Abstract
Long interspersed element (LINE) 1 retrotransposons are major genomic parasites that represent ≈17% of the human genome. The LINE-1 ORF2 protein is also responsible for the mobility of Alu elements, which constitute a further ≈11% of genomic DNA. Representative members of each element class remain mobile, and deleterious retrotransposition events can induce spontaneous genetic diseases. Here, we demonstrate that APOBEC3A and APOBEC3B, two members of the APOBEC3 family of human innate antiretroviral resistance factors, can enter the nucleus, where LINE-1 and Alu reverse transcription occurs, and specifically inhibit both LINE-1 and Alu retrotransposition. These data suggest that the APOBEC3 protein family may have evolved, at least in part, to defend the integrity of the human genome against endogenous retrotransposons.
Keywords: APOBEC3 protein, mutagenesis, retrotransposon, genome stability, intrinsic immunity
Retroelements can be divided into at least three classes: exogenous retroviruses, retrotransposons containing LTRs, and retrotransposons lacking LTRs (non-LTR retrotransposons). Non-LTR retrotransposons can be subdivided further into long interspersed elements (LINEs) and short interspersed elements (SINEs). LINE-1, the most common human LINE, is transcribed by RNA polymerase II to give an ≈6-kb mRNA that encodes two proteins, ORF1p (or p40) and ORF2p, which are required for retrotransposition (1, 2). In contrast, the most common human SINE, Alu, is transcribed by RNA polymerase III to give an ≈300-nt noncoding RNA (3). Alu retrotransposition depends on the LINE-1 ORF2p (4).
Humans are subject to infection by a limited number of exogenous retroviruses and contain few, if any, biologically active LTR retrotransposons, although a small number of structurally similar endogenous retroviruses may remain viable (5, 6). However, humans contain ≈100 intact LINE-1 elements (7, 8), and de novo LINE-1 and short interspersed element retrotransposition events occur in ≈10% of human genomes per generation (9, 10). Although they are frequently innocuous, these retrotransposition events account for ≈0.2% of all spontaneous deleterious human mutations (11, 12). Moreover, LINE-1 and Alu have accumulated to very high levels in the human genome. LINE-1 elements now constitute ≈17% of the human genome, and the >106 copies of Alu constitute a further ≈11% (5).
The ability of human APOBEC3G (A3G) to function as an innate inhibitor of exogenous retroviruses was first noted during studies analyzing the HIV type 1 (HIV-1) Vif protein (6, 13). These experiments revealed that A3G is a potent inhibitor of Vif-deficient, but not wild-type, HIV-1 replication. In the absence of Vif, A3G is specifically packaged into progeny virion particles and then interferes with reverse transcription during subsequent infections (6, 13). Although the mechanisms underlying this inhibition are not fully defined, A3G is a cytidine deaminase (CDA) that edits dC residues to dU on nascent DNA minus strands during reverse transcription (14–16). This activity induces extensive mutagenesis of the HIV-1 provirus and may destabilize incomplete reverse transcripts.
The human APOBEC3 protein family consists of at least five active members that contain one or two consensus CDA active sites (6, 17). Two sites are found in APOBEC3B (A3B, 382 aa), APOBEC3F (A3F, 373 aa), and A3G (384 aa), and one is found in the smaller APOBEC3A (A3A, 199 aa) and APOBEC3C (A3C, 190 aa) proteins. Although A3B, A3F, and A3G can all inhibit Vif-deficient HIV-1 replication, A3A is not active against HIV-1; A3C is only weakly active but does inhibit Vif-deficient simian immunodeficiency virus (6, 18, 19).
The human APOBEC3 proteins are undergoing rapid adaptive evolution, implying that these gene products are in an evolutionary race with some form of deleterious retroelement(s) (20, 21). Human APOBEC3 proteins can inhibit exogenous retroviruses of non-human origin as well as several LTR retrotransposons, thus suggesting that these retroelements could be a source of selective pressure (6, 22–24). Other potential drivers of this adaptive evolution include the human non-LTR retrotransposons and, in particular, LINE-1 and Alu. Here, we demonstrate that two members of the human APOBEC3 family, A3A and A3B, can indeed inhibit both LINE-1 and Alu mobility.
Results
Subcellular Localization of APOBEC3 Proteins.
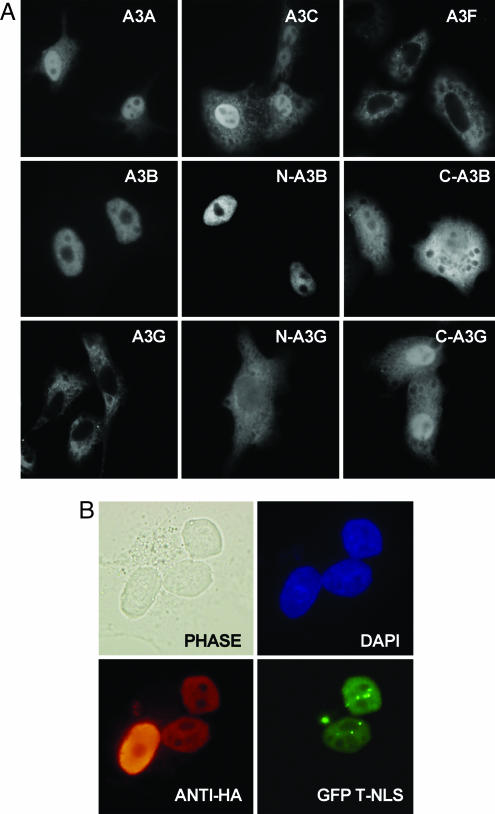
Although human A3G can inhibit several exogenous retroviruses and LTR retrotransposons (6), it has no effect on LINE-1 mobility (24, 25). Unlike retroviruses and LTR retrotransposons, which undergo cytoplasmic reverse transcription, LINE-1 RNA is reverse-transcribed in the nucleus (26, 27), and A3G has previously been reported to be restricted to the cytoplasm (14). If the inability of A3G to inhibit LINE-1 retrotransposition reflects this compartmentalization, then APOBEC3 proteins that enter the nucleus might be more effective inhibitors of LINE-1 retrotransposition. Because the exclusion size for passive diffusion through the nuclear pore complex is ≈40 kDa (28), we asked whether A3A and A3C, which fall below this limit, would enter the nucleus. As shown in Fig. 1A, although A3G and A3F are excluded from nuclei, A3A and A3C are indeed found in both the nucleus and cytoplasm. This result is consistent with the hypothesis that A3A and A3C can enter nuclei by passive diffusion. Unexpectedly, A3B appeared nuclear at steady state. The 382-aa A3B protein is too large to diffuse into the nucleus (28), implying that A3B contains a nuclear localization signal (NLS).
Fig. 1.
Subcellular localization of human APOBEC3 proteins. (A) HeLa cells were transfected with plasmids expressing the indicated wild-type or mutant APOBEC3 protein bearing a C-terminal HA epitope tag. At 48 h after transfection, cells were fixed and incubated with a mouse monoclonal anti-HA antibody, followed by tetramethylrhodamine B isothiocyanate-conjugated goat anti-mouse antiserum, and visualized by fluorescence. (B) Nucleocytoplasmic shuttling was visualized by using heterokaryons (45). Briefly, COS7 cells were transfected with a plasmid expressing A3B-HA. Two days later, the COS7 cells were mixed with HeLa-GSN cells, which express GFP fused to a NLS (31). The mixed culture was then treated with cycloheximide and fused by using polyethylene glycol. A3B-HA was detected as described above. Nuclei were visualized by staining with DAPI; GFP was detected by intrinsic fluorescence.
As a first step toward mapping the A3B NLS, we divided A3B into two nonoverlapping segments, extending from amino acid 1 to 192 and from amino acid 193 (a methionine) to 382, that resemble single CDA domain APOBEC3 proteins. As shown in Fig. 1A, the N-terminal half of A3B, termed N-A3B, localized to the nuclei of expressing cells, whereas the C-terminal half, termed C-A3B, gave the same diffuse subcellular localization seen with A3A and A3C. Therefore, we conclude that the A3B NLS is located in the N-terminal 192-aa segment. The analogous N-terminal (N-A3G, 196 aa) and C-terminal (C-A3G, 188 aa) segments of A3G both showed a diffuse subcellular localization (Fig. 1A). This result is consistent with the hypothesis that A3G lacks a NLS and suggests that the cytoplasmic localization of full-length A3G (Fig. 1A) indeed results from the inability of the ≈42-kDa A3G protein to enter nuclei by passive diffusion.
At first glance, the nuclear localization of A3B appears inconsistent with reports that demonstrate that A3B, like A3G, can assemble into the virions of several exogenous retroviruses in the cytoplasm and then inhibit their replication (19, 29, 30). To test whether A3B is actually a nucleocytoplasmic shuttle protein, we fused COS7 cells expressing hemagglutinin (HA) epitope-tagged A3B to HeLa-GSN cells that express a fusion protein consisting of GFP linked to the SV40 T antigen NLS (31). As shown in Fig. 1B, the HA-tagged A3B protein moved from COS7 nuclei to HeLa-GSN nuclei that were present in the resultant heterokaryons. In contrast, the GFPT-NLS fusion protein, which lacks a nuclear export signal (NES), failed to move from HeLa-GSN nuclei to COS7 nuclei. We therefore conclude that A3B is a nucleocytoplasmic shuttle protein that contains a NLS and a NES.
A3A and A3B Inhibit LINE-1 Retrotransposition.
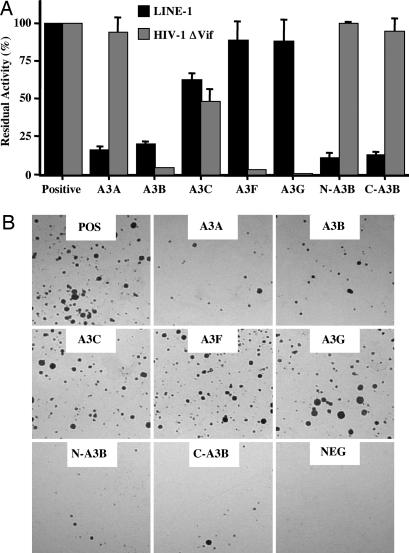
The ability of A3A, A3B, and A3C to enter nuclei suggested that one or more of these proteins might be able to interfere with the retrotransposition of human LINE-1. To explore this hypothesis, we tested whether an engineered LINE-1 element harboring a retrotransposition indicator (pJM101/L1.3) (32, 33) was affected by APOBEC3 protein expression. The retrotransposition indicator cassette consists of a neo gene in the antisense orientation (relative to LINE-1) that is disrupted by an intron in the sense orientation (32, 34). Therefore, neo expression requires LINE-1 transcription, removal of the intron by splicing, reverse transcription, and integration followed by expression of the now intact neo gene. Cotransfection of HeLa cells with pJM101/L1.3 and a plasmid encoding an APOBEC3 protein revealed that A3A and A3B are effective inhibitors of LINE-1 retrotransposition (Fig. 2). The A3C protein exerted a modest but significant inhibitory effect on LINE-1 mobility, whereas A3G and A3F had little effect on retrotransposition. The observed inhibition was not due to nonspecific toxicity, because we have previously shown that the APOBEC3 proteins do not reduce the number of G418-resistant colonies obtained after cotransfection into HeLa cells with a neo expression plasmid (23). Comparison of the effect of APOBEC3 proteins on LINE-1 retrotransposition with their effect on HIV-1ΔVif infectivity (Fig. 2A) showed that there is essentially no correlation. All APOBEC3 proteins were expressed at comparable levels (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 2.
Inhibition of LINE-1 retrotransposition and HIV-1ΔVif infectivity by APOBEC3 proteins. (A) The effect of APOBEC3 proteins on HIV-1ΔVif infectivity was quantified as described in ref. 41. Data are presented relative to a culture lacking any APOBEC3 protein (positive), which was designated as 100% activity. Data are the average of at least three experiments, with standard deviation indicated. The relative expression of each APOBEC3 protein was comparable (Fig. 6). Because the effect of APOBEC3 proteins on the infectivity of Vif-deficient HIV-1 was measured by using 293T cells, although their effect on LINE-1 retrotransposition was measured in HeLa cells, these results are only qualitatively comparable. (B) Representative experiment showing the relative number of G418-resistant colonies obtained after selection of HeLa cells transfected with the LINE-1 retrotransposition indicator construct pJM101/L1.3 or the negative (NEG) control construct pJM105/L1.3 (lacking a functional ORF2 gene) in the presence or absence (POS) of the indicated APOBEC3 proteins.
Inhibition of LINE-1 Retrotransposition Occurs in the Absence of Hypermutation.
Inhibition of retrovirus or LTR retrotransposon replication by wild-type A3G is associated with hypermutation of any resultant integrated proviruses (6, 14–16, 24). In contrast, inhibition of hepatitis B virus replication by A3G seems to occur in the absence of dC deamination (36, 37), and mutants of A3G that are unable to edit retain substantial inhibitory activity against HIV-1ΔVif (38). Extensive sequencing of LINE-1 elements that retrotransposed in the presence of A3A, A3B, A3C, or A3G failed to detect G-to-A hypermutation (Fig. 7, which is published as supporting information on the PNAS web site). Therefore, inhibition of LINE-1 retrotransposition by A3A and A3B does not involve editing, or edited nuclear reverse transcripts are rapidly degraded.
Although A3G, like A3B and A3F, contains two consensus CDA active sites, only the C-terminal site is enzymatically active; the N-terminal site appears to be involved in mediating selective incorporation of A3G into virion particles (38–40). To determine whether an enzymatically active CDA consensus site is required for inhibition of LINE-1 retrotransposition by A3A or A3B, we used mutagenesis to inactivate the CDA active site that is present in A3A or one of the two CDA active sites that are present in A3B. The introduced mutations, which changed the glutamic acid residue in the CDA consensus active site (His-X-Glu-X25–28-Pro-Cys-X2–4-Cys) (6) to glutamine, have previously been shown to block CDA enzyme activity (38). We also asked whether the N- or C-terminal halves of A3B (Fig. 1A) could function as CDAs and/or inhibit LINE-1 retrotransposition.
To address whether the various A3A and A3B mutants have, in fact, lost the ability to function as CDAs, we used a previously described DNA mutator assay in bacteria (40). This assay measures the ability of an expressed gene product to mutate the RNA polymerase B gene (rpoB) in Escherichia coli. Mutations in rpoB can be detected by screening for the frequency of rifampicin-resistant (rifR) colonies. Expression of A3G or A3F has previously been shown to enhance the frequency of such mutations (40).
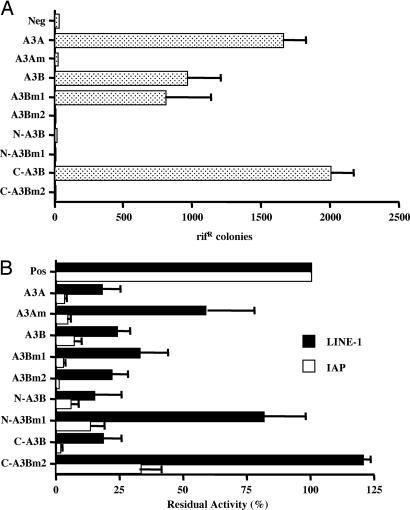
As shown in Fig. 3A, wild-type A3A and A3B both acted as potent mutators in E. coli. Sequencing the rpoB gene in the resistant colonies that were obtained in the presence of either A3A or A3B revealed selective C-to-T mutagenesis with a consensus target sequence of TC* (data not shown). This consensus is the same one previously reported for A3B and A3F but differs from the consensus target sequence for A3G, which is CC* (19, 29, 40, 41).
Fig. 3.
Effect of mutations on the CDA activity and retrotransposition inhibitory activity of A3A and A3B. (A) This assay measures the ability of the indicated wild-type or mutant A3B and A3A proteins to enhance mutagenesis in bacteria (40). Plasmids encoding the indicated proteins were introduced into E. coli, and their expression was activated by using isopropyl β-d-thiogalactoside. The level of mutation induced by each protein was then assessed by plating the bacteria on plates containing rifampicin and counting the number of resistant colonies. (B) Effect of the indicated wild-type or mutant A3A and A3B proteins on retrotransposition of human LINE-1 or the murine LTR retrotransposon IAP. This analysis of IAP retrotransposition frequency was performed as described in ref. 23. The average of three independent experiments is shown.
Mutation of the single CDA consensus site in A3A (A3Am) blocked CDA activity as expected. Of interest, mutation of the N-terminal CDA consensus site in A3B (A3Bm1) had no effect on editing by A3B, whereas mutation of the C-terminal consensus site (A3Bm2) entirely blocked A3B editing (Fig. 3A). Consistent with the hypothesis that the C-terminal A3B consensus site is necessary and sufficient for CDA enzyme activity, the N-terminal 192-aa segment of A3B (N-A3B) did not display any mutator activity in bacteria, whereas the C-terminal 190-aa segment (C-A3B) was highly active unless the CDA active site was mutated (Fig. 3A). Importantly, all of these proteins were expressed at comparable levels in bacteria (Fig. 8A, which is published as supporting information on the PNAS web site). Therefore, we conclude that A3A and A3B both contain a single enzymatically active CDA consensus site and that, in A3B, this site is located in the C-terminal half. A3B is therefore similar to A3G, which also contains only a single enzymatically active CDA consensus site located in the C-terminal half of the protein (38, 39).
Having constructed several mutants of A3A and A3B that lack (A3Am, A3Bm2, N-A3B, N-A3Bm1, and C-A3Bm2) or retain (A3Bm1 and C-A3B) CDA activity, we next asked whether these proteins would inhibit LINE-1 retrotransposition. As a control, we also measured their ability to inhibit the mobility of the mouse LTR retrotransposon intracisternal A particle (IAP) and, in the case of N-A3B and C-A3B, the infectivity of HIV-1 virions. We recently demonstrated that A3A is a potent inhibitor of IAP retrotransposition and that inhibition of IAP by A3A does not require an intact CDA active site (23). As shown in Fig. 8B, all of these mutant proteins were expressed at comparable levels in transfected human cells.
The most important point to emerge from these studies is that two of the mutant APOBEC3 proteins that lack any detectable CDA activity (i.e., the full-length A3B mutant A3Bm2 and the N-terminal 192-aa segment of A3B termed N-A3B) nevertheless remain fully active against both LINE-1 and IAP (Fig. 3B). Indeed, both the front (N-A3B) and back (C-A3B) halves of A3B are at least as active as full-length A3B in inhibiting LINE-1 retrotransposition, although both these “half-proteins” differ from full-length A3B in being entirely inactive against HIV-1ΔVif (Fig. 2A).
Because the division of the A3G ORF into analogous N-terminal (N-A3G) and C-terminal (C-A3G) segments also resulted in the production of stable half-proteins (Figs. 1A and 8 C and D), we also asked whether these truncated proteins were biologically active. As expected, wild-type A3G and C-A3G enhanced mutagenesis in bacteria to an equivalent degree, whereas the N-A3G protein was inactive (Fig. 9A, which is published as supporting information on the PNAS web site). However, neither N-A3G nor C-A3G could inhibit either LINE-1 retrotransposition or HIV-1 infectivity (Fig. 9B). The inability of both C-A3G (Fig. 9B) and wild-type A3C (Fig. 2A) to effectively inhibit LINE-1 retrotransposition argues that expression of an enzymatically active CDA in the nucleus is not sufficient to inhibit LINE-1 mobility.
Although the above data argue that CDA activity is neither necessary nor sufficient for effective inhibition of LINE-1 retrotransposition by an APOBEC3 protein, it appears that an intact CDA consensus site nevertheless plays an important role in mediating this effect. In particular, introduction of the E68Q mutation into N-A3B, a protein that already lacks detectable CDA enzyme activity (Fig. 3A), nevertheless alleviated inhibition of LINE-1 retrotransposition (Fig. 3B), although inhibition of IAP retrotransposition was less affected. Similarly, mutation of the CDA active sites that are present in A3A (A3Am) and C-A3B (C-A3Bm2) also strongly attenuated the observed inhibition of LINE-1 mobility, although, again, inhibition of IAP was less affected. The N-terminal CDA consensus site of A3G is not enzymatically active yet appears to be required for efficient packaging of A3G into HIV-1 virion particles (38, 39). Similarly, it appears that an intact CDA consensus site plays an important role in mediating the inhibition of LINE-1 retrotransposition by A3A and A3B, perhaps by allowing a specific interaction with LINE-1 ribonucleoprotein complexes, although enzymatic activity per se is not important.
Inhibition of Alu Retrotransposition by Human A3A and A3B.
Alu elements represent a highly abundant family of human short interspersed elements and constitute ≈11% of the entire human genome (5). Retrotransposition of Alu elements is mediated by the LINE-1 ORF2 protein (4), which has reverse transcriptase and endonuclease activities (35, 42) but does not require the LINE-1 ORF1 RNA-binding protein (4).
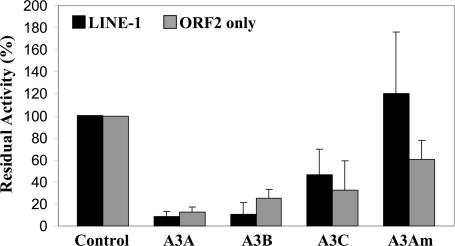
To test whether human APOBEC3 proteins can inhibit Alu mobility, we cotransfected HeLa cells with a previously described Alu construct tagged with a retrotransposition indicator cassette (4) together with a plasmid expressing either full-length wild-type LINE-1 or only LINE-1 ORF2p. Finally, the cells were also cotransfected with plasmids expressing A3A, A3B, A3C, or the A3Am mutant. A3A and A3B both acted as potent inhibitors of Alu retrotransposition regardless of whether both ORF1p and ORF2p, or only ORF2, were coexpressed (Fig. 4; see also Fig. 10, which is published as supporting information on the PNAS web site). As in the case of LINE-1 (Figs. 2 and 3B), the A3Am mutant bearing an inactive CDA consensus sequence failed to effectively inhibit Alu retrotransposition, whereas A3C exhibited a moderate inhibitory effect. We therefore conclude that A3A and A3B can inhibit retrotransposition of both LINE-1 and Alu and that, at least in the latter case, this inhibition does not involve an interaction with the LINE-1 ORF1 protein. Sequence analysis of Alu elements that had retrotransposed in the presence of A3A, A3B, or A3C failed to reveal an increase in the level of C-to-T or G-to-A mutagenesis when compared with control cultures (Fig. 11, which is published as supporting information on the PNAS web site). These data suggest that inhibition of Alu retrotransposition, like inhibition of LINE-1 retrotransposition, does not result from editing of reverse transcripts.
Fig. 4.
A3A and A3B inhibit Alu retrotransposition. HeLa cells were cotransfected with an Alu construct tagged with a retrotransposition indicator cassette, an APOBEC3 expression plasmid, or a control plasmid (pK/β-arr) and expression plasmids encoding wild-type LINE-1 or a LINE-1 lacking ORF1. Three days after transfection, cells were subjected to G418 selection, and neo-resistant colonies were stained and counted 12 days later. Data are presented relative to the culture cotransfected with pK/β-arrestin (Control), which was set at 100%. Data are the average of two experiments performed in duplicate, with standard deviation indicated.
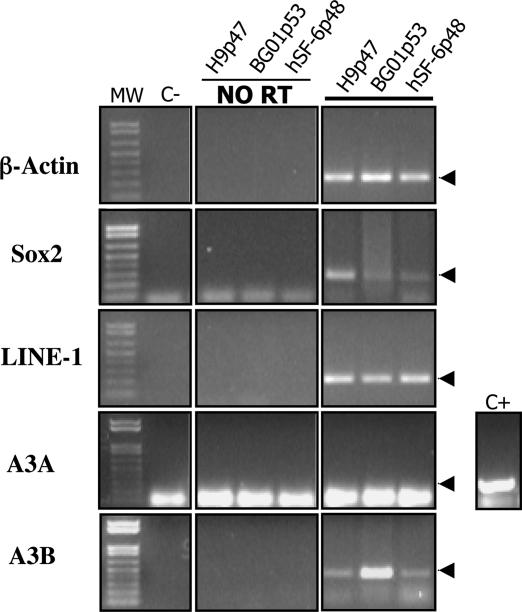
A3B mRNA Is Expressed During Early Human Development.
Although LINE-1 and Alu can retrotranspose in some somatic tissues, to propagate they must retrotranspose in germ cells and/or during the earliest stages in human embryonic development (1, 2). Because mice do not encode A3A and A3B homologs (17), inhibition of LINE-1 and Alu mobility can be investigated only by using human tissue samples. Moreover, because an antibody specific for A3A or A3B does not exist yet, analysis of A3A or A3B expression can currently be performed only at the mRNA level. Previous analysis of A3B mRNA expression has shown that this mRNA is present at a low to very low level in a wide range of human somatic tissues (17, 18, 29). In contrast, A3A mRNA expression has been reported in peripheral blood lymphocytes (PBLs) but not elsewhere (17). These data were confirmed by RT-PCR analysis, which showed a low level of A3B mRNA expression in a wide range of somatic tissues, including ovary and testis, whereas A3A mRNA was detected only in PBLs and in spleen (Fig. 12, which is published as supporting information on the PNAS web site).
We also analyzed mRNA samples derived from three distinct undifferentiated hES cell lines (H9p47, BG01p53, and hSF-6p48). RT-PCR analysis detected both A3B mRNA and LINE-1 mRNA in these hES cell lines, although A3A mRNA was not detected. We therefore conclude that A3B mRNA is expressed in human tissues, including very early embryonic tissues, where novel LINE-1 and Alu retrotransposition events could lead to new heritable insertions.
Discussion
Although the human APOBEC3 gene family is undergoing rapid adaptive evolution, the driving force(s) behind this evolution has remained unclear (20, 21). Given that the only known function of the APOBEC3 proteins is as inhibitors of retroelements, the most likely sources of positive selection for novel APOBEC3 sequences and activities are exogenous retroviruses, endogenous retroviruses, the closely related LTR retrotransposons, and, finally, non-LTR retrotransposons.
A3G was initially identified as a potent inhibitor of Vif-deficient HIV-1 (13), and at least four human APOBEC3 gene products (A3G, A3B, A3F, and A3C) have subsequently been shown to inhibit various non-human exogenous retroviruses as well as hepatitis B virus (6). The APOBEC3 proteins may therefore play an important role in preventing the zoonotic transmission of animal retroviruses into the human population, but this hypothesis is difficult to test.
The second possibility (i.e., that APOBEC3 proteins play a role in controlling endogenous human retroviruses and LTR retrotransposons) seems unlikely because the human genome does not contain any known biologically active LTR retrotransposons and few if any functional endogenous retroviruses (1, 5). Therefore, although A3A, A3B, and A3G can inhibit the mobility of LTR retrotransposons of murine origin (23, 24), human endogenous retroviruses and LTR retrotransposons seem unlikely of being capable of exerting significant selective pressure, at least at this stage in human and primate evolution.
Unlike LTR retrotransposons, the human genome does contain a substantial number of biologically active non-LTR retrotransposons, and novel germ-line non-LTR retrotransposition events are believed to occur in ≈10% of all humans (7–10). As a result, non-LTR retrotransposons, of which the most common are LINE-1 and Alu, have accumulated over time to comprise ≈30% of the entire human genome and continue to influence the evolution of that genome (5). The question then is: Do the human APOBEC3 proteins play a role in restricting non-LTR retrotransposon mobility to the low level that is currently seen?
In this study, we present evidence arguing that at least two members of the human APOBEC3 protein family, A3A and A3B, can indeed act as effective inhibitors of LINE-1 and Alu retrotransposition (Figs. 2 and 4). Moreover, at least in the case of A3B, mRNA encoding this protein is readily detectable in both hES cells (Fig. 5) and in human testis and ovary (Fig. 12), thus suggesting that the A3B protein is expressed in the right places to exert a physiologically relevant inhibitory effect on the appearance of novel heritable insertions of LINE-1 or Alu.
Fig. 5.
Analysis of A3A and A3B mRNA expression in hES cells. RT-PCR was used to assess the expression of LINE-1, Sox2, A3A, and A3B mRNA in three undifferentiated hES cell lines (H9p47, BG01p53, and hSF-6p48). A positive control for the RT-PCR is shown beside the gel panel for A3A. The procedures used for hES cell culture and RT-PCR analyses are described in Supporting Methods, which is published as supporting information on the PNAS web site.
The APOBEC3 proteins are CDAs, and inhibition of exogenous retrovirus replication is associated with hypermutation of retroviral proviruses (6, 14–16). However, recent data demonstrate that certain mutants of A3G or A3A retain the ability to inhibit HIV-1 or LTR retrotransposon replication, respectively, even in the absence of a functional CDA active site (23, 38). Here, we report that inhibition of LINE-1 or Alu retrotransposition by A3A or A3B does not result in detectable hypermutation (Figs. 7 and 11), and we have also identified A3B mutants that are fully able to inhibit LINE-1 retrotransposition yet are inactive as CDAs (Fig. 3). The mechanism(s) underlying inhibition of LINE-1 mobility by A3B in the absence of editing, or the inhibition of HIV-1 infectivity by enzymatically inactive forms of A3G, currently remains unclear. However, the observation that A3A and A3B are effective inhibitors of Alu retrotransposition mediated solely by the LINE-1 ORF2 protein (i.e., in the absence of the LINE-1 ORF1 RNA-binding protein) suggests that this inhibition is not mediated by specific recruitment of A3A or A3B by a protein that is functionally equivalent to the retroviral nucleocapsid domain of Gag. This result is surprising, because this domain of Gag has been shown to be critical for the recruitment of APOBEC3 proteins into retroviral virion particles, where they then exert their inhibitory effect (6). It remains possible that an unknown cellular RNA-binding protein(s) may play a role in mediating both Alu retrotransposition and APOBEC3 recruitment.
Although the A3A, A3B, and, to a lesser extent, A3C proteins inhibit both LINE-1 and Alu retrotransposition (Figs. 2 and 4), A3G and A3F have little or no effect on LINE-1 mobility, although these proteins do function as potent inhibitors of Vif-deficient HIV-1 (Fig. 2). This distinction correlates with the observation that A3A, A3B, and A3C can enter the cell nucleus, where LINE-1 reverse transcription occurs, whereas A3F and A3G are restricted to the cytoplasm (Fig. 1). In the case of A3A and A3C, nuclear entry appears to be due to passive diffusion, but A3B clearly contains both a NLS and a nuclear export signal (NES), thus implying that it functions in both the nucleus and cytoplasm of expressing cells. Once the NES and NLS in A3B have been defined, it will be of interest to see whether these protein sorting signals play a role in mediating the inhibition of LINE-1 mobility and HIV-1 infectivity that are characteristic of A3B (Fig. 2A). In the interim, it is interesting to speculate about how cells avoid the problem of random mutagenesis of their DNA genome by nuclear APOBEC3 proteins.
Materials and Methods
Molecular Clones.
The LINE-1 retrotransposition indicator constructs pJM101/L1.3 and pJM105/L1.3, the wild-type LINE-1 expression vector pJM101/L1.3Δneo, and the LINE-1 ORF2 expression plasmid pCep 5′UTR ORF2Δneo are described in refs. 32, 33, and 44. Also previously described are the Alu retrotransposition indicator construct AluneoTet (4) and the HIV-1 proviral indicator construct pHIV-LucΔVif (41). pK-based expression plasmids expressing C-terminally HA-tagged β-arrestin, A3A, A3B, A3C, A3F, and A3G are described in ref. 23. A3B and A3G cDNAs encoding C-terminally HA-tagged N-A3B (amino acids 1–192), C-A3B (amino acids 193–382), N-A3G (amino acids 1–196), and C-A3G (amino acids 197–384) were generated by PCR and cloned into the pK vector. All A3A and A3B point mutants were generated by site-directed mutagenesis and verified by DNA sequencing.
Bacterial Mutator Assay.
APOBEC3 cDNAs (wild type and mutant), including the C-terminal HA tag, were excised from the relevant pK-based plasmid by cleavage with Asp-718 and XhoI and subcloned into Asp718 and SalI sites present in the bacterial expression plasmid pTrc99A (AP Biosciences, Piscataway, NJ). The uracil DNA glycosylase-deficient E. coli strain BW310 (40) was transformed with the pTrc99A parental plasmid and the various wild-type and mutant A3A and A3B derivatives. Transformed bacteria were then selected overnight on LB plates containing ampicillin. Twenty colonies were pooled into 2 ml of LB plus ampicillin plus 1 mM isopropyl β-d-thiogalactoside (IPTG), and cultures were grown overnight at 37°C. One hundred microliters of the saturated culture was then plated on LB plates containing 100 μg/ml rifampicin, and the total number of rifR colonies per plate was counted 24 h later. To verify protein expression, 100 μl of the saturated IPTG-induced culture was lysed, subjected to gel electrophoresis, and analyzed by Western blot as previously described.
Note.
While this report was under review, Chen et al. (43) reported that A3A can inhibit LINE-1 retrotransposition.
Supplementary Material
Acknowledgments
We thank Michael Malim (King’s College, London), Reuben Harris (University of Minnesota, Minneapolis), Bryce Paschal (University of Virginia, Charlottesville), and Thierry Heidmann (Institute Gustave Roussy, Villejuif, France) for reagents and Brian Doehle for help with the construction of mutants. This work was supported by National Institutes of Health Grants AI65301 (to B.R.C.), GM60518 (to J.V.M.), and GM69985 (to K.S.O.); Ministerio de Educación y Ciencia de España/Fulbright Postdoctoral Grant EX-200300881 (to J.L.G.-P.); and National Institutes of Health Michigan Predoctoral Training Grant 5T32GM07544 (to A.E.H.).
Abbreviations
- LINE
long interspersed element
- A3X
APOBEC3X
- CDA
cytidine deaminase
- HIV-1
HIV type 1
- NLS
nuclear localization signal
- HA
hemagglutinin
- IAP
intracisternal A particle.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Moran J. V., Gilbert N. In: Mobile DNA II. Craig N., Craggie R., Gellert M., Lambowitz A., editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 836–869. [Google Scholar]
- 2.Moran J. V., Holmes S. E., Naas T. P., DeBerardinis R. J., Boeke J. D., Kazazian H. H., Jr Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 3.Batzer M. A., Deininger P. L. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 4.Dewannieux M., Esnault C., Heidmann T. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 5.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Cullen B. R. J. Virol. 2006;80:1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sassaman D. M., Dombroski B. A., Moran J. V., Kimberland M. L., Naas T. P., DeBerardinis R. J., Gabriel A., Swergold G. D., Kazazian H. H., Jr. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 8.Brouha B., Schustak J., Badge R. M., Lutz-Prigge S., Farley A. H., Moran J. V., Kazazian H. H., Jr. Proc. Natl. Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazazian H. H., Jr. Nat. Genet. 1999;22:130. doi: 10.1038/9638. [DOI] [PubMed] [Google Scholar]
- 10.Li X., Scaringe W. A., Hill K. A., Roberts S., Mengos A., Careri D., Pinto M. T., Kasper C. K., Sommer S. S. Hum. Mutat. 2001;17:511–519. doi: 10.1002/humu.1134. [DOI] [PubMed] [Google Scholar]
- 11.Kazazian H. H., Jr., Moran J. V. Nat. Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 12.Ostertag E. M., Kazazian H. H., Jr. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 13.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 14.Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H., Yang B., Pomerantz R. J., Zhang C., Arunachalam S. C., Gao L. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 17.Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 18.Yu Q., Chen D., König R., Mariani R., Unutmaz D., Landau N. R. J. Biol. Chem. 2004;279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- 19.Bishop K. N., Holmes R. K., Sheehy A. M., Davidson N. O., Cho S.-J., Malim M. H. Curr. Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 20.Sawyer S. L., Emerman M., Malik H. S. PLoS Biol. 2004;2:1278–1285. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J., Webb D. M. Hum. Mol. Genet. 2004;13:1785–1791. doi: 10.1093/hmg/ddh183. [DOI] [PubMed] [Google Scholar]
- 22.Dutko J. A., Schäfer A., Kenny A. E., Cullen B. R., Curcio M. J. Curr. Biol. 2005;15:661–666. doi: 10.1016/j.cub.2005.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogerd H. P., Wiegand H. L., Doehle B. P., Lueders K. K., Cullen B. R. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esnault C., Heidmann O., Delebecque F., Dewannieux M., Ribet D., Hance A. J., Heidmann T., Schwartz O. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 25.Turelli P., Vianin S., Trono D. J. Biol. Chem. 2004;279:43371–43373. doi: 10.1074/jbc.C400334200. [DOI] [PubMed] [Google Scholar]
- 26.Feng Q., Schumann G., Boeke J. D. Proc. Natl. Acad. Sci. USA. 1998;95:2083–2088. doi: 10.1073/pnas.95.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cost G. J., Feng Q., Jacquier A., Boeke J. D. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Görlich D., Kutay U. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 29.Doehle B. P., Schäfer A., Cullen B. R. Virology. 2005;339:281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Doehle B. P., Schäfer A., Wiegand H. L., Bogerd H. P., Cullen B. R. J. Virol. 2005;79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Black B. E., Lévesque L., Holaska J. M., Wood T. C., Paschal B. M. Mol. Cell. Biol. 1999;19:8616–8624. doi: 10.1128/mcb.19.12.8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei W., Morrish T. A., Alisch R. S., Moran J. V. Anal. Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- 33.Wei W., Gilbert N., Ooi S. L., Lawler J. F., Ostertag E. M., Kazazian H. H., Boeke J. D., Moran J. V. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freeman J. D., Goodchild N. L., Mager D. L. BioTechniques. 1994;17:46–52. 48–49. [PubMed] [Google Scholar]
- 35.Mathias S. L., Scott A. F., Kazazian H. H., Jr., Boeke J. D., Gabriel A. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 36.Turelli P., Mangeat B., Jost S., Vianin S., Trono D. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 37.Rosier C., Kock J., Kann M., Malim M. H., Blum H. E., Baumert T. F., von Weizsacker F. Hepatology. 2005;42:301–309. doi: 10.1002/hep.20801. [DOI] [PubMed] [Google Scholar]
- 38.Newman E. N. C., Holmes R. K., Craig H. M., Klein K. C., Lingappa J. R., Malim M. H., Sheehy A. M. Curr. Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 39.Navarro F., Bollman B., Chen H., König R., Yu Q., Chiles K., Landau N. R. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 40.Haché G., Liddament M. T., Harris R. S. J. Biol. Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 41.Wiegand H. L., Doehle B. P., Bogerd H. P., Cullen B. R. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Q., Moran J. V., Kazazian H. H., Jr., Boeke J. D. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 43.Chen H., Lilley C. E., Yu Q., Lee D. V., Chou J., Narvaiza I., Landau N. R., Weitzman M. D. Curr. Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 44.Alisch R. S., Garcia-Perez J. L., Muotri A. R., Gage F. H., Moran J. V. Genes Dev. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiegand H. L., Coburn G. A., Zeng Y., Kang Y., Bogerd H. P., Cullen B. R. Mol. Cell. Biol. 2002;22:245–256. doi: 10.1128/MCB.22.1.245-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.