Abstract
Pathogen recognition by Toll-like receptors (TLRs) initiates innate immune responses that are essential for inhibiting pathogen dissemination and for the development of acquired immunity. The TLRs recognize pathogens with their N-terminal ectodomains (ECD), but the molecular basis for this recognition is not known. Recently we reported the x-ray structure for unliganded TLR3-ECD; however, it has proven difficult to obtain a crystal structure of TLR3 with its ligand, dsRNA. We have now located the TLR3 ligand binding site by mutational analysis. More than 50 single-residue mutations have been generated throughout the TLR3-ECD, but only two, H539E and N541A, resulted in the loss of TLR3 activation and ligand binding functions. These mutations locate the dsRNA binding site on the glycan-free, lateral surface of TLR3 toward the C terminus and suggest a model for dsRNA binding and TLR3 activation.
Keywords: double-stranded RNA, innate immunity, leucine-rich repeat
The innate recognition of pathogens by vertebrate hosts triggers an immediate inflammatory response that prevents pathogen dissemination and plays a crucial role in the development of adaptive immunity (1). Recent studies have indicated that the Toll-like receptors (TLRs) are essential for the host response to a wide variety of pathogen-derived substances (reviewed in ref. 2). The TLRs belong to a family of type I transmembrane glycoproteins that, in contrast to the receptors of the adaptive immune system, are encoded in the germ line. Ten human TLR paralogs recognize evolutionarily conserved ligands including lipids, proteins, carbohydrates, and nucleic acids from bacteria, fungi, viruses, and parasites. Ligand recognition by TLRs is mediated through their N-terminal ectodomains (ECD), which contain 18–25 tandem copies of a short (≈24 residues) motif known as the leucine-rich repeat (LRR). Recently, we (3) and others (4) reported the first x-ray structures of a TLR ligand binding domain, that of human TLR3-ECD. TLR3-ECD resembles a horseshoe-shaped solenoid of 23 LRRs, with each LRR forming one turn of the solenoid. Notable features of the TLR3-ECD include a large parallel β-sheet on the concave surface, two large insertions protruding from the lateral and convex surfaces of LRRs 12 and 20, respectively, and a single, extensive, glycan-free surface in an otherwise highly glycosylated molecule.
TLR3 is activated by dsRNA, the genetic material of some viruses, and a viral replication intermediate in others (5). Ligand recognition by the TLR3-ECD results in recruitment of the adapter molecule Toll/IL-1 domain-containing adapter inducing IFN-β (TRIF) to the cytoplasmic Toll–IL-1 receptor homology (TIR) domain of TLR3. The recruitment of TRIF, in turn, triggers a kinase cascade ultimately leading to IRF-3 activation and the transcription of type I IFN genes (6). As a critical viral sensor, TLR3 is expressed in immune cells, such as conventional dendritic cells (7, 8), natural killer cells (9–11), and mast cells (12, 13) as well as fibroblasts and epithelial cells (2, 14). We have demonstrated that TLR3-ECD binds directly to short dsRNA and polyinosinic-polycytidylic acid (pI:pC), a dsRNA surrogate (3). When bound to these ligands, TLR3-ECD formed large oligomers, suggesting that receptor multimerization may initiate the cytosolic signaling cascade.
Cocrystals of TLR3-ECD with its ligand are currently unavailable; thus, the molecular details of ligand binding are not yet known. However, several potential binding sites have been proposed based on the apo structure. Choe et al. (4) postulated that dsRNA might bind at one or both of two patches of positively charged residues on the glycan-free face of the TLR3-ECD. We noted the presence of two sulfate molecules from the crystallization medium stably bound to residues in LRRs 12 and 20, the two LRRs that contain large insertions (3). Because sulfate ions share the same atomic arrangement as phosphate groups, such as those in the dsRNA backbone, we proposed that phosphates from the dsRNA backbone might bind at one or both of the sulfate binding sites. To determine whether any of the proposed ligand binding sites was responsible for TLR3 ligand recognition, we mutated residues around the two sulfate binding sites and at the positively charged patches. Here we demonstrate that ligand binding depends on residues in a region encompassing the LRR20 sulfate binding site and extending to the glycan-free surface, thus providing the first indication of how TLRs interact with their cognate ligands.
Results
H539E Mutation Abolishes the Ligand-Dependent Activation of TLR3.
To determine whether either of the two bound sulfate ions mark ligand binding sites, the coordinating residues (Fig. 1) were mutated to either similarly sized hydrophobic residues or residues with opposing charge. All mutants were expressed at comparable levels by Western blot analysis (Fig. 7, which is published as supporting information on the PNAS web site). To test whether the mutated proteins were active, human embryonic kidney (HEK) 293 cells were transiently transfected with each TLR3 mutant and stimulated with pI:pC. Mutation of residues Y326, H359, and N361 within sulfate binding site 1 (Fig. 1B) had no significant effect on TLR3 stimulation (Fig. 2A), indicating that these residues are not necessary for ligand recognition. At site 2 (Fig. 1C), mutation of three residues that directly coordinate the sulfate ion, R488, N515, and Q538, also had no effect on TLR3 stimulation. Pairwise mutation of these residues could reveal underlying compensation among these residues in ligand coordination. However, mutation of His-539 to glutamic acid led to a complete loss of function. The H539E mutant, when overexpressed, activated NF-κB constitutively (Fig. 2B), indicating that the mutant protein is expressed and that signaling components remain intact. However, at no level of expression did H539E exhibit ligand-dependent activation. Interestingly, substitution of H539 with an alanine failed to significantly block the activation of TLR3 by pI:pC (Fig. 2), indicating that the loss of function in the H539E mutant is due to the introduction of a negative charge at the position of the H539 imidazole.
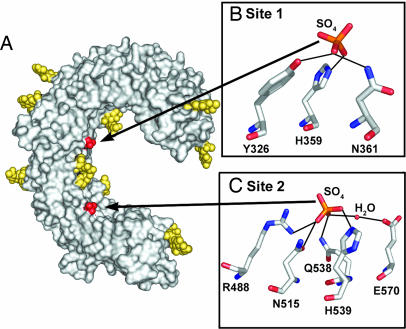
Fig. 1.
Two sulfate ions bind to TLR3-ECD. (A) View of the concave and glycan-free lateral surfaces of TLR3-ECD (Protein Data Bank ID code 2A0Z). The molecular surface is shown in gray, N-linked glycan is in yellow, and two bound sulfate ions are in red. (B and C) Detailed views of residues coordinating the sulfate ions with hydrogen bonds indicated by dotted lines. (B) At site 1, the sulfate ion is coordinated by N361, Y326, and H359. (C) At site 2, the sulfate is coordinated directly by R488, N515, Q538, and H539 and by a water-mediated hydrogen bond (W52) to E570.
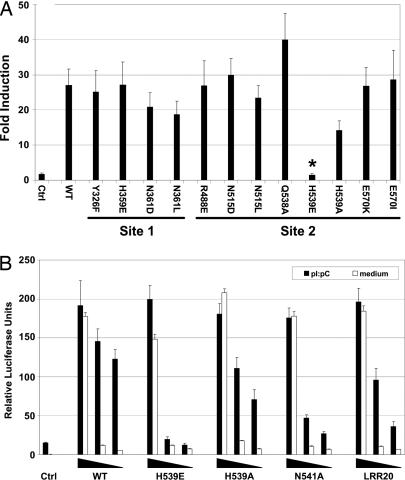
Fig. 2.
Mutation of His-539 to Glu in sulfate site 2 abrogates TLR3 signaling. (A) Mutation of residues coordinating sulfate 1 (site 1) and sulfate 2 (site 2). HEK293 cells, transfected with WT or mutant TLR3, were stimulated or not with p(I):p(C) and assayed for NF-κB activation. Results (±SEM) are reported as fold induction relative to unstimulated cells. Only H539E was significantly different from WT (∗, P < 0.05). (B) HEK293 cells were cotransfected with 40, 4, or 0.4 ng of TLR3 DNA and stimulated (filled bars) or not (open bars) with p(I):p(C). NF-κB activation measurements, reported as relative luciferase units (±SEM), are averaged from at least three separate experiments. Note that transfection with 40 ng of DNA gives constitutive activation with all plasmids except for the empty vector control.
N541 Is Essential for dsRNA Recognition.
The imidazole side chain of H539 lies exposed to the medium between the concave and glycan-free lateral surfaces of the TLR3 horseshoe in LRR20. To define other dsRNA:TLR3 interactions, we mutated several residues proximal to H539 on both the concave and glycan-free lateral surfaces of the TLR3-ECD (Fig. 3A and B). Mutation of six residues on the concave surface (colored green in Fig. 3 A and B) and five residues on the lateral surface (colored blue) had no effect on activity (Fig. 3C). In striking contrast, mutation of N541 (colored magenta) on the lateral face almost totally abrogated activity, and increased expression of the N541A mutant only minimally restored ligand-dependent activation (Fig. 2B). The pivotal roles played by H539 and N541 in ligand recognition suggest that dsRNA interacts with TLR3 on the glycan-free lateral surface and predicts that glycans are not directly involved in ligand binding. To confirm the lack of involvement of glycan in ligand binding, we mutated each of the 15 asparagine residues in consensus N-linked glycosylation motifs (Asn-X-Ser/Thr; X is any residue) to aspartic acid. None of these mutations had a significant effect on TLR3 expression or activity (Fig. 8, which is published as supporting information on the PNAS web site).
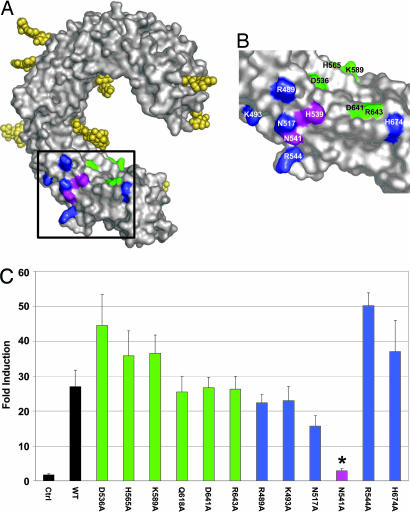
Fig. 3.
Mutation of residues near H539 further define the dsRNA binding site. (A and B) Residues mutated on the concave and lateral surfaces proximal to H539 are highlighted in green and blue, respectively. The two residues important for function, H539 and N541, are colored magenta. The boxed area in A is shown in detail in B. (C) Transfected cells were examined for responsiveness to pI:pC as in Fig. 2A. Bars are color-coded as in A. Only N541A showed significant loss of responsiveness (∗, P < 0.05).
H539E and N541A Mutations Abrogate dsRNA Binding.
We next asked whether the lack of TLR3 function in the H539E and N541A mutants was due to a loss of ligand-binding capacity. WT or mutant TLR3-ECD, from small-scale transient transfection of insect cells, was incubated with p(I):p(C), and the mixture was separated by size-exclusion chromatography. As previously reported (3), WT TLR3-ECD migrates as a single peak (Fig. 4 Top, fraction 11). However, in the presence of p(I):p(C), a major portion of WT TLR3-ECD elutes in the void volume, at the same position as pI:pC (fraction 4), indicating that WT TLR3-ECD directly binds pI:pC. The H539E and N541A mutants, in the absence of ligand, exhibit an elution pattern similar to WT TLR3-ECD (Fig. 4 Middle and Bottom), signifying a lack of gross mutant-induced structural alterations. In striking contrast to the WT protein, the H539E and N541A mutants show only negligible binding to p(I):p(C). We conclude that the loss of activating capacity of the H539E and N541A mutants is due to their inability to bind ligand and that ligand binding occurs in the patch defined by H539 and N541 on the glycan-free lateral surface at LRR20.
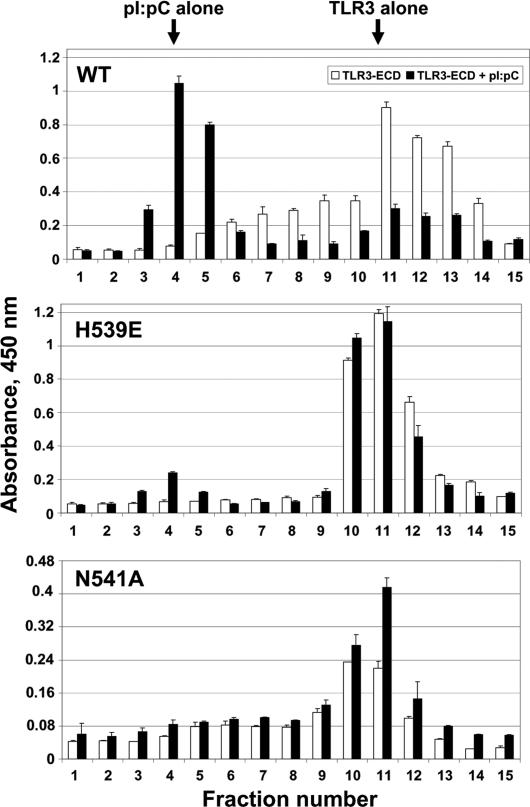
Fig. 4.
H539E and N541A fail to bind ligand. TLR3-ECD (WT or mutant) protein was incubated with p(I):p(C) or buffer and analyzed by size-exclusion chromatography. Column fractions were assayed for TLR3-ECD by ELISA at 450 nm. Data are representative of two separate experiments. Elution profiles of WT (Top), H539E (Middle), and N541A (Bottom) TLR3-ECD protein are shown. Open bars, protein alone; filled bars, protein plus p(I):p(C). Arrows indicate the elution volumes of pI:pC and TLR3-ECD when run alone.
Roles of LRR Insertions and Positively Charged Patches in Ligand Recognition.
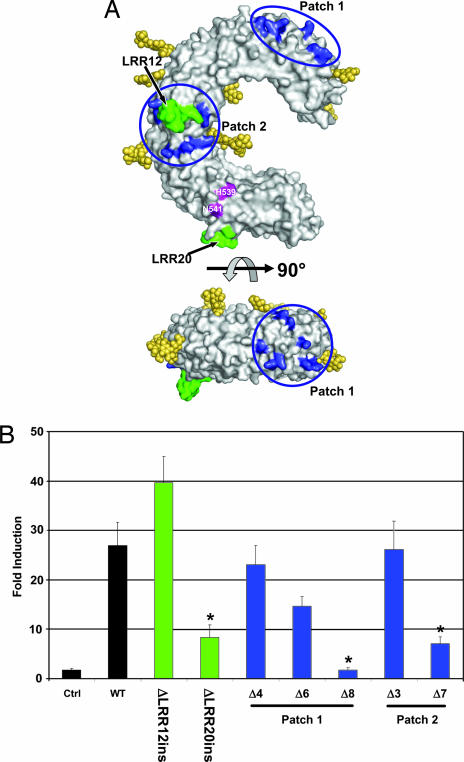
The TLRs are distinct from other LRR proteins in that their LRR consensus motifs are often interrupted by extended insertions (15). TLR3 contains two such insertions, located in LRRs 12 and 20, respectively (Fig. 5A). Previously, we suggested that such insertions might play a crucial role in ligand recognition. To test this hypothesis, we deleted the insertions from LRRs 12 and 20, thereby returning them to the consensus motif. Although removal of the LRR12 insertion had no effect on TLR3 stimulation, deletion of the LRR20 insertion produced a significant, but not total, loss in activity (Figs. 2B and 5B). It is possible that the LRR20 insertion interacts directly with dsRNA, but we cannot exclude the alternative explanation that removal of the LRR20 insertion (residues 546–553) disrupts the local structure near residues H539 and N541 and their interaction with ligand. In either case, this result provides further evidence that ligand interacts with TLR3 on the glycan-free face of LRR20.
Fig. 5.
Role of LRR insertions and basic residue patches in dsRNA recognition. (A) Location of mutated residues. Groups of positively charged residues in patches 1 and 2, shown in blue, were mutated to alanine. The inserts in LRRs 12 and 20, shown in green, were deleted. (B) Responsiveness of TLR3 mutants to pI:pC. Deletions of LRR inserts are shown in green, and mutants in the positively charged patches are color-coded blue. In patch 1, mutant Δ4 contains four mutations, R65A, K89A, K117A, and K147A; Δ6 contains six mutations, R65A, K89A, K117A, K137A, K139A, and K147A; and Δ8 contains eight mutations, R65A, K89A, K117A, K137A, K139A, K145A, K147A, and K165A. In patch 2, Δ3 contains three mutations, K335A, K345A, K371A; and Δ7 contains seven mutations, R331A, K335A, K345A, K371A, R394A, K416A, and K418A. ∗, significant loss in activity (P < 0.05).
TLR3 also contains two patches of basic residues, one near the N terminus and one on the lateral, glycan-free face (patches 1 and 2, respectively, in Fig. 5A), that could offer electrostatically favorable docking sites for the negatively charged phosphate backbone of dsRNA (4). Of the 15 residues that comprise these two patches, positive charge is conserved only at positions 137 and 335 in TLR3 orthologs ranging from zebrafish to humans. In patch 1, mutation of either four or six positively charged residues to alanine had no significant effect on ligand recognition, although mutation of an additional two positively charged residues resulted in a loss of function (Fig. 5B). However, when these latter two residues were mutated individually, no loss in activity was detected (Fig. 9, which is published as supporting information on the PNAS web site). Similarly, mutation of three positively charged residues in patch 2 had no effect on activity, whereas removal of seven positive charges resulted in a significant loss in function (Fig. 5B). Because loss of activity is observed only when large numbers of positively charged residues are mutated, and because this occurs in two widely separated patches, it is possible that the loss of activity reflects secondary effects, such as structural alterations within the TLR3 horseshoe, rather than direct alterations of ligand binding residues. Because such secondary effects are difficult to verify, we cannot absolutely exclude these basic patches as potential sites of ligand interaction, but we are unable to identify a subset of basic residues essential for ligand recognition. Mutation of four other positively charged residues, including K27, H39, K41, and H60, which lie diagonally across the TLR3-ECD horseshoe from H539/N541, had no effect on activity (Fig. 9).
Discussion
To locate the ligand binding site on TLR3, we analyzed >50 mutations within the TLR3-ECD. Remarkably, only 2 of the 50 residues tested resulted in abrogation of both the activation of TLR3 by pI:pC and the direct binding of pI:pC to purified TLR3-ECD protein. These two residues, H539 and N541, are conserved from zebrafish to humans (Fig. 10, which is published as supporting information on the PNAS web site) and position the ligand binding site on the glycan-free surface of the ECD at LRR20.
Replacing His-539 with an alanine has little effect on TLR3 responsiveness, whereas substitution of a negatively charged carboxyl group for an imidazole ring at this site results in a total loss of function. This finding suggests that a negative charge from a backbone phosphate group on dsRNA occupies a position in close proximity to residue 539 in the ligand–receptor complex. In the WT protein a protonated imidazole ring of His-539 would neutralize the negative charge of the phosphate, but in the H539E mutant electrostatic repulsion from the negative charge of the glutamate carboxyl would prevent the ligand from approaching the TLR3 surface. A histidine imidazole ring can be protonated under mildly acidic conditions, suggesting that TLR3 would signal best at acidic pH. This prediction is supported by recent data from de Bouteiller et al. (16), who reported that the optimal pH range for TLR3 stimulation fell between pH 5.7 and 6.7. Nevertheless, the interaction between the His-539 imidazole and the backbone phosphate is not necessary for function, because simply removing the imidazole group (in H539A) does not lead to a loss of function. This lack of an effect is in contrast to the situation with Asn-541, where replacing the Asn-541 with an alanine was sufficient to negate TLR3 stimulation and ligand binding, indicating that an interaction between the Asn-541 amido group with the ligand is essential for ligand binding. Amido groups, in dsRNA binding domains, often coordinate the 2′ hydroxyl of the ribose or the phosphodiester group of dsRNA (17, 18).
Although TLR3 is one of the most heavily glycosylated of the TLRs (19), it nevertheless contains a lateral face that is totally devoid of glycan and therefore free to interact with ligand or another protein molecule. Not surprisingly, His-539 and Asn-541 are located on this face. The glycan-free face is characteristic of TLR3, because most other human TLRs contain fewer glycosylation sites than TLR3 and, according to their predicted placement on the LRRs (19), do not have an identifiable interaction surface. It has been reported that mutation of 2 of the 15 glycosylation sites on TLR3 (N247 and N413) results in a complete loss of function (16, 20), although we failed to detect a significant effect on activity resulting from an Asp-for-Asn replacement at any single glycosylation site (Fig. 8). Because N-linked glycan is usually involved in either protein folding or trafficking (21), further examination of these glycosylation mutants is necessary to define whether loss of receptor function could be attributed to misfolding or inappropriate localization.
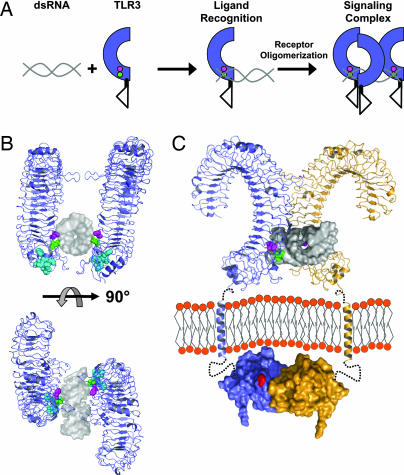
Based on the results of our mutational analyses, we developed a model for the binding of ligand to TLR3-ECD (Fig. 6 A and B). Several orientations of ligand are possible given only the restraint of direct interaction with H539 and N541, but some could be excluded because mutations, such as H60 and H39 for a vertical dsRNA orientation or deletion of the LRR12 insertion and lysines on the glycan-free surface, did not significantly alter ligand recognition. In our best model, H539 is positioned within close proximity to the nonbridging oxygens of a phosphodiester group within the backbone of the dsRNA ligand, consistent with a negative charge at position 539 disrupting ligand binding. Interestingly, with this positioning, the amide side chain of N541, which is required for ligand binding, is within hydrogen bonding distance of a 2′ hydroxyl located two nucleotides downstream of the phosphate interaction (Fig. 11, which is published as supporting information on the PNAS web site). This interaction could explain why pdI:pdC, which lacks 2′ hydroxyl groups, fails to bind or activate TLR3 (ref. 5 and J.K.B., unpublished observation). Both of these interactions are within the minor groove of the dsRNA helix (Fig. 6B). By satisfying direct ligand interactions to both H539 and N541, it is not possible to position the phosphate backbone of dsRNA to occupy the sulfate ion site (Fig. 1C). Previously, we and others showed that the association of dsRNA with recombinant TLR3-ECD or TLR3 chimeras leads to receptor oligomerization (3) and signal transduction (16). With our model for dsRNA recognition, a second TLR3-ECD, related by an ≈180° rotation and a short translation, could interact identically with the opposite strand of the dsRNA molecule, taking advantage of the inherent twofold symmetry within the dsRNA molecule. Additional TLR3 molecules could bind in a similar manner to long dsRNA oligomers. This mode of receptor aggregation differs from the crystallographic TLR3-ECD dimer previously proposed as a possible signaling structure (3, 4). The crystallographic dimer does not allow ligand access to the two key residues, His-539 and Asn-541, directly involved in ligand binding.
Fig. 6.
Model of TLR3 recognition of dsRNA and signaling complex. (A) Diagram of TLR3 ligand recognition and signaling complex. dsRNA is depicted in gray, and TLR3-ECD is shown in blue. H539 and N541 are shown as magenta and green circles, respectively. (B) A 19-bp dsRNA molecule (gray surface, Protein Data Bank ID code 1QC0) was docked onto the TLR3-ECD structure (blue). In the model, H539 (magenta) coordinates a phosphate group in the minor groove whereas N541 (green) forms a hydrogen bond to the 2′OH of the ribose also in the minor groove. The LRR20 insertion is shown in cyan. Two TLR3-ECD molecules, related by an ≈180° rotation, bind to one dsRNA ligand. Multiple TLR3 molecules could bind to a longer dsRNA as illustrated in A. (C) A proposed signaling complex of full-length TLR3 includes the oligomer model of ligand interaction from B rotated 45° with 22-residue transmembrane helices and TIR domains (Protein Data Bank ID code 1FYV). Single receptors are shown in blue or orange. Linker regions between the LRR-CT and TM (7 residues) and the TM and TIR domain (19 residues) are denoted by black dotted lines. The homologous position of the Lpsd mutation in the BB loop of the TLR4 TIR domain (25), postulated to interact with MyD88 (26), is indicated in red. Notably, the separation of the TLR3-ECD in the proposed oligomer easily accommodates the formation of a TIR dimer (27).
In contrast to T cell, B cell, and Fc receptors, where receptor crosslinking per se is sufficient to trigger a response, TLR activation appears to require the correct positioning of receptor molecules in ligand–receptor complexes. This finding is supported by the observation that anti-TLR antibodies, unlike antibodies against T cell, B cell, and Fc receptors, usually fail to trigger activation and, in fact, often serve as antagonists (e.g., refs. 14 and 22). According to our model, the bringing together of two TLR3-ECDs at their glycan-free surfaces by dsRNA, as depicted in Fig. 6C, could position the TIR domains in the correct orientation for signaling. Precise orientation of cytoplasmic domains in TLRs would be facilitated by the lack of long, flexible linkers between the receptor and transmembrane domain; in the case of TLR3, just seven residues act as a linker between these two domains.
With the first structure of a TLR ligand binding domain now solved, the question remains of how the LRR-based solenoid structure has adapted to recognize ligands varying from small chemical compounds such as imiquimod and Taxol, to bacterial lipids, nucleic acids, and proteins. Insertions within the consensus LRR sequences of TLRs contribute unique features to each receptor and have been previously suggested to play important roles in ligand recognition (15). In the case of TLR3, neither of the two insertions was essential for the response to pI:pC, although the LRR20 insertion may contribute to ligand recognition. In contrast to TLR3, it is highly likely that insertions in LRRs 2, 5, and 8 of TLR9 play an important role in the binding of CpG DNA, because these insertions contain known CpG DNA binding motifs (15). These two examples suggest that insertions play different roles among the 10 human TLRs, much like the degree of glycosylation may distinguish potential ligand recognition sites as noted above. Thus, although this study offers the first indication of how a pathogen-associated molecular pattern can interact with a TLR, it is likely that each new TLR structure will reveal a unique mode of pathogen recognition.
Experimental Procedures
Site-Directed Mutagenesis.
Human TLR3 cDNA [National Center for Biotechnology Information (NCBI) accession no. NM_003265] in pUNO-hTLR3 (InvivoGen, San Diego) was mutated by using a QuikChange site-directed mutagenesis kit (Stratagene). Primers for each mutation are listed in Table 1, which is published as supporting information on the PNAS web site. Individual mutations included K27A, H39A, K41A, H60A, Y326F, H359E, N361D, N361L, R488E, R489A, K493A, N515D, N515L, N517A, D536A, Q538A, H539E, H539A, N541A, R544A, H565A, E570K, E570I, K589A, Q618A, D641A, R643A, and H674A. Two basic patches containing residues R331, K335, K345, K371, R394, K416, and K418 (Patch 1) and R65, K89, K117, K137, K139, K145, K147, and K163 (Patch 2) were mutated to alanine in groups defined in the legend to Fig. 5. The ΔLRR12 insertion deleted residues 335–342 and a P344S substitution, and the ΔLRR20 insertion deleted residues 546–553. WT TLR3-ECD and the H539E and N541A mutants, including residues 22–703 followed by a C-terminal tobacco etch virus protease cleavage site, FLAG, and His6 tags were cloned into pIEX3 (EMD Biosciences, San Diego) for transient expression in insect cells. All mutations were confirmed by DNA sequencing of the entire ORF.
Cell Culture.
HEK293 cells (American Type Culture Collection) were cultured in DMEM containing 2 mM l-glutamine, 50 units ml−1 penicillin, 50 μg ml−1 streptomycin, and 10% low-endotoxin FBS. Sf9 cells (Invitrogen) were cultured in BacVector medium (EMD Biosciences).
NF-κB Activity.
NF-κB activity was measured as described previously (23). Briefly, adherent HEK293 cells in 96-well plates were cotransfected with WT or mutated TLR3 DNA (1.3 ng per well) and Ig-κB-luciferase and β-galactosidase reporter plasmids (6.0 ng of each per well) by using calcium phosphate precipitation (Stratagene). Empty pUNO-mcs vector (InvivoGen) served as the negative control. In titration experiments, each well received 6.0 ng of the two reporter plasmids, increasing doses of TLR3 vector (0.4, 4, or 40 ng per well), and empty pUNO-mcs DNA to bring the total amount of DNA to 220 ng. To initiate the assay, medium was removed and 100 μl of medium containing 5 μg ml−1 of p(I):p(C) was added to the wells. After 6 h of stimulation, cells were lysed, luciferase and β-galactosidase activities were determined, and the ratios of luciferase to β-galactosidase activities were calculated. Each sample was measured in triplicate, and each mutant was assayed in a minimum of three separate experiments. Statistical analyses were performed by David Venzon (Biostatistics and Data Management Section, Center for Cancer Research, National Cancer Institute, Bethesda) as described in Supporting Text, which is published as supporting information on the PNAS web site.
Ligand Binding.
Adherent Sf9 cells in T75 flasks were transfected with WT or mutant TLR3-ECD DNA (12 μg) in pIEX by using Insect GeneJuice (Novagen) as per the manufacturer's protocol. Supernatants were harvested after 48 h, and TLR3-ECD was purified by absorption to anti-FLAG M2 agarose (Sigma) and elution with FLAG peptide. Purified protein was incubated with 20 μg of p(I):p(C) in Hepes buffer (0.02 M Hepes, pH 7.5/0.1 M NaCl/1 mM 2-mercaptoethanol) or with Hepes buffer alone overnight at 4°C. Samples (50 μl) were eluted on a 3.2-ml Superdex 200 size-exclusion chromatography column in Hepes buffer with the SMART System (Amersham Pharmacia), and 80-μl fractions were collected and analyzed by ELISA as described (24). Briefly, duplicate samples were incubated overnight at 4°C in 96-well anti-FLAG M2-coated plates. After washing, anti-His-6 conjugated to horseradish peroxidase was added, and absorbance at 450 nm was developed by using TMB substrate solution (R & D Systems).
Supplementary Material
Acknowledgments
We thank Dr. David Venzon for valuable assistance with the statistical analysis of our data and Drs. Alison Hickman, Steven Shaw, and Paul Roche for critical comments on the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health (National Cancer Institute and National Institute of Diabetes and Digestive and Kidney Diseases) and by a National Institutes of Health/Federal Drug Administration Intramural Biodefense Award from the National Institute of Allergy and Infectious Diseases.
Abbreviations
- TLR
Toll-like receptor
- ECD
ectodomain
- LRR
leucine-rich repeat
- pI:pC
polyinosinic-polycytidylic acid
- TIR
Toll–IL-1 receptor; HEK, human embryonic kidney.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Janeway C. A., Jr. Proc. Natl. Acad. Sci. USA. 2001;98:7461–7468. doi: 10.1073/pnas.131202998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S., Uematsu S., Takeuchi O. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Bell J. K., Botos I., Hall P. R., Askins J., Shiloach J., Segal D. M., Davies D. R. Proc. Natl. Acad. Sci. USA. 2005;102:10976–10980. doi: 10.1073/pnas.0505077102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choe J., Kelker M. S., Wilson I. A. Science. 2005;309:581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 5.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 6.Schroder M., Bowie A. G. Trends Immunol. 2005;26:462–468. doi: 10.1016/j.it.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Visintin A., Mazzoni A., Spitzer J. H., Wyllie D. H., Dower S. K., Segal D. M. J. Immunol. 2001;166:249–255. doi: 10.4049/jimmunol.166.1.249. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto M., Funami K., Tanabe M., Oshiumi H., Shingai M., Seto Y., Yamamoto A., Seya T. J. Immunol. 2003;171:3154–3162. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 9.Sivori S., Falco M., Della C. M., Carlomagno S., Vitale M., Moretta L., Moretta A. Proc. Natl. Acad. Sci. USA. 2004;101:10116–10121. doi: 10.1073/pnas.0403744101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pisegna S., Pirozzi G., Piccoli M., Frati L., Santoni A., Palmieri G. Blood. 2004;104:4157–4164. doi: 10.1182/blood-2004-05-1860. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt K. N., Leung B., Kwong M., Zarember K. A., Satyal S., Navas T. A., Wang F., Godowski P. J. J. Immunol. 2004;172:138–143. doi: 10.4049/jimmunol.172.1.138. [DOI] [PubMed] [Google Scholar]
- 12.Kulka M., Alexopoulou L., Flavell R. A., Metcalfe D. D. J. Allergy Clin. Immunol. 2004;114:174–182. doi: 10.1016/j.jaci.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 13.Matsushima H., Yamada N., Matsue H., Shimada S. J. Immunol. 2004;173:531–541. doi: 10.4049/jimmunol.173.1.531. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto M., Kikkawa S., Kohase M., Miyake K., Seya T. Biochem. Biophys. Res. Commun. 2002;293:1364–1369. doi: 10.1016/S0006-291X(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 15.Bell J. K., Mullen G. E. D., Leifer C. A., Mazzoni A., Davies D. R., Segal D. M. Trends Immunol. 2003;24:528–533. doi: 10.1016/s1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 16.de Bouteiller O., Merck E., Hasan U. A., Hubac S., Benguigui B., Trinchieri G., Bates E. E., Caux C. J. Biol. Chem. 2005;280:38133–38145. doi: 10.1074/jbc.M507163200. [DOI] [PubMed] [Google Scholar]
- 17.Ryter J. M., Schultz S. C. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang K. Y., Ramos A. FEBS J. 2005;272:2109–2117. doi: 10.1111/j.1742-4658.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- 19.Weber A. N., Morse M. A., Gay N. J. J. Biol. Chem. 2004;279:34589–34594. doi: 10.1074/jbc.M403830200. [DOI] [PubMed] [Google Scholar]
- 20.Sun J., Duffy K. E., Ranjith-Kumar C. T., Xiong J., Lamb R. J., Santos J., Masarapu H., Cunningham M., Holzenburg A., Sarisky R. T., et al. J. Biol. Chem. 2006;281:11144–11151. doi: 10.1074/jbc.M510442200. [DOI] [PubMed] [Google Scholar]
- 21.Helenius A., Aebi M. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 22.Shimazu R., Akashi S., Ogata H., Nagai Y., Fukudome K., Miyake K., Kimoto M. J. Exp. Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mullen G. E., Kennedy M. N., Visintin A., Mazzoni A., Leifer C. A., Davies D. R., Segal D. M. Proc. Natl. Acad. Sci. USA. 2003;100:3919–3924. doi: 10.1073/pnas.0630495100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy M. N., Mullen G. E., Leifer C. A., Lee C., Mazzoni A., Dileepan K. N., Segal D. M. J. Biol. Chem. 2004;279:34698–34704. doi: 10.1074/jbc.M405444200. [DOI] [PubMed] [Google Scholar]
- 25.Poltorak A., He X., Smirnova I., Liu M. Y., Huffel C. V., Du X., Birdwell D., Alejos E., Silva M., Galanos C., et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y., Tao X., Shen B., Horng T., Medzhitov R., Manley J. L., Tong L. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 27.Tao X., Xu Y., Zheng Y., Beg A. A., Tong L. Biochem. Biophys. Res. Commun. 2002;299:216–221. doi: 10.1016/s0006-291x(02)02581-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.