor, tRNA, quadruplet codon, stop codon, protein engineering
The site-specific incorporation of unnatural amino acids (UAAs) into proteins biosynthetically is a powerful methodology that is seeing increasing use. The primary approach has been stop codon (nonsense) suppression using a specially designed tRNA with an anticodon that recognizes the stop codon. A wide range of in vitro translation systems has been used, along with expression in Escherichia coli and, to a lesser extent, yeast. Nonsense suppression in higher eukaryotes has, for the most part, been limited to the Xenopus oocyte, where microinjection of the required mRNA and aminoacyl tRNA is straightforward and electrophysiology provides a sensitive probe of UAA incorporation (1, 2). Other experiments in higher eukaryotes have relied on the evolution of a unique tRNA and a complementary aminoacyl-tRNA synthetase (aaRS) to insert a UAA in response to the UAG or UGA stop codon, but currently, only 3-iodo-tyrosine (3), p-benzoyl-phenylalanine (4), and 5-hydroxy-tryptophan (5) have been incorporated.
A remarkable variant of this approach is the use of quadruplet codons, a process that is termed frameshift suppression and was pioneered by Sisido and coworkers (6, 7). The success of this approach opens up the possibility of developing multiple additional codons, thus incorporating several different UAAs into a protein. This multiple incorporation, in turn, would enable the use of innovative biophysical approaches such as incorporating FRET pairs, structural probes such as unique cross-linking approaches, and more detailed structure–function studies.
To date, frameshift suppression in vivo has been performed only in E. coli by using a unique tRNA/aaRS pair, and homoglutamine is the only UAA incorporated by this method. Frameshift suppression was used simultaneously with nonsense suppression to incorporate two UAAs in E. coli (8). It has yet to be established whether frameshift suppression by chemically aminoacylated tRNA can be effective in vivo in general and in eukaryotic cells such as the Xenopus oocyte in particular. In fact, a previous attempt to perform frameshift suppression in Xenopus oocytes showed very poor suppression efficiency (9). Here, we show that with appropriately designed frameshift suppressor (FS) tRNAs, frameshift suppression is a viable approach to UAA incorporation in eukaryotic cells. Also, the efficiency of frameshift suppression can be substantially improved by “masking” the mRNA of all in-frame quadruplet sequences that match the frameshift suppression site. In particular, we describe two tRNAs with four-base anticodons that can deliver UAAs in response to the quadruplet codons CGGG and GGGU. When directly compared with an amber suppressor (AS) tRNA (THG73, Tetrahymena thermophila G73) that has been used extensively in Xenopus oocytes, the FS tRNAs are less efficient at delivering UAAs. However, both FS tRNAs are more “orthogonal” than THG73, producing much less incorporation of undesired natural amino acids at promiscuous sites. We also show that suppression by FS tRNAs increases nonlinearly with the amount of injected tRNA. To illustrate the potential of this methodology, we have successfully incorporated two and three different UAAs simultaneously into a neuroreceptor expressed in a Xenopus oocyte.
Results
Testing Frameshift Suppression Viability in Vivo.
To determine whether frameshift suppression is viable in Xenopus oocytes, we chose to use a tRNA that can be aminoacylated in vivo. We selected the human serine AS (HSAS), because it is aminoacylated (with serine) in eukaryotic cells and the seryl-tRNA synthetase does not recognize the anticodon (10–12). The CUA anticodon of HSAS was replaced with CCCG and ACCC to create the human serine FSs (HSFSCCCG and HSFSACCC) (cloverleaf structures shown in Fig. 1A), which recognize the quadruplet codons CGGG and GGGU. Prior research showed that these four-base codons are efficient in vitro (7). Injection of wild-type muscle nicotinic acetylcholine receptor (nAChR) mRNA and either HSFSCCCG or HSFSACCC (2.5 or 10 ng per oocyte; no amino acid ligated to the tRNA) into Xenopus oocytes resulted in no detectable channel expression. The addition of the original AS HSAS with wild-type nAChR mRNA did show channel expression with 2.5 ng of tRNA per oocyte but not with 10 ng. These results suggested that the HSFS tRNAs were causing +1 frameshifts, resulting in undesirable truncation of wild-type protein and thus a lack of detectable current. Analysis of the four nAChR subunits revealed four CGGG and one GGGU in-frame quadruplet codons, which were mutated to degenerate codons (see Materials and Methods) to avoid suppression. We refer to the resulting mRNAs as the “masked” constructs. Other groups have similarly removed undesired in-frame quadruplets (7, 9, 13). Injection of 2.5 ng per oocyte of either unligated HSFS plus the masked nAChR mRNAs resulted in functional channels with the same EC50 as channels expressed without tRNA (data not shown). Unless otherwise noted, all subsequent experiments used such masked constructs.
Fig. 1.
tRNAs and UAAs. (A) The AS tRNAs are shown, with the CUA anticodon and the FS anticodons shown at the bottom. YFaFS tRNA acceptor stem mutations are shown next to the YFFS tRNA body (italicized). (B) The three UAAs used in this study.
To test whether a naturally occurring amino acid (serine) could be incorporated in response to a quadruplet codon, we probed a highly conserved leucine of the nAChR M2 domain, a site designated Leu-9′. This site is a promiscuous site in the nicotinic receptor, and replacement of the native leucine with essentially any natural amino acid produces a functional receptor, usually with a quite noticeable shift in EC50. In particular, prior research showed that a leucine-to-serine mutation in the β-subunit (β9′) resulted in an ≈33-fold increased sensitivity to acetylcholine (ACh) (14). This site was mutated to UAG, CGGG, or GGGU. When mutant mRNA was injected into Xenopus oocytes along with 2.5 ng of unligated HSAS or HSFS tRNA, which should be aminoacylated with serine by the endogenous seryl-tRNA synthetase, significant channel expression was seen. However, the EC50 values varied depending on the incubation time (Table 1). This finding suggested that natural amino acids other than serine were being placed at the β9′ site with 2-day incubations, because the conventional mutant, β9′Ser, shows no change in EC50 (Table 1). The variability in EC50 between 1- and 2-day incubations suggests that the tRNAs are being modified to accept other amino acids. Modification of yeast phenylalanine tRNA in Xenopus oocytes has been shown to increase greatly from 1- to 2- day incubation times (15). Thus, we avoid this complication by incubation for 1 day. Amber suppression is highly efficient when the average maximal peak current (Imax) is measured at 1.25 ng of tRNA per oocyte and decreases slightly when 2.5 ng is added (Table 2). CGGG shows lower suppression than GGGU, in agreement with previous in vitro studies (7, 16). CGGG suppression is highly nonlinear, with a 330% increase in current when twice as much tRNA is injected (Table 2). GGGU, however, shows an almost linear relationship, with an increase of 86% in response to doubling (Table 2). These data suggest that HSFSACCC is a more efficient tRNA at recognizing its cognate quadruplet codon and/or has less competition with endogenous triplet tRNA in Xenopus oocytes than HSFSCCCG. These experiments establish that frameshift suppression is viable in Xenopus oocytes and that UAA incorporation should be feasible when using the appropriate FS tRNA.
Table 1.
HSAS and HSFS suppression experiments at the β9′ site
| β9′X | tRNA, 2.5 ng | EC50, 1 day* | nH | n | EC50, 2 days* | nH | n |
|---|---|---|---|---|---|---|---|
| AGC (serine) | none | 1.5 ± 0.04 | 1.7 ± 0.07 | 5 | 1.5 ± 0.2 | 1.9 ± 0.3 | 3 |
| UAG | HSAS | 1.7 ± 0.06 | 1.7 ± 0.09 | 6 | 0.70 ± 0.008 | 1.9 ± 0.07 | 14 |
| CGGG | HSFSCCCG | 2.1 ± 0.09 | 1.7 ± 0.1 | 8 | 1.3 ± 0.1 | 1.9 ± 0.3 | 13 |
| GGGU | HSFSACCC | 1.9 ± 0.08 | 1.5 ± 0.08 | 9 | 0.68 ± 0.1 | 1.7 ± 0.04 | 5 |
*Incubation time.
Table 2.
HSAS and HSFS suppression experiments at the β9′ site (1-day incubation)
| β9′X | tRNA | Imax ± SE,* 1.25† | n | Imax ± SE,* 2.5† | n | % HSAS, 1.25† | % HSAS, 2.5† | % change‡ |
|---|---|---|---|---|---|---|---|---|
| UAG | HSAS | −19 ± 2 | 12 | −14 ± 3 | 11 | 100 | 100 | −26 |
| CGGG | HSFSCCCG | −1.3 ± 0.3 | 10 | −5.6 ± 1 | 12 | 6.8 | 40 | 330 |
| GGGU | HSFSACCC | −8.6 ± 3 | 10 | −16 ± 3 | 12 | 45 | 110 | 86 |
*Average Imax (in μA) recorded at 50 μM ACh.
†Nanograms of tRNA.
‡Between 1.25 and 2.5 ng.
UAA Incorporation by Frameshift Suppression.
THG73 is an AS tRNA (cloverleaf structure shown in Fig. 1A) (17) that is used extensively for incorporating UAAs into various ion channels expressed in Xenopus oocytes (2). Initially, a FS derived from THG73 that recognizes the quadruplet codon CGGG (THG73FSCCCG) was tested for UAA incorporation. Attempts to suppress β9′CGGG with THG73FSCCCG-L, where L was chemically aminoacylated onto the tRNA, showed no current in vivo. This result is consistent with data from Voss and coworkers (9), who saw very little UAA incorporation with THG73FSACCC in Xenopus oocytes.
We then chose to screen yeast phenylalanine FS (YFFS) tRNA, which was used successfully by Sisido and colleagues (7, 16) in vitro. We studied both YFFSCCCG and YFaFSACCC (yeast phenyalanine containing acceptor stem mutations FS); Fig. 1A shows cloverleaf structures. The latter contains acceptor stem mutations (denoted by the “a”) incorporated to reduce glycyl-tRNA synthetase recognition (7). We first evaluated a nonpromiscuous position of the nAChR, α149W, an agonist-binding site tryptophan that makes a cation-π interaction with ACh (18). Wild-type recovery (i.e., suppressing the α149 quadruplet codons with YFFSCCCG-W or YFaFSACCC-W) resulted in functional, wild-type channels (Table 3). To demonstrate UAA incorporation, we relied on previous work using the AS THG73 that established that 5-fluoro-tryptophan (WF1) (structure shown in Fig. 1B) incorporated at α149 decreased the cation-π interaction and caused an ≈4-fold increase in EC50 (18). YFFSCCCG-WF1 suppression at α149CGGG resulted in a comparable increase in EC50 (Table 3), establishing the successful incorporation of the UAA WF1.
Table 3.
Wild-type recovery and UAA incorporation by frameshift suppression in vivo
We next considered the previously mentioned Leu-9′ residue. Suppression at β9′GGGU and δ9′GGGU with YFaFSACCC-Aba (where Aba is α-aminobutyric acid) and YFaFSACCC-Nval (where Nval is norvaline) (UAA structures shown in Fig. 1B), respectively, resulted in reductions in EC50 (Table 3) that were consistent with previous studies that used the same UAAs and the AS THG73 (14). All frameshift suppression experiments had an Imax between −1.6 and −4.4 μA, which is more than adequate for UAA studies in vivo and should allow for the incorporation of multiple UAAs. In all cases, injection of full-length tRNA that had no amino acid attached to the 3′ end resulted in no detectable currents in response to added ACh, directly showing a lack of aminoacylation by endogenous Xenopus aaRSs.
Masking Effects on Frameshift Suppression.
Experiments with HSFS required the masking of the nAChR subunits to avoid protein truncation caused by +1 frameshifts. To demonstrate the effect on UAA incorporation, suppression experiments were performed with wild-type and masked constructs. The quadruplet codon GGGU was chosen because there was only one in-frame quadruplet in the signaling sequence of the nAChR β-subunit and none in the α-, γ-, or δ-subunits. Wild-type recovery was performed by suppressing at α149GGGU with YFaFSACCC-W and adding either wild-type or masked β mRNA to the injection mixture. Table 6, which is published as supporting information on the PNAS web site, shows the dramatic effect of masking one position on frameshift suppression. With a 1:1:1:1 ratio of α:β:γ:δ, the masked construct gives a 2.7-fold increase in Imax relative to wild type. As the amount of α-subunit (which contains the suppression site) is increased, the masking effect decreases to 1.5-fold and 1.2-fold with subunit ratios of 5:1:1:1 and 10:1:1:1, respectively. Calculations that assume two equally efficient quadruplet codons reproduce this trend (Table 6), suggesting that the α149GGGU and the GGGU present in the β-subunit have similar suppression efficiencies.
Comparison of Frameshift and Nonsense Suppression Efficiencies.
To compare frameshift and nonsense suppression, the α149 and β9′ sites were studied in more detail. Suppression of α149CGGG or GGGU with 10 ng of YFFSCCCG-W or YFaFSACCC-W resulted in 38% and 48%, respectively, of the current from 10 ng of THG73-W suppression at α149UAG (Table 4). Suppression of β9′UAG with 2 ng of THG73-L resulted in the largest Imax (Table 4). Suppression at β9′CGGG or GGGU with 2 ng of YFFSCCCG-L or YFaFSACCC-L resulted in 14% and 36%, respectively, of the current from THG73-L (Table 4). We conclude that amber suppression is more efficient than frameshift suppression, in agreement with a trend previously seen in a eukaryotic cell-free translation system (16). In particular, the suppression efficiency observed here follows the order: THG73 > YFaFSACCC > YFFSCCCG.
Table 4.
Comparison of suppression efficiency, aminoacylation, and read-through in vivo
| mRNA | tRNA | tRNA, ng | n | Imax ± SE* | % THG73 or % UAG |
|---|---|---|---|---|---|
| α149UAG | THG73-W | 10 | 18 | −4.8 ± 2 | 100† |
| α149CGGG | YFFSCCCG-W | 10 | 20 | −1.8 ± 0.3 | 38† |
| α149GGGU | YFaFSACCC-W | 10 | 13 | −2.3 ± 0.9 | 48† |
| β9′UAG | THG73-L‡ | 2 | 15 | −6.1 ± 2 | 100† |
| β9′CGGG | YFFSCCCG-L‡ | 2 | 12 | −0.84 ± 0.2 | 14† |
| β9′GGGU | YFaFSACCC-L‡ | 2 | 9 | −2.2 ± 0.5 | 36† |
| β9′CGGG | YFFSCCCG-L‡ | 6 | 13 | −8.8 ± 0.9 | NA† |
| β9′GGGU | YFaFSACCC-L‡ | 6 | 13 | −16 ± 2 | NA† |
| β9′UAG | THG73-dCA | 2 | 13 | −4.8 ± 1 | 100† |
| β9′CGGG | YFFSCCCG-dCA | 2 | 13 | −0.42 ± 0.8 | 8.8† |
| β9′GGGU | YFaFSACCC-dCA | 2 | 13 | −0.092 ± 0.02 | 1.9† |
| β9′UAG | THG73-dCA | 6 | 13 | −8.2 ± 1 | 100† |
| β9′CGGG | YFFSCCCG-dCA | 6 | 12 | −1.2 ± 0.3 | 15† |
| β9′GGGU | YFaFSACCC-dCA | 6 | 11 | −0.27 ± 0.09 | 3.3† |
| β9′UAG | — | — | 13 | −0.37 ± 0.1 | 100§ |
| β9′CGGG | — | — | 13 | −0.085 ± 0.03 | 23§ |
| β9′GGGU | — | — | 13 | −0.078 ± 0.02 | 21§ |
NA, not applicable; —, no tRNA.
*Average Imax (in μA) recorded at 1 mM ACh.
†THG73.
‡Currents in response to 10 μM and 1 mM ACh displayed a ratio of 0.1, as anticipated from the Hill equation fit for one wild-type receptor.
§UAG.
Interestingly, the yield of receptors from frameshift suppression at the β9′ site was substantially improved by increasing the amount of tRNA injected. Suppression with 6 ng of YFFSCCCG-L or YFaFSACCC-L gave dramatic increases in Imax, with a percentage change of 950% and 630%, respectively (Table 4). This large change in Imax in response to a modest increase in tRNA concentration implicates a competition with endogenous triplet tRNA that responds nonlinearly to the amount of injected FS tRNA. A comparable increase in the amount of injected THG73-L led to complications due to reacylation of the tRNA by endogenous aaRSs (undesired) and incorporation of natural amino acids other than leucine, an issue that is addressed in detail in the following section and in Discussion.
Comparison of Aminoacylation of Suppressor tRNA and Read-Through of Suppression Sites.
To evaluate aminoacylation in vivo, which is undesirable for any tRNA used to incorporate UAAs, the β9′ site was again studied, because most amino acids produce functional receptors when substituted at this position (14). In all experiments, tRNAs that had been ligated to dinucleotide deoxyCA (dCA) but did not contain an amino acid at the 3′ end were injected to more closely mimic the biologically active, full-length tRNA. To maximize the potential for aminoacylation by endogenous aaRSs, 2-day incubations and relatively large mRNA injections (16.5 ng) were used. Surprisingly, THG73-dCA, which has been used extensively for UAA incorporation in Xenopus oocytes, showed significant aminoacylation in vivo, with an Imax of −4.8 and −8.2 μA for 2 and 6 ng of tRNA, respectively (Table 4). Note that under other conditions (less mRNA and shorter incubations), previous work has found no complications from aminoacylation using THG73-dCA in Xenopus oocytes (9, 14, 17). Still, the present results establish that THG73 is susceptible to aminoacylation by aaRSs, which is undesired. No aminoacylation was seen with 2 ng of THG73-L, suggesting that aminoacylation by endogenous aaRSs is more likely when nonaminoacylated THG73 is injected, as noted previously (17). Both FS tRNAs show much lower amounts of aminoacylation by aaRSs, as evidenced by the decrease in Imax (Table 4). YFFSCCCG-dCA shows only 8.8% and 15% of the Imax of THG73-dCA at 2 and 6 ng, respectively. The most orthogonal suppressor is YFaFSACCC-dCA, with 1.9% and 3.3% of the Imax of THG73-dCA at 2 and 6 ng, respectively. The orthogonality trend thus follows the order: YFaFSACCC-dCA > YFFSCCCG-dCA > THG73-dCA. YFaFSACCC is the most orthogonal and efficient FS tRNA, and it therefore offers a viable replacement for THG73, especially when aminoacylation by aaRSs poses a problem in vivo.
Read-through at the β9′ site was also assessed by injection of mRNA only (Table 4). β9′UAG showed the most read-through, presumably because there is only one in-frame stop codon before desired termination. β9′CGGG and β9′GGGU show 23% and 21% read-through, respectively, relative to the UAG stop codon. This finding is consistent with the idea that an endogenous triplet tRNA recognizing the first three bases of a quadruplet codon causes a −1 frameshift, which then presents multiple stop codons. Again, we designed this experiment to enhance signals from read-through by injecting large amounts of mRNA (50 ng). No current was detectable after injection of mRNA containing UAG, CGGG, or GGGU at position α149, confirming that this site is much less promiscuous than β9′.
Incorporation of Two UAAs.
To investigate the simultaneous incorporation of two UAAs, we again built on previous work that used THG73 to incorporate UAAs into the nAChR at positions α149, β9′, and δ9′. Importantly, EC50 changes associated with mutations at these sites are independent of one another (18, 19), allowing one to qualitatively anticipate the consequences of multiple mutations. In particular, both β9′Aba and δ9′Nval produce predictable reductions in EC50 that should be reproduced when combined with mutations at α149 (14). That is, the previously noted 4-fold increase in EC50 that is seen when the native tryptophan at α149 is replaced by WF1 should persist when in combination with β9′Aba or δ9′Nval.
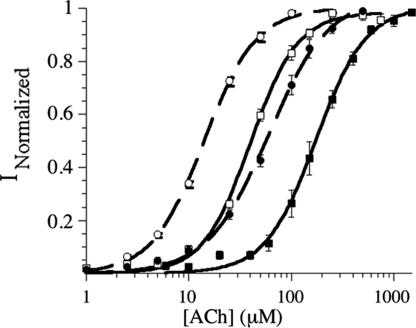
Successful incorporation of two UAAs to produce large ACh-induced currents could be seen when a 5-fold excess of mutant to wild-type mRNA was used. Suppression with α149UAG/THG73-W and β9′CGGG/YFFSCCCG-L is a wild-type recovery experiment that gave the expected EC50 for ACh of 50 μM (Table 5). Maintaining β9′CGGG/YFFSCCCG-L but substituting α149UAG/THG73-WF1 resulted in the anticipated 4-fold increase in EC50 (Table 5) (18). For incorporation of two UAAs, α149UAG/THG73-W or WF1 was combined with either β9′CGGG/YFFSCCCG-Aba or δ9′GGGU/YFaFSACCC-Nval (Table 5 and Fig. 2; representative traces are shown in Fig. 4, which is published as supporting information on the PNAS web site). The α149 WF1:W EC50 ratios are 4.4 for the β9′ and δ9′ mutants. These experiments establish that frameshift suppression can be combined with nonsense suppression to incorporate two UAAs in a eukaryotic system.
Table 5.
Incorporation of two UAAs
| α149 | tRNA | β or δ | tRNA | EC50 (theoretical)* | nH | n |
|---|---|---|---|---|---|---|
| UAG | THG73-W | β9′CGGG | YFFSCCCG-Aba | 14 ± 0.4 (16)† | 1.7 ± 0.06 | 9 |
| UAG | THG73-W | δ9′GGGU | YFaFSACCC-Nval | 41 ± 2 (36)† | 1.9 ± 0.1 | 9 |
| UAG | THG73-W | β9′CGGG | YFFSCCCG-L | 50 ± 3 (50)‡ | 1.4 ± 0.08 | 20 |
| UAG | THG73-WF1 | β9′CGGG | YFFSCCCG-Aba | 61 ± 3 | 1.5 ± 0.08 | 7 |
| UAG | THG73-WF1 | δ9′GGGU | YFaFSACCC-Nval | 180 ± 7 | 1.8 ± 0.1 | 6 |
| UAG | THG73-WF1 | β9′CGGG | YFFSCCCG-L | 200 ± 7 (200)‡ | 1.3 ± 0.04 | 9 |
Fig. 2.
Fits to the Hill equation (for incorporation of two UAAs). Data correspond to the rows in Table 5 as follows: row 1, open circles; row 2, open squares; row 4, filled circles; row 5, filled squares. Rows 3 and 6 are left out for clarity and have previously been reported (Table 3 and ref. 18).
Incorporation of Three UAAs.
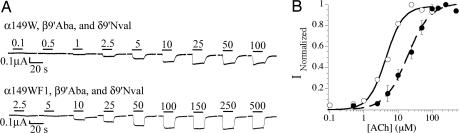
To demonstrate the incorporation of three UAAs, we combined the two-UAA incorporation experiments described above, taking advantage of the knowledge that EC50 is lowered monotonically by appropriate 9′ mutations at multiple subunits (20). Thus, one expects a lower EC50 when β9′Aba and δ9′Nval are incorporated simultaneously. Suppression of α149UAG:β9′CGGG:γ:δ9′GGGU by using an mRNA ratio of 5:5:1:5 with THG73-W, YFFSCCCG-Aba, and YFaFSACCC-Nval resulted in functional channel expression with an EC50 of 4.5 μM ACh (Fig. 3), which is lower than either of the two UAAs (Aba or Nval) incorporated separately. However, the same conditions with THG73-WF1 yielded only small currents. To obtain more expression, α149UAG mRNA and THG73-WF1 were initially injected, and, 24 h later, β9′CGGG:γ:δ9′GGGU (5:1:5) was injected with YFFSCCCG-Aba and YFaFSACCC-Nval (final mRNA ratio of 5:5:1:5). This strategy resulted in adequate expression and an EC50 of 19 μM ACh (Fig. 3). The ratio of the EC50s (α149 WF1:W) is 4.2, confirming that three UAAs were simultaneously incorporated in vivo.
Fig. 3.
Simultaneous incorporation of three UAAs. (A) Representative current traces from oocytes incorporating three UAAs. (B) Dose–response curves are as follows: α149W, β9′Aba, and δ9′Nval, open circles; α149WF1, β9′Aba, and δ9′Nval, filled circles. EC50 = 4.5 ± 0.4 and nH = 1.7 ± 0.3, and EC50 = 19 ± 2 and nH = 1.3 ± 0.1, respectively. The ratio of the EC50s is 4.2.
Discussion
The present results establish that frameshift suppression is viable in a eukaryotic, vertebrate cell and that it can be used to incorporate multiple UAAs in a single experiment. Previous work in Xenopus oocytes found that UAA incorporation using THG73FSACCC was inefficient, and it was proposed that either the Xenopus translational machinery was not compatible with frameshift suppression or that THG73FSACCC was a poor template for quadruplet recognition (9). Our results support the second rationalization, and a second FS derived from THG73, THG73FSCCCG, is also not viable. It thus appears that THG73-derived FS tRNAs are misfolded, not recognized by the elongation factor EF-Tu, or not accepted by other components of the translational machinery.
However, frameshift suppression is viable in the Xenopus oocyte by using either HSFS or YFFS tRNAs. We find that, in Xenopus oocytes, the quadruplet GGGU is suppressed more efficiently by both HSFSACCC and YFaFSACCC than the corresponding CGGG/tRNA pairs. This difference is seen despite the fact that in Xenopus, the GGG triplet is used twice as frequently (12.9 per 1,000) as the CGG triplet (21). Frameshift suppression must compete with endogenous triplet-recognizing tRNAs. Codon usage is apparently not a perfect predictor of frameshift suppression efficiency.
We have evaluated three different tRNAs: the AS THG73 and the FSs YFFSCCCG and YFaFSACCC. For UAA incorporation in the Xenopus oocyte, both YFFS tRNAs are less efficient than the AS THG73. This finding parallels results from earlier in vitro studies (16). Apparently, the competition between release factors and the AS tRNA is less detrimental than the competition between FS tRNAs and endogenous, triplet-recognizing tRNA. This view is supported by the rapid, nonlinear rise in suppression efficiency when the amount of YFFS tRNA is increased (Table 4). CGGG-recognizing tRNAs are more sensitive to the amount injected than GGGU-recognizing tRNAs. Increasing the amount of FS tRNA for UAA incorporation is essential to maximize suppression efficiency.
The incorporation of UAAs site-specifically into proteins requires the suppressor tRNA to be orthogonal to the endogenous aaRSs. Read-through of the suppression site or aminoacylation of the suppressor tRNA (once the chemically ligated UAA has been removed) can result in the undesired incorporation of natural amino acids at the suppression site. The two YFFS tRNAs studied here exhibit much more orthogonality than THG73 under the extreme conditions (extended incubation time and increased mRNA) used in Table 4. However, THG73 is an orthogonal suppressor tRNA to the Xenopus oocyte when used properly; THG73 has been used to successfully incorporate >100 residues at scores of sites in 20 different proteins (1, 2). Even promiscuous sites, such as the β9′UAG, can be efficiently suppressed by THG73-UAA when using less tRNA, mRNA, and incubation time (14). β9′UAG injected with THG73-dCA shows no greater current than mRNA alone with similar conditions. The small current is <1% of typical UAA incorporation experiments and is caused by read-through of the UAG codon (17). Voss and coworkers (9) found that THG73 incorporated three UAAs and Phe with efficiencies of 93.5–99.5% (determined by THG73-UAA incorporation relative to natural amino acids placed by read-through or aminoacylation of THG73-dCA) using luciferase expressed in Xenopus oocytes. The current results show that the YFFS tRNAs are even more orthogonal, and so the efficiency of UAA incorporation (relative to natural amino acids) should be greater than with THG73.
An important contributor to our ability to efficiently incorporate two and three UAAs is the masking of undesired quadruplets to prevent loss of UAA. In general, the requirement for masking of mRNA to remove undesirable quadruplet codons does complicate the frameshift suppression approach. The only previous examples of UAA incorporation in higher eukaryotes were performed by nonsense suppression (1–5, 10). Frameshift suppression may be limited in vivo to cells that are dormant (such as Xenopus oocytes), express large quantities of the target mRNA, or come from genetically engineered organisms. Also, suppressor tRNAs may be limited to rare codons because of possible toxicity arising from undesired suppression in other proteins (22).
The combination of nonsense and frameshift suppression allows one to incorporate multiple UAAs site-specifically into proteins expressed in Xenopus oocytes. These methods are compatible with our entire library of UAAs (2, 23) and will allow for multiple UAAs to be incorporated into other ion channels for structure–function studies, cross-linking, and FRET experiments. In principle, further quadruplet codons could be used to simultaneously incorporate more than three UAAs.
Materials and Methods
Materials.
All oligonucleotides were synthesized by the California Institute of Technology Biopolymer Synthesis facility or Integrated DNA Technologies (Coralville, IA) (sequences are listed in Table 7, which is published as supporting information on the PNAS web site). NotI was purchased from Roche Applied Science (Indianapolis). BamHI, EcoRI, FokI, T4 DNA ligase, and T4 RNA ligase were purchased from NEB (Beverly, MA). Kinase Max, T7 MEGAshortscript, and T7 mMessage mMachine kits were from Ambion (Austin, TX). dCA and 6-nitroveratryloxycarbonyl-protected dCA-UAA were prepared as reported in refs. 14, 18, and 24. ACh chloride was purchased from Sigma-Aldrich.
Gene Construction and RNA Preparation.
The α-, β-, γ-, and δ-subunits of nAChR were previously subcloned in the pAMV vector (25). All four in-frame CGGGs were mutated (shown in italics) to degenerate codons (α182CGC, β23AGG, β402AGG, and δ195AGG), and one GGGT was mutated at the fourth position (β1AGC); these constructs are termed “masked.” α149TAG, CGGG, GGGT; β9′TAG, CGGG, GGGT; and δ9′GGGT mutations were placed on masked constructs by QuikChange site-directed mutagenesis (Stratagene). Mutations were verified by DNA sequencing (at the California Institute of Technology Sequencing/Structure Analysis Facility). Template DNA was linearized with NotI and mRNA prepared with the T7 mMessage mMachine kit. mRNA was purified by using the RNeasy Mini kit (Qiagen, Valencia, CA) and quantified by absorption at 260 nm.
THG73 and HSAS in pUC19 vector were previously made (10, 17). Genes for HSFSCCCG, THG73FSCCCG, and YFFSCCCG (sequence from ref. 6) with flanking EcoRI and BamHI overhangs were phosphorylated by using the Kinase Max kit, annealed, and ligated with T4 DNA ligase into EcoRI and BamHI linearized pUC19 vector as described in ref. 24. A73G; C2G, G3C, G4C; and C69G, C70G, G71C mutations (from ref. 7) were sequentially placed by QuikChange mutagenesis on the YFFSCCCG construct to obtain YFaFSCCCG (“a” refers to acceptor stem mutations). HSFSACCC and YFaFSACCC (sequence from ref. 7) were prepared by replacing the anticodon of HSFSCCCG and YFaFSCCCG with ACCC by using QuikChange. All mutations were verified by DNA sequencing (at the California Institute of Technology Sequencing/Structure Analysis Facility). Template DNA for tRNA lacking the 3′CA was prepared by FokI digestion, and tRNA was transcribed by using the T7 MEGAshortscript kit. tRNA was desalted by using CHROMA SPIN-30 DEPC-H2O columns (BD Biosciences), and concentration was determined by absorption at 260 nm.
dCA and dCA-UAA Ligation to Suppressor tRNA.
dCA and 6-nitroveratryloxycarbonyl-protected dCA-UAA were coupled to suppressor tRNA by using T4 RNA ligase for 30 min as described in refs. 24 and 26, desalted by using CHROMA SPIN-30 DEPC-H2O columns, and quantified by absorption at 260 nm. tRNA ligation efficiency was determined by MALDI mass spectrometry (26), and all tRNA dCA or dCA-UAA ligations were >75%.
In Vivo Suppression Experiments.
Stage VI oocytes of Xenopus laevis were prepared as described in ref. 27. All tRNA were refolded at 65°C for 2 min, and tRNA-UAA were deprotected for 5 min by UV irradiation before injection (17). The injection volume for all experiments was 50 nl, and the incubation time was 44–52 h unless otherwise noted. Suppression of HSAS and HSFS (1.25 or 2.5 ng of tRNA) with 20 ng of mRNA in a subunit ratio of 2:5:1:1 for α:β9′(UAG, CGGG, or GGGU):γ:δ was recorded after 1 or 2 days. Single UAA incorporation was performed by using 20–30 ng of mRNA in a subunit ratio of 10:1:1:1 for α149(CGGG or GGGU):β:γ:δ, 2:5:1:1 for α:β9′GGGU:γ:δ, or 2:1:1:5 for α:β:γ:δ9′GGGU with 4.8–16.5 ng of YFFSCCCG/YFaFSACCC-UAA. Comparison of β-masked and wild-type suppression contained 25 ng of total mRNA injected in the subunit ratio listed in Table 6 with 1:1 γ:δ and 10 ng of YFaFSACCC-W. For comparison of suppression efficiency and aminoacylation of tRNA in vivo, all mRNA was normalized to the same concentration, and 16.5 ng of mRNA was injected in the subunit ratio 10:1:1:1 for α149(UAG, CGGG, or GGGU):β:γ:δ or 2:5:1:1 for α:β9′(UAG, CGGG, or GGGU):γ:δ with tRNA amounts listed in Table 4. For read-through experiments, 50 ng of mRNA in the ratio 2:5:1:1 for α:β9′(UAG, CGGG, or GGGU):γ:δ was injected. Two UAA experiments were performed by injection of 20–30 ng of mRNA in a subunit ratio of 5:5:1:1 for α149UAG:β9′CGGG:γ:δ or 5:1:1:5 for α149UAG:β:γ:δ9′GGGU with 10–25 ng of each suppressor tRNA-UAA. For three UAAs, α149W, β9′Aba, and δ9′Nval, 26 ng of mRNA in a ratio of 5:5:1:5 for α149UAG:β9′CGGG:γ:δ9′GGGU was injected with 20 ng of each suppressor tRNA-UAA, and a second injection of 33 ng of each tRNA-UAA was done 24 h later. For α149WF1, β9′Aba, and δ9′Nval, 8 ng of α149UAG mRNA with 50 ng of THG73-WF1 was injected, and a second injection of 18 ng of mRNA with a subunit ratio of 5:1:5 for β9′CGGG:γ:δ9′GGGU with 25 ng of each YFFSCCCG-Aba and YFaFSACCC-Nval was performed 24 h later; oocytes were recorded 3 days after the first injection.
Electrophysiology.
Recordings employed two-electrode voltage clamp on the OpusXpress 6000A (Axon Instruments, Union City, CA). ACh was stored at −20°C as a 1 M stock, diluted in Ca2+-free ND96, and delivered to oocytes by computer-controlled perfusion system. For HSAS and HSFS experiments, the holding potential was −60 mV, and all UAA experiments were performed at either −60 or −80 mV. Dose–response data were obtained from at least nine ACh concentrations, and comparisons were tested at one drug concentration, except β9′(UAG, CGGG, or GGGU) with tRNA-L, for which two concentrations, 10 μM and 1 mM, were used to check for aminoacylation (Table 4). Dose–response relations were fit to the Hill equation to determine EC50 and the Hill coefficient (nH). All reported values are represented as a mean ± SE of the tested oocytes [number (n) is listed with each table].
Supporting Information.
Masking experiments, representative traces for two UAA experiments (α149W or WF1 and β9′Aba), and oligonucleotides used in this study are detailed in Fig. 4 and Tables 6 and 7.
Supplementary Material
Acknowledgments
E.A.R. is a National Science Foundation Predoctoral Fellow. This work was supported by National Institutes of Health Grants NS-34407 and NS-11756.
Abbreviations
- UAA
unnatural amino acid
- aaRS
aminoacyl-tRNA synthetase
- AS
amber suppressor
- FS
frameshift suppressor
- HSAS
human serine AS
- HSFS
human serine FS
- YFFS
yeast phenylalanine FS
- WF1
5-fluoro-tryptophan
- Aba
α-aminobutyric acid
- Nval
norvaline
- ACh
acetylcholine
- nAChR
nicotinic ACh receptor
- dCA
dinucleotide deoxyCA.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dougherty D. A. Curr. Opin. Biotechnol. 2000;4:645–652. doi: 10.1016/s1367-5931(00)00148-4. [DOI] [PubMed] [Google Scholar]
- 2.Beene D. L., Dougherty D. A., Lester H. A. Curr. Opin. Neurobiol. 2003;13:264–270. doi: 10.1016/s0959-4388(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 3.Sakamoto K., Hayashi A., Sakamoto A., Kiga D., Nakayama H., Soma A., Kobayashi T., Kitabatake M., Takio K., Saito K., et al. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hino N., Okazaki Y., Kobayashi T., Hayashi A., Sakamoto K., Yokoyama S. Nat. Methods. 2005;2:201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z., Alfonta L., Tian F., Bursulaya B., Uryu S., King D. S., Schultz P. G. Proc. Natl. Acad. Sci. USA. 2004;101:8882–8887. doi: 10.1073/pnas.0307029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hohsaka T., Ashizuka Y., Murakami H., Sisido M. J. Am. Chem. Soc. 1996;118:9778–9779. [Google Scholar]
- 7.Hohsaka T., Ashizuka Y., Taira H., Murakami H., Sisido M. Biochemistry. 2001;40:11060–11064. doi: 10.1021/bi0108204. [DOI] [PubMed] [Google Scholar]
- 8.Anderson J. C., Wu N., Santoro S. W., Lakshman V., King D. S., Schultz P. G. Proc. Natl. Acad. Sci. USA. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shafer A. M., Kalai T., Bin Liu S. Q., Hideg K., Voss J. C. Biochemistry. 2004;43:8470–8482. doi: 10.1021/bi035542i. [DOI] [PubMed] [Google Scholar]
- 10.Monahan S. L., Lester H. A., Dougherty D. A. Chem. Biol. 2003;10:573–580. doi: 10.1016/s1074-5521(03)00124-8. [DOI] [PubMed] [Google Scholar]
- 11.Anderson J. C., Magliery T. J., Schultz P. G. Chem. Biol. 2002;9:237–244. doi: 10.1016/s1074-5521(02)00094-7. [DOI] [PubMed] [Google Scholar]
- 12.Saks M. E., Sampson J. R., Abelson J. N. Science. 1994;263:191–197. doi: 10.1126/science.7506844. [DOI] [PubMed] [Google Scholar]
- 13.Murakami H., Kourouklis D., Suga H. Chem. Biol. 2003;10:1077–1084. doi: 10.1016/j.chembiol.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Kearney P. C., Zhang H. Y., Zhong W., Dougherty D. A., Lester H. A. Neuron. 1996;17:1221–1229. doi: 10.1016/s0896-6273(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 15.Grosjean H., Droogmans L., Giege R., Uhlenbeck O. C. Biochim. Biophys. Acta. 1990;1050:267–273. doi: 10.1016/0167-4781(90)90179-6. [DOI] [PubMed] [Google Scholar]
- 16.Taira H., Fukushima M., Hohsaka T., Sisido M. J. Biosci. Bioeng. 2005;99:473–476. doi: 10.1263/jbb.99.473. [DOI] [PubMed] [Google Scholar]
- 17.Saks M. E., Sampson J. R., Nowak M. W., Kearney P. C., Du F. Y., Abelson J. N., Lester H. A., Dougherty D. A. J. Biol. Chem. 1996;271:23169–23175. doi: 10.1074/jbc.271.38.23169. [DOI] [PubMed] [Google Scholar]
- 18.Zhong W. G., Gallivan J. P., Zhang Y. O., Li L. T., Lester H. A., Dougherty D. A. Proc. Natl. Acad. Sci. USA. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kearney P. C., Nowak M. W., Zhong W., Silverman S. K., Lester H. A., Dougherty D. A. Mol. Pharmacol. 1996;50:1401–1412. [PubMed] [Google Scholar]
- 20.Labarca C., Nowak M. W., Zhang H., Tang L., Deshpande P., Lester H. A. Nature. 1995;376:514–516. doi: 10.1038/376514a0. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura Y., Gojobori T., Ikemura T. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magliery T. J., Anderson J. C., Schultz P. G. J. Mol. Biol. 2001;307:755–769. doi: 10.1006/jmbi.2001.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.England P. M. Biochemistry. 2004;43:11623–11629. doi: 10.1021/bi048862q. [DOI] [PubMed] [Google Scholar]
- 24.Nowak M. W., Gallivan J. P., Silverman S. K., Labarca C. G., Dougherty D. A., Lester H. A. Methods Enzymol. 1998;293:504–529. doi: 10.1016/s0076-6879(98)93031-2. [DOI] [PubMed] [Google Scholar]
- 25.Nowak M. W., Kearney P. C., Sampson J. R., Saks M. E., Labarca C. G., Silverman S. K., Zhong W., Thorson J., Abelson J. N., Davidson N., et al. Science. 1995;268:439–442. doi: 10.1126/science.7716551. [DOI] [PubMed] [Google Scholar]
- 26.Petersson E. J., Shahgholi M., Lester H. A., Dougherty D. A. RNA. 2002;8:542–547. doi: 10.1017/s1355838202026146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quick M. W., Lester H. A. In: Ion Channels of Excitable Cells. Narahashi T., editor. Vol. 19. San Diego: Academic; 1996. pp. 261–279. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.