Abstract
d-amino acids are much less common than their l-isomers but are widely distributed in most organisms. Many d-amino acids, including those necessary for bacterial cell wall formation, are synthesized from the corresponding l-isomers by α-amino acid racemases. The important class of pyridoxal phosphate-independent racemases function by an unusual mechanism whose details have been poorly understood. It has been proposed that the stereoinversion involves two active-site cysteine residues acting in concert as a base (thiolate) and an acid (thiol). Although crystallographic structures of several such enzymes are available, with the exception of the recent structures of glutamate racemase from Bacillus subtilis and of proline racemase from Trypanosoma cruzi, the structures either are of inactive forms (e.g., disulfide) or do not allow unambiguous modeling of the substrates in the active sites. Here, we present the crystal structures of diaminopimelate (DAP) epimerase from Haemophilus influenzae with two different isomers of the irreversible inhibitor and substrate mimic aziridino-DAP at 1.35- and 1.70-Å resolution. These structures permit a detailed description of this pyridoxal 5′-phosphate-independent amino acid racemase active site and delineate the electrostatic interactions that control the exquisite substrate selectivity of DAP epimerase. Moreover, the active site shows how deprotonation of the substrates’ nonacidic hydrogen at the α-carbon (pKa ≈ 29) by a seemingly weakly basic cysteine residue (pKa ≈ 8–10) is facilitated by interactions with two buried α-helices. Bacterial racemases, including glutamate racemase and DAP epimerase, are potential targets for the development of new agents effective against organisms resistant to conventional antibiotics.
Keywords: racemase, stereochemical mechanism
The increase in microbial resistance to conventional antibiotics has rekindled intense interest in new methods of inhibiting bacterial cell wall biosynthesis (1), including blocking formation or utilization of d-amino acids such as d-alanine (2), d-glutamate (3), and meso-diaminopimelic acid (4) present in peptidoglycan (5). The inclusion ofd-amino acids in the peptidoglycan layer of the cell wall is thought to provide bacteria with protection from the action of host proteases (2). Glutamate racemase (6, 7) and diaminopimelate (DAP) epimerase (EC 5.1.1.7) (8), as well as aspartate racemase (9) and proline racemase (10, 11), are examples of pyridoxal 5′-phosphate (PLP)-independent amino acid racemases that invert the configuration at the α-carbon of an amino acid without the use of cofactors, metals, or reducible keto or imino functionalities. The proposed “two-base” mechanism (11) for these enzymes involves one active-site cysteine thiolate acting as a base to deprotonate the α-carbon, while a second cysteine thiol functions as an acid to reprotonate the resulting planar carbanionic intermediate from the opposite face (Scheme 1). This process is not trivial, because the pKa of the α-proton is ≈21 for the fully protonated form of an amino acid, whereas the corresponding pKa of the zwitterionic form predominant in neutral aqueous solution is ≈29, partially due to the presence of the carboxylate anion (12, 13). The proposed use of cysteine (typical pKa of ≈8–10) as a base is surprising; thiolate anions are poor catalysts of α-carbon deprotonation in aqueous solution, with an estimated half-time for deprotonation of proline zwitterions of >1,500 years at pH 7 (13). Furthermore, in a fully protonated amino acid, the hydrogen at the α-carbon is much less acidic than the protons on the carboxylic acid and ammonium functionalities and would not normally be removed by a general base pKa of ≈10. The debate surrounding the mechanism by which the significant energetic barrier for α-deprotonation in these enzymes is overcome has been hindered by the lack of detailed structural information on the mode of substrate binding in the active site (7, 13).
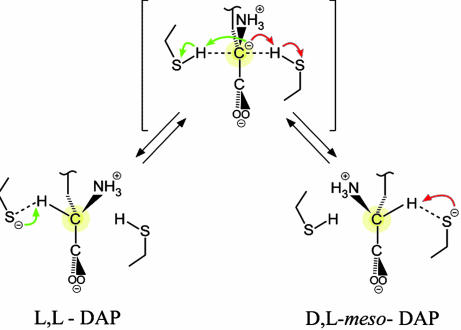
Scheme 1.
Mechanism of catalysis and inhibition. (Upper) The proposed “two-base” mechanism for the interconversion of ll- and dl-(meso)-DAP. (Lower) Inhibition of DAP epimerase by ll-aziridino (Azi)DAP (1) and dl-AziDAP (2).
Among the more mechanistically interesting PLP-independent amino acid racemases is the DAP epimerase, which interconverts ll-DAP to meso-DAP (Scheme 1) in the biosynthetic pathway leading from l-aspartate to l-lysine (14, 15). Because meso-DAP is a precursor to l-lysine and an essential building block of the peptidoglycan layer of the bacterial cell wall, specific inhibitors of the enzymes in the DAP pathway may lead to a new class of antibiotics (4). Also, because mammals lack this metabolic pathway and require l-lysine as a dietary component, inhibitors of the meso-DAP/lysine biosynthetic pathway offer the additional advantage of potentially providing selective toxicity against bacteria. DAP epimerase is a unique member of this family of racemases, because its substrates (ll-DAP and meso-DAP) contain two stereocenters. dd-DAP is not a substrate for DAP epimerase, and, therefore, the stereochemistry at the nonreacting (distal) carbon is critical for substrate recognition. The three-dimensional structure of an inactive form of DAP epimerase from Haemophilus influenza identified a previously unrecognized fold for this 274-aa enzyme, consisting of two structurally similar α/β domains, with the active site lying at the interface between the two domains (16, 17). A disulfide linkage between the two active-site cysteines (Cys-73 and Cys-217) resulted in a disrupted active site, thereby preventing any reliable modeling of a bound substrate or inhibitor.
Crystal structures of glutamate racemase from Aquifex pyrophilus (3) and of aspartate racemase from Pyrococcus horikoshii (9) have also been determined and reveal two similar domain (α/β) organizations for these enzymes. The x-ray crystal structure of glutamate racemase has been reported with the substrate analogue d-glutamine (3), but the lack of any strong interactions with the enzyme allows for a variety of possible binding modes in molecular modeling studies and precludes convincing mechanistic deductions (18). Recently, Rice and coworkers (19) have reported that the crystal structure of d-glutamate bound to glutamate racemase from Bacillus subtilis; this structure revealed a “closed conformation” for this enzyme upon ligand binding. The authors suggested that, in glutamate racemase, an aspartate (Asp-10) and/or histidine (His-187) could act as a base catalyst but that the epimerization mechanism utilizes a neutral thiol. The recently reported crystal structure of proline racemase from Trypanosoma cruzi also reveals a monomer with two symmetric α/β domains separated by a deep crevice (20). The binding of a transition state analogue, inhibitor pyrrole-2-carboxylic acid, to proline racemase close to two symmetrically located cysteine residues at the bottom of the crevice is in agreement with their proposed acid–base-catalyzed mechanism. The authors suggest that the cysteine residues are assisted by a histidine (His-132) and an aspartate (Asp-296), as seen in the glutamate racemase active site. In contrast, we propose that the active site of DAP epimerase exists as a thiol–thiolate pair at neutral pH and does not use any neighboring active-site bases to assist the deprotonation process.
On the basis of sequence and structural similarity, it appears that the glutamate and aspartate racemases constitute one homologous family of enzymes, whereas DAP epimerase and proline racemase both belong to a different family.
To gain insight into the racemization mechanism of these unique enzymes, we have synthesized aziridino (Azi) analogues of DAP and shown that they produce irreversible inhibition of DAP epimerase (21). We report here the crystal structures of DAP epimerase from H. influenza in complex with ll-AziDAP and dl-AziDAP to resolutions of 1.35 and 1.7 Å, respectively. These crystal structures of a PLP-independent amino acid racemase with specific inhibitors provide detailed insight into the mechanism of this unique protein catalyst and also assist in the understanding of other members of this important family of enzymes.
Results and Discussion
Structures of DAP Epimerase Inactivated with ll-AziDAP and dl-AziDAP.
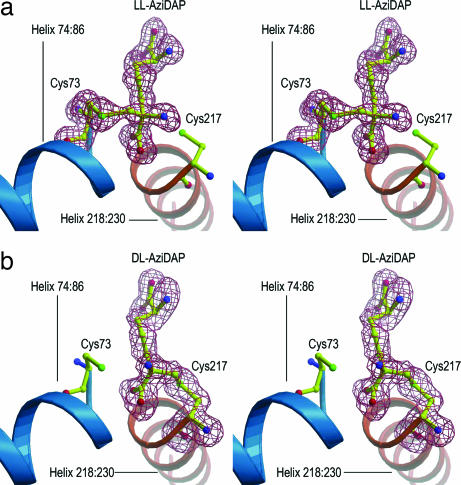
Isomorphous cocrystals of the ll-AziDAP- and dl-AziDAP-inhibited forms of DAP epimerase belong to space group C2221. Both structures were solved by molecular replacement and refined to respectable R factors. DAP epimerase has a two-domain α/β structure, with residues 1–117 and 263–274 forming the N-terminal domain and residues 118–262 forming the C-terminal domain; each domain contains eight β-strands and two α-helices. An internal pseudodyad relates the two domains of DAP epimerase, with an rms deviation (rmsd) of 1.72 Å for 99 Cα pairs of the two domains. In the initial stages of refinement, both the 2|Fo| − |Fc| and the |Fo| − |Fc|, αc-phased electron density maps revealed the location of the ll-AziDAP covalently bound to Cys-73 in the ll-AziDAP-inhibited enzyme and the location of the dl-AziDAP covalently bound to Cys-217 in the dl-AziDAP-inhibited enzyme. Omit maps of the refined models confirmed the placement of these inhibitors in the active site of DAP epimerase (Fig. 1). The two diastereomeric inhibitors 1 and 2 (Scheme 1) are bound to the respective cysteine residues with little difference in the DAP epimerase backbone structure; the rmsd for the 274 Cα atom pairs is 0.15 Å. The inhibitor ll-AziDAP (1) is bound at the DAP epimerase active site by a covalent bond between the methylene carbon of the aziridine ring and the thiolate (Sγ) of Cys-73 that resulted from the nucleophilic ring opening of the three-membered aziridine ring (Scheme 1) (21). Similarly, the structure of DAP epimerase inhibited by dl-AziDAP (2) shows analogous alkylation of the thiolate (Sγ) of Cys-217. These complexes are excellent mimics for the substrate bound in the active site; the covalent bond between cysteine sulfur and the methylene carbon atom offers an approximation to the approach of the thiol base during the α-deprotonation of a DAP substrate.
Fig. 1.
Stereoviews of omit Fo − Fc map (flesh) contoured at 3σ in the active site of DAP epimerase in the ll-AziDAP–epimerase complex (a) and the dl-AziDAP–epimerase complex (b). The two inhibitors are represented in as balls and sticks, with carbon shown in yellow, oxygen in red, and nitrogen in blue. The catalytic cysteines Cys-73 and Cys-217 are represented as balls and sticks, with carbon shown in yellow, sulfur in green, oxygen in red, and nitrogen in blue. The two active-site helices, A (74:86) and B (218:230), are shown in teal and orange, respectively.
Inhibitor Binding and Active-Site Restructuring.
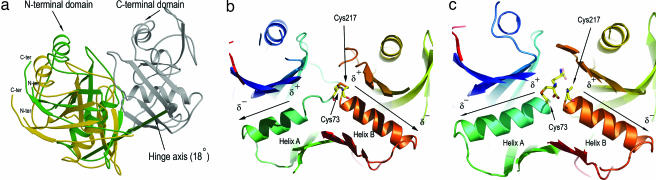
The binding of the AziDAP inhibitors to DAP epimerase induces a large conformational change in the enzyme. The most striking difference between the structure of the native and inhibitor-bound DAP epimerase is a clear decrease in the distance separating the two domains in the latter (Fig. 2a). Superposition of the entire ll-AziDAP-bound enzyme onto the native enzyme yields an rmsd of 2.45 Å for the 274 Cα atom pairs. However, the individual domains are more similar, with an rmsd of 1.05 Å for the 129 Cα atom pairs between the N-terminal domains and an rmsd of 0.32 Å for the 145 Cα atom pairs constituting the C-terminal domains. Analysis of this conformational change reveals an oyster shell-like clamping of the two domains, which can be represented by an ≈18° rotation of the N-terminal domain relative to the C-terminal domain of the inactive form of DAP epimerase (16, 17) (Fig. 2a), thereby displacing water from the active-site cleft and burying the inhibitors in an anhydrous cylindrical cavity ≈15 Å in length (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 2.
Conformational changes on inhibitor binding. (a) Shell-like domain closure upon binding AziDAP inhibitors. The hinge rotation axis (≈18°), represented by an arrow, was determined by superimposition of the C-terminal domain of the native, inactive DAP epimerase (PDB ID code 1GQZ) with the C-terminal domain of the ll-AziDAP-inhibited DAP epimerase (rmsd = 0.32 Å for 145 Cα atoms). The N-terminal domain of the unbound enzyme is shown in gold, and that of the inhibited DAP epimerase is shown in green. The arrow coloring indicates the direction of rotation of the N-terminal domain (green) relative to the C-terminal domain (gray). (b) Environments of the two opposed α-helices in the native unbound DAP epimerase (PDB ID code 1GQZ). The N-terminal four-helix A residues, Gly-74 to Ala-77, are partially unwound in this structure. Helix A is colored teal; helix B (Gly-218 to Met-230) in the C-terminal domain of DAP epimerase is fully formed and colored orange. The two catalytic cysteines (Cys-73 and Cys-217) are involved in a disulfide link. (c) Helices A and B, in the presence of the bound inhibitor AziDAP, covalently linked to Sγ of Cys-73. The carbon atoms of the inhibitor are colored yellow (nitrogen atoms, blue; oxygen, red). Helix A (Gly-74 to Leu-86) is on the left, and helix B is on the right. The opposing dipole moments are directed at the α-carbon carboxylate.
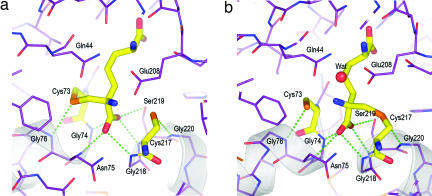
In addition to the N-terminal domain movement, there is an important coil-to-helix transformation involving residues Gly-74, Asn-75, and Gly-76. The acquisition of this additional turn at the N terminus of helix A (Gly-74 to Leu-86) has a major influence on the active form of the enzyme (Fig. 2 b and c). This loop reorganization creates a unique catalytic site in which two α-helices (A and B) related by the pseudo-twofold axis between the two domains face each other, with the two catalytic cysteines mounted at the N termini of each of these helices. The dipole moment of helix A opposes that of helix B (Gly-218 to Met-230), and both N-terminal partial positive charges (≈+0.5 e) are directed at the α-carboxylate moieties of the inhibitors. In addition to the dipole moments directed at the carboxylate group on the α-carbon undergoing racemization, this carboxylate is the recipient of four hydrogen bonds from the main-chain nitrogens of residues Gly-74, Asn-75, Gly-218, and Ser-219, as well as a fifth hydrogen bond from the highly conserved Ser-219 Oγ (Fig. 3). A similar hydrogen-bonding network to the α-carboxylate group of pyrrole-2-carboxylate is seen in the proline racemase structure (20), with the exception that the H bond from Ser-219 in DAP epimerase comes from a threonine in proline racemase. Apart from the H bond from Ser-219 Oγ, the four main-chain hydrogen bonds to the carboxylate oxygens in DAP epimerase approach from a direction of ≈45° to the plane of the carboxylate. This unusual hydrogen bonding to the carboxylate suggests sp3 hybridization of the oxygen atoms and thereby plays an important role in the catalytic mechanism.
Fig. 3.
Hydrogen bonding at the catalytic center. (a) Close-up of the atomic details of the active-site DAP epimerase in the presence of the ll-AziDAP inhibitor covalently bound to Cys-73 Sγ. The α-carboxylate is the recipient of five hydrogen bonds (green dotted lines). Cys-73 Sγ and Cys-217 Sγ are both recipients of main-chain N···H hydrogen bonds from Gly-76 and Gly-220, respectively. Carbon atoms of the enzyme are shown in violet, nitrogen in blue, and oxygen in red. The carbon atoms of the inhibitor are shown in yellow, as are the carbon atoms of the catalytic cysteines. (b) Hydrogen-bonding interactions (green dotted lines) in the active site of DAP epimerase in the presence of the inhibitor dl-AziDAP. The atom coloring is the same as in a. dl-AziDAP is covalently bonded to Cys-217 Sγ.
A comparison of the native and inhibitor-bound DAP epimerase also reveals a marked rearrangement of the amino acid side chains lining the active-site cavity. The residues lining the active-site cavity are largely conserved across all of the DAP epimerase sequences isolated from different bacterial species and include: Asn-11, Phe-13, Gln-44, Tyr-60, Asn-64, Val-70, Glu-72, Cys-73, Gly-74, Asn-75, Asn-157, Asn-190, Glu-208, Arg-209, Cys-217, Gly-218, and Ser-219 (17). Some of these residues in the inhibitor-bound enzyme form an extensive hydrogen-bonded network surrounding the inhibitor (Fig. 7, which is published as supporting information on the PNAS web site). The highly structured epimerase active site (Figs. 3 and 7) is absent in the inactive disulfide form of the enzyme (16, 17) and results from changes in side-chain conformations of the active site residues accompanying the substrate binding and domain closure.
Sterochemical Control in Catalysis.
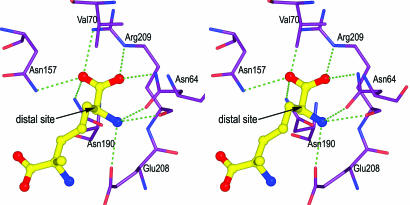
Our structural results also clarify why DAP epimerase has very strict requirements for its substrates, accepting only DAP isomers having the l configuration at the distal ε-carbon (4). The carboxyl group at the distal site of both inhibitors forms a salt bridge with the positively charged side chain of Arg-209 along with three hydrogen bonds from side-chain Nδ2 atoms of residues Asn-64, Asn-157, and Asn-190 (Fig. 4). The distal positively charged amino group of the inhibitors forms three hydrogen bonds with side-chain oxygen atoms of Asn-64 and Glu-208 and the carbonyl oxygen of Arg-209. The asymmetric disposition of side chains of Asn-64 and Asn-157 situated on opposite sides of the ε-carbon of the inhibitor, coupled with tight packing of other active-site residues surrounding the distal site, prevents the binding of a d-isomer at that position. The binding interactions of the distal l-stereocenter of ll- and dl-AziDAP help in positioning the substrate carbon skeleton correctly for forming the covalent bond from the cysteine sulfur and the methylene carbon of AziDAP.
Fig. 4.
Stereo representation of hydrogen-bonding interactions at the distal center of ll-AziDAP. The inhibitor and enzyme model are shown as balls and sticks; the carbon atoms of ll-AziDAP are shown in yellow, and the carbon atoms of the enzyme are shown in violet (nitrogen, blue; oxygen, red). Hydrogen bonds are represented by dotted green lines.
Substrate Activation and Catalysis.
The elaborate machinery required to catalyze epimerization at the α-carbon atom of DAP becomes evident from a comparison of both the enzyme–inhibitor structures. Superimposition of the two inhibitor–epimerase complexes reveals a remarkable degree of overlap, thereby indicating that the residues of the active site are held rigidly (Fig. 3 a and b). We have identified six structural features of the epimerase that contribute to catalysis of this otherwise unfavorable reaction.
Dispersal of the negative charge on the α-carboxylate of the zwitterionic substrate by the opposing α-helical dipole moments and the hydrogen-bonding environment of the α-carboxylates.
The α-carboxylates of both inhibitors are located directly on the vectors defining the dipole moments of helices A and B (Fig. 2c). Also, each oxygen of the carboxylate group is the recipient of two hydrogen bonds from the main-chain amides at the N termini of each of these helices (Fig. 3). As mentioned earlier, calculations have indicated that protonated α-carboxyl groups of α-amino acids show dramatically reduced (factor of ≈1010) pKas for the α-proton (13). The observed electrostatic environment of the α-carboxylate likely disperses its negative charge far more effectively than protonation, thereby amplifying this effect (22).
Concomitant orientation of the carboxylate to maximize orbital overlap between its π system and the breaking σ bond of the hydrogen-to-α-carbon (i.e., stereoelectronic alignment).
Both enzyme–inhibitor structures possess an identical network of hydrogen bonds to the carboxylate. Its minimal movement indicates that this delocalization of anionic charge would also be maintained during catalysis. Simultaneously, as these electrostatic interactions align the carboxyl π system to the Cα–H bond, hydrophobic binding of the substrate central chain sets the dihedral angles for Cβ–Cα–C–O at −161° (Cys-217-meso-AziDAP) and at 175° (Cys-73-ll-AziDAP). This conformation represents an ideal alignment of the σ-orbital of the forming anion with the π-orbitals of the carbonyls (Scheme 1 and Fig. 5) (23), a phenomenon also known to increase the acidity of α-carbon protons (24).
Fig. 5.
Proposed mechanism for the interconversion of ll-DAP and dl-meso-DAP by DAP epimerase. The transition state is a planar carbanion, with the six atoms, NH3+, Cβ, Cα, C, and the two carboxyl oxygens forming the plane. This transition state can be approached from either direction with equal frequency. The two catalytic cysteine residues Cys-73 and Cys-217 are poised to repeatedly deprotonate and reprotonate the Cα of the bound substrate from opposite faces until it is released.
Full protonation of the α-amino group with a switch in the hydrogen-bonding partner from Glu-208 to Gln-44 during epimerization.
When the meso-DAP mimic (dl-AziDAP) is linked to Cys-217, the fully protonated α-amino group at the α-carbon forms hydrogen bonds with Gln-44 Oε1, Asn-11 Oδ1, water 535, and Cys-73 Sγ, whereas, with the ll mimic (ll-AziDAP) bonded to Cys-73, the protonated α-amino functionality donates hydrogen bonds to Glu-208 Oε1, Asn-11 Oδ1, and Cys-217 Sγ. The switch in hydrogen-bonding partners from Glu-208 to Gln-44 during epimerization is not only necessary for stereochemical inversion of the Cα atom, it is likely that temporary release from the hydrogen-bonding partner in the planar α-anionic intermediate enhances the electron-withdrawing capability of the ammonium moiety. It would also stabilize the α-carbanion as the transition state or an intermediate. It could be argued that the acidic ammonium group might participate in acid/base chemistry in the active site (e.g., proton transfer from ammonium to the anionic α-carbon), but this seems unlikely. During the transitory switch in hydrogen bonding partners of the ammonium species, it is probable that both cysteines are protonated and relatively close to the anionic α-carbon of the intermediate and thereby preferred over the α-ammonium group for delivery of a proton.
Catalytic cysteines mounted at the N termini of active site helices.
Both Cys-73 and Cys-217 in the inhibitor-bound enzyme are mounted at the N terminus of the two active-site helices. The negative charge on the thiolate base of either of these residues could be stabilized with the effective positive charge at the N termini of the α-helix dipole. The electrostatic field associated with an α-helix pointing with its N terminus toward the cysteine residue has been implicated to lower the thiol pKa value by up to 5 pH units in glutaredoxin and DsbA (25). Also, the thiols of Cys-73 and Cys-217 are within hydrogen-bonding distance of the amide nitrogens of the residues Gly-76 and Gly-220, respectively, of the two active-site helices. These observations are in agreement with kinetic studies on DAP epimerase, which suggest a reduced pKa of ≈6–7 for the two cysteines Cys-73 and Cys 217 in the forward and reverse reactions, respectively (8). Therefore, at neutral pH, the cysteines exist as a rapidly equilibrating thiolate–thiol pair in the presence of the substrate. Indeed, no basic residue available for deprotonation of the thiol is present in the active site, in contrast to the recently reported structures of glutamate racemase from B. subtilis with d-glutamate (19) and of proline racemase from T. cruzi with pyrrole-2-carboxylic acid (20).
Formation of a rigidly held thiolate–thiol pair from the two cysteines in the active site with exclusion of solvent to enhance basicity for removal of the α-hydrogen.
The binding of the two inhibitors is accompanied by a major clam-like domain closure (Fig. 2a) and the exclusion of water molecules from the active site at the domain interface. In the ll-AziDAP-bound enzyme, there are no water molecules within 4.5 Å of the inhibitor atoms, whereas, in the dl-AziDAP-bound structure, there is only one water molecule, which is bound between Glu-208 and Gln-44, within 4.5 Å of the inhibitor atoms. The resultant low dielectric constant in the buried active site enhances the strength of the electrostatic interactions between the enzyme and the inhibitor (13). As mentioned above, the acidity of one cysteine thiol results in the formation of a rigidly held thiolate–thiol pair (Cys-73 and Cys-217), which, combined with a concomitant increase of thiolate basicity due to water exclusion, facilitates removal of the α-hydrogen (13).
Minimal movement of the substrate during catalysis, with mobility primarily at the α-carbon, attached ammonium group, and β-carbon.
The acid–base partners Cys-73 and Cys-217 are on different domains related by the pseudo-twofold axis and are arranged in line on opposite sides of the α-carbon of the inhibitor. Both Cys-73 and Cys-217 are ideally located to accept and donate the α-proton, with the Sγ(Cys-73)–Cα–Sγ(Cys-217) distances in the two structures being 2.9/3.8 Å (ll-AziDAP) and 2.8/3.6 Å (dl-AziDAP). Furthermore, the other polar active-site residues are involved in hydrogen bonding to the substrate/inhibitor carboxyl, and amino groups and are rigidly held away from the α-hydrogen to prevent interference in the epimerization reaction by donation of a proton to the thiolate base. Comparison of the two AziDAP–epimerase structures indicates that, once the substrate is bound, epimerization takes place with minimal molecular motion (Fig. 5). The cysteines are poised to repeatedly deprotonate and reprotonate from opposite faces of the bound substrate until it is released, analogous to the action of Newton’s cradle of balls.
Comparison with Other PLP-Independent Amino Acid Racemases.
Crystal structures of PLP-independent amino acid racemases reveal two structurally similar α/β domains, with the active site lying between the two domains and containing a pair of conserved cysteine residues that are directly involved in catalysis. The overall structure of DAP epimerase is strikingly similar to that of the recently reported crystal structure of the inhibitor-bound proline racemase from T. cruzi, with an rmsd of 1.73 Å for 217 Cα atom pairs (20). However, the α/β domains of DAP epimerase (composed of mainly β-strands) are very different from those of aspartate racemase and glutamate racemase. A recent report on the crystal structure of glutamate racemase from B. subtilis in complex with d-glutamate reveals a similar conformational rearrangement in the enzyme upon substrate binding (19). Interestingly, despite the enzyme being cocrystallized in the presence of l-glutamate, the product d-glutamate was the species trapped in the crystal structure. However, the observation of a highly ordered d-glutamate in the crystal structure is surprising, because the enzyme forms a racemic mixture of l- and d-glutamate under the conditions of crystallization. Also, on the basis of the proximity of the catalytic cysteines (Cys-74 and Cys-185) to residues of the enzyme that could function as base catalysts and to groups on the substrate, Rice and coworkers (19) suggest a mechanism using a neutral cysteine thiol. However, a clear role for these groups in the activation of the catalytic cysteines could not be identified from their structure. A similar general base-assisted mechanism employing residues His-132 and Asp-296 has also been proposed for proline racemase (20).
The corresponding residues in the active site of DAP epimerase are Glu-208 and His-159, but the closest contact between these residues and the catalytic cysteines is with the Sγ of Cys-217, 4.1 and 5.6 Å, respectively. The residue His-159 is engaged in hydrogen bonds with active-site residues Asn-157 and Ser-219, which in turn interact with the ε- and α-carboxyl groups, respectively, of AziDAP. Although Glu-208 is located within 4.5 Å of the Sγ of Cys-217, its side chain carboxylate forms hydrogen bonds with the α-amino group of AziDAP, the side chain Nε2 of Gln-44, and the Oγ of Ser-219. In the dl-AziDAP-bound enzyme, the interaction of Glu-208 with the α-amino group of AziDAP is mediated through a water molecule. On the basis of these interactions, we conclude that Glu-208 has as its major role the binding of the α-amino group of the substrate. In the glutamate racemase structure, Asp-10 and His-187 are involved in binding the α-amino and the γ-carboxyl groups, respectively, of the bound product d-glutamate. Also, the residues His-132 and Asp-296 in proline racemase are engaged in hydrogen bonds to the pyrrole nitrogen of the bound pyrrole-2-carboxylic acid. Hence, the suggested possible involvement in activating the cysteine thiol during epimerization seems rather unlikely.
Our studies clearly suggest that the active site of DAP epimerase utilizes a rapidly equilibrating thiolate–thiol pair in catalysis, with the Cys-73(S−)/Cys-217(SH) pair responsible for binding ll-AziDAP and the Cys-73(SH)/Cys-217(S−) pair responsible for binding dl-AziDAP. Also, comparison of the structure of the product-bound glutamate racemase from B. subtilis with that of the unliganded enzyme structure from A. pyrophilus reveals a coil-to-helix transformation at the active site upon ligand binding, resulting in the two helices facing each other around the α-carboxyl of the substrate, as we have observed for the DAP epimerase. A similar arrangement of the two active-site α-helices around the α-carboxyl of the bound inhibitor is also seen in the crystal structure of proline racemase (20). These results emphasize the important roles played by the active-site helices in substrate activation and catalysis by this family of enzymes.
Conclusion
Crystal structures of DAP epimerase in complex with diastereomers of AziDAP irreversibly bound in the active site provide a detailed picture of the mechanism of PLP-independent racemases, which are potential antibacterial targets. These enzymes have an unusual mechanism that is the target of numerous on-going studies and recent publications; the mechanism cannot be easily duplicated in the absence of the protein. The key factors that enable catalysis are now revealed to be rigid localization/separation of acidic and basic functionality in the active site, extensive hydrogen bonding of the substrate’s carboxylate moiety with concomitant orientation to favor α-deprotonation, and desolvation of the thiolate base in a hydrophobic environment. Also, recent structural results from proline racemase and glutamate racemase have shown these enzymes to employ similar features in catalysis.
Materials and Methods
Inhibition of DAP Epimerase for X-Ray Studies.
The stereospecific preparation of aziridine analogues of diaminopimelic acid, ll- and dl-AziDAP, and details of inhibition studies with DAP epimerase have been reported (21). The H. influenza DAP epimerase was isolated and purified as described in ref. 16. A solution of ll-AziDAP (1–5 mg/ml) was added in excess (10–100 eq) to a solution of DAP epimerase (0.54 mg/ml) in a buffered solution (100 mM Tris·HCl/1 mM EDTA/1 mM DTT, pH 7.8) and incubated at room temperature for 16 h. To ensure complete inhibition, DAP epimerase activity was monitored spectrophotometrically by using a coupled assay with DAP dehydrogenase that detects the production of NADPH at 340 nm. An aliquot was removed and purified by RP-HPLC using a Jupiter (Torrance, CA) reverse-phase C18 column. The purified enzyme was then dialyzed against Hepes buffer (pH 8.0, 25 mM Hepes/5 mM DTT) and concentrated by using a Millipore Ultrafree Biomax 10K concentrator to a concentration of 10 mg/ml. A similar procedure was followed to prepare dl-AziDAP complexed to DAP epimerase.
Crystallization and Data Collection.
Cocrystals of the inhibitors ll- and dl-AziDAP with DAP epimerase from H. influenzae were grown at room temperature by the hanging-drop vapor-diffusion method. Crystals of both complexes were obtained in 2.8 M sodium acetate/0.1 M Hepes (pH 7.0) at a protein concentration of ≈10 mg/ml in 25 mM Hepes/5 mM DTT (pH 8.0). Cryoprotection was achieved by washing the crystals in mother liquor that contained 25% (vol/vol) glycerol. Intensity data from flash-frozen crystals were collected at Beamline 8.3.1 at the Advanced Light Source (Berkeley, CA) to resolutions of 1.35 Å (ll-AziDAP) and 1.7 Å (dl-AziDAP). The data were processed with programs denzo and scalepack (26).
Structure Solution and Refinement.
The crystal structure of ll-AziDAP-inhibited DAP epimerase was solved by molecular replacement with the program molrep (27) of the ccp4 program suite (28) using the coordinates of the unliganded DAP epimerase [Protein Databank (PDB) ID code 1GQZ] as the search model. A solution with a single molecule in the asymmetric unit was found (R = 43.8%; correlation coefficient = 53.8%). This solution was optimized by using rigid-body refinement in cns (29), with a random 5% of the starting data set aside for crossvalidation. The structure was refined further by using the simulated annealing protocol in cns. ll-AziDAP was modeled in a region of closely fitting positive |Fo| − |Fc|, αc-phased electron density, in the vicinity of Cys-73 (Fig. 1a). Repeated rounds of model building and refinement were performed by using o (30) and cns, respectively. Stereochemical parameters for ll-AziDAP were generated by using prodrg (31). During the final stages of refinement, water molecules were inserted automatically by using arp/warp (32) of the ccp4 suite, followed by visual inspection using o. From this stage onward, refmac (33) of the ccp4 suite was used to refine the structure, allowing for refinement of anisotropic B factors of individual atoms. Cycles of model building, water additions, and refmac (33) refinement have resulted in a final model for the 274 amino acids of the epimerase with excellent sterochemistry as verified by procheck (34).
The structure of DAP epimerase in complex with dl-AziDAP was determined by molecular replacement with molrep (27) using the atomic coordinates of ll-AziDAP–epimerase complex as the search model. dl-AziDAP was modeled in a region of positive |Fo| − |Fc|, αc-phased, electron density in the vicinity of Cys-217 (Fig. 1b). Stereochemical parameters for dl-AziDAP were generated by using prodrg (31). The structure was refined by using refmac (33) to obtain a final model containing all of the 274 amino acids having excellent stereochemistry verified by procheck (34). Data collection and refinement statistics for the two crystal structures are summarized in Table 1. Conformational changes were calculated by using dyndom (35). Figures were generated by using pymol (http://pymol.sourceforge.net), bobscript (36), and molscript (37). The PDB accession codes for these inhibitor-bound structures are 2GKE (ll-AziDAP) and 2GKJ (dl-AziDAP).
Table 1.
Summary of data collection and refinement statistics
| Crystallographic parameters* | ll-AziDAP epimerase | dl-AziDAP epimerase |
|---|---|---|
| Data collection | ||
| X-ray | ALS, Beamline 8.3.1 | ALS, Beamline 8.3.1 |
| Wavelength, Å | λ = 1.115869 | λ = 1.115869 |
| Space group | C2221 | C2221 |
| Unit cell dimension, Å | ||
| a | 96.39 | 96.52 |
| b | 104.02 | 104.09 |
| c | 59.94 | 59.99 |
| Resolution, Å | 30–1.35 (1.40–1.35) | 30–1.7 (1.76–1.70) |
| Observed/unique reflections | 193,306/63,229 | 130,209/33,374 |
| Redundancy | 3.1 (2.2) | 3.9 (3.8) |
| Completeness, % | 95.2 (86.4) | 99.2 (98.1) |
| Rmerge | 5.3 (33.0) | 6.2 (41.2) |
| 〈I/σ(I)〉 | 18.2 (2.0) | 22.5 (3.3) |
| Refinement | ||
| Resolution, Å | 30–1.35 | 30–1.70 |
| Rcryst, % | 13.8 | 17.4 |
| Rfree, % | 16.8 | 20.9 |
| No. of atoms | ||
| Protein | 2,122 | 2,122 |
| Water | 394 | 279 |
| Inhibitor | 14 | 14 |
| rmsd from ideal | ||
| Bonds, Å | 0.012 | 0.010 |
| Angles, ° | 1.50 | 1.32 |
| Ramachandran outliers | None | None |
Rmerge = Σ(|I − 〈I〉|)/ΣI; Rcryst/Rfree = Σ(‖Fo |−| Fc‖)/Σ|Fo|. ALS, Advanced Light Source.
*Values in parentheses indicate the highest resolution shell.
Supplementary Material
Acknowledgments
We thank Joanne Lemieux and Ernst Bergmann for help with the data collection; the staff at Beamline 8.3.1 at the Advanced Light Source and the Alberta Synchrotron Institute for support and funding; and Perry D’Obrennan for his help with Fig. 5. Financial support was provided by the Natural Sciences and Engineering Research Council of Canada, Merck Frosst Inc., the Canada Research Chair in Bioorganic and Medicinal Chemistry (J.C.V.), the Canada Research Chair in Protein Structure and Function (M.N.G.J.), the Canadian Institutes of Health Research (M.N.G.J.), and the Alberta Heritage Foundation for Medical Research.
Abbreviations
- Azi
aziridino
- DAP
diaminopimelate
- PLP
pyridoxal 5′-phosphate
- rmsd
rms deviation.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2GKE (ll-AziDAP) and 2GKJ (dl-AziDAP)].
References
- 1.Katz A. H., Caufield C. E. Curr. Pharm. Design. 2003;9:857–866. doi: 10.2174/1381612033455305. [DOI] [PubMed] [Google Scholar]
- 2.Walsh C. T. J. Biol. Chem. 1989;264:2393–2396. [PubMed] [Google Scholar]
- 3.Hwang K. Y., Cho C. S., Kim S. S., Sung H. C., Yu Y. G., Cho Y. Nat. Struct. Biol. 1999;6:422–426. doi: 10.1038/8223. [DOI] [PubMed] [Google Scholar]
- 4.Cox R. J., Sutherland A., Vederas J. C. Bioorg. Med. Chem. Lett. 2000;8:843–871. doi: 10.1016/s0968-0896(00)00044-4. [DOI] [PubMed] [Google Scholar]
- 5.van Heijenoort J. Nat. Prod. Rep. 2001;18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 6.Glavas S., Tanner M. E. Biochemistry. 2001;40:6199–6204. doi: 10.1021/bi002703z. [DOI] [PubMed] [Google Scholar]
- 7.Mobitz H., Bruice T. C. Biochemistry. 2004;43:9685–9694. doi: 10.1021/bi049580t. [DOI] [PubMed] [Google Scholar]
- 8.Koo C. W., Blanchard J. S. Biochemistry. 1999;38:4416–4422. doi: 10.1021/bi982911f. [DOI] [PubMed] [Google Scholar]
- 9.Liu L., Iwata K., Kita A., Kawarabayasi Y., Yohda M., Miki K. J. Mol. Biol. 2002;319:479–489. doi: 10.1016/S0022-2836(02)00296-6. [DOI] [PubMed] [Google Scholar]
- 10.Albery W. J., Knowles J. R. Biochemistry. 1986;25:2572–2577. doi: 10.1021/bi00357a043. [DOI] [PubMed] [Google Scholar]
- 11.Williams G., Maziarz E. P., Amyes T. L., Wood T. D., Richard J. P. Biochemistry. 2003;42:8354–8361. doi: 10.1021/bi0345992. [DOI] [PubMed] [Google Scholar]
- 12.Richard J. P., Amyes T. L. Curr. Opin. Chem. Biol. 2001;5:626–633. doi: 10.1016/s1367-5931(01)00258-7. [DOI] [PubMed] [Google Scholar]
- 13.Richard J. P., Amyes T. L. Bioorg. Chem. 2004;32:354–366. doi: 10.1016/j.bioorg.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Anita M., Hoare D. S., Work E. Biochem. J. 1957;3:448–459. doi: 10.1042/bj0650448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiseman J. S., Nichols J. S. J. Biol. Chem. 1984;259:8907–8914. [PubMed] [Google Scholar]
- 16.Cirilli M., Zheng R., Scapin G., Blanchard J. S. Biochemistry. 1998;37:16452–16458. doi: 10.1021/bi982138o. [DOI] [PubMed] [Google Scholar]
- 17.Lloyd A. J., Huyton T., Turkenburg J., Roper D. I. Acta Crystallogr. D. 2004;60:397–400. doi: 10.1107/S0907444903027999. [DOI] [PubMed] [Google Scholar]
- 18.Puig E., Garcia-Viloca M., Gonzalez-Lafont A., Lopez I., Daura X., Lluch J. M. J. Chem. Theory Comput. 2005;1:737–749. doi: 10.1021/ct049881g. [DOI] [PubMed] [Google Scholar]
- 19.Ruzheinikov S. N., Taal M. A., Sedelnikova S. E., Baker P. J., Rice D. W. Structure (London) 2005;13:1707–1713. doi: 10.1016/j.str.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 20.Buschiazzo A., Goytia M., Schaeffer F., Degrave W., Shepard W., Gregoire C., Chamond N., Cosson A., Berneman A., Coatnoan N., Alzari P. M., Minoprio P. Proc. Natl. Acad. Sci. USA. 2006;103:1705–1710. doi: 10.1073/pnas.0509010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaper C. M., Sutherland A., Pillai B., James M. N., Semchuk P., Blanchard J. S., Vederas J. C. Org. Biomol. Chem. 2005;3:4402–4411. doi: 10.1039/b513409a. [DOI] [PubMed] [Google Scholar]
- 22.Zhong Z., Postnikova B. J., Hanes R. E., Lynch V. M., Anslyn E. V. Chem. Eur. J. 2005;11:2385–2394. doi: 10.1002/chem.200400396. [DOI] [PubMed] [Google Scholar]
- 23.Corey E. J., Sneen R. A. J. Amer. Chem. Soc. 1956;78:6269–6278. [Google Scholar]
- 24.Gerlt J. A., Kozarich J. W., Kenyon G. L., Gassman P. G. J. Amer. Chem. Soc. 1991;113:9667–9669. [Google Scholar]
- 25.Kortemme T., Creighton T. E. J. Mol. Biol. 1995;253:799–812. doi: 10.1006/jmbi.1995.0592. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Vagin A., Teplyakov A. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 28.Collaborative Computational Project, Number 4. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 29.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Gros-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Schuettelkopf A. W., Van Aalten D. M. F. Acta Crystallogr. D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 32.Lamzin V. S., Wilson K. S. Methods Enzymol. 1997;277:269–305. doi: 10.1016/s0076-6879(97)77016-2. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov G. N., Vagin A. A., Dodson E. J. Acta Crystallogr. D. 1993;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R. A., MacArthur M. W., Moss S. D., Thornton J. M. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 35.Hayward S., Berendsen H. J. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 36.Esnouf R. M. J. Mol. Graph. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 37.Kraulis P. J. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.