Abstract
Activation-induced cytidine deaminase (AID) initiates Ig class switch recombination and somatic hypermutation by producing U:G mismatches in DNA. These mismatches also have the potential to induce DNA damage including double-stranded breaks and chromosome translocations; therefore, strict regulation of AID is important for maintaining genomic stability. In addition to transcriptional regulation, it has been proposed that phosphorylation can also modulate AID activity. Using a combination of MS and immunochemical approaches we found that 5–15% of the AID expressed in activated B cells was phosphorylated at serine-38 (p38AID). This form of AID was enriched in the chromatin fraction in activated B cells, suggesting a role for phosphorylation in targeting AID to DNA. Consistent with this idea, serine-38 to alanine mutant AID (AIDS38A) showed diminished somatic hypermutation activity on artificial and physiological DNA targets. We conclude that a small fraction of AID is phosphorylated in activated B cells and that the modified form contributes disproportionately to hypermutation.
Keywords: B cells, class switch recombination, protein kinase A, somatic hypermutation
Somatic diversification of antibody genes produces a large repertoire of receptors for recognition of pathogen-associated antigens. Antibody gene diversification occurs in two separate stages during B lymphocyte development. In early developing B cells, random recombination of germ-line-encoded variable (V), diversity (D), and joining (J) gene fragments leads to assembly of antibodies that can recognize most non-self antigens, but antigen binding is usually of relatively low affinity (1). In mature B cells responding to antigen, antibodies are further diversified by somatic hypermutation (SHM), which introduces point mutations or occasional gaps and duplications in Ig heavy and light chain genes (2). B cells carrying somatically mutated antibodies with high affinity for antigen are positively selected to produce plasma cells that secrete the high-affinity antibodies (3).
SHM is initiated by activation-induced cytidine deaminase (AID) (4–6), an enzyme that deaminates cytidine residues in single-stranded DNA exposed during Ig gene transcription (7–13). U:G mismatches created by AID can be processed by a combination of uracil-N-glycosylase, mismatch repair enzymes, and error-prone polymerases to produce a diverse set of mutations in Ig gene DNA (11, 14–22). Alternatively, the mismatches can be processed to produce double-strand breaks that lead to class switch recombination (CSR) or, rarely, to chromosome translocations (15, 16, 20, 21, 23–27). Although AID initiates both SHM and CSR, the molecular requirements for the two reactions are distinct. For example, the carboxyl-terminal domain of AID is required for CSR but not for SHM (28).
How AID is regulated to minimize genomic damage is not well understood but may involve active nuclear export of AID (29, 30) and specific targeting to Ig genes (reviewed in ref. 31). In addition, it has been suggested that posttranslational modification of AID by cAMP-dependent protein kinase A (PKA) may regulate the ability of AID to mediate CSR (8, 32, 33). Here we report that in B cells only a fraction of the AID expressed is phosphorylated, but this form of AID is chromatin-associated and required for efficient hypermutation.
Results
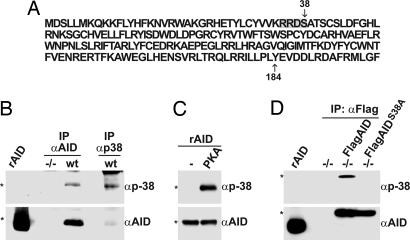
Posttranslational modification of AID was investigated by using MS. AID was affinity-purified from B cells cultured in LPS and IL-4 and subjected to hypothesis-driven tandem MS (34). We found single phosphorylation modifications on AID peptides containing serine-38, a consensus PKA phosphorylation site, and tyrosine-184 (Fig. 1A). To confirm AID serine-38 phosphorylation we produced specific antibodies to AID phospho-serine-38 (anti-p38). Anti-p38 did not react with recombinant AID produced in bacteria but showed reactivity with AID purified from activated B cells in Western blots (Fig. 1B) and with recombinant AID that had been phosphorylated in vitro by recombinant PKA (Fig. 1C). Anti-p38 antibody also recognized Flag-tagged AID (Flag-AID) but not serine-38 to alanine mutant Flag-tagged AID (Flag-AIDS38A) purified from retrovirally transduced AID−/− B cells stimulated with LPS and IL-4 (Fig. 1D). The slightly slower migrating form of AID detected by both anti-AID and anti-p38 (Fig. 1B) could reflect an additional modification such as tyrosine-184 phosphorylation. We conclude that AID is phosphorylated on serine-38 and that anti-p38AID antibodies specifically recognize serine-38-phosphorylated AID purified from B cells.
Fig. 1.
AID is phosphorylated at amino acid position 38. (A) Amino acid sequence of AID showing the location of residue 38 within a consensus PKA phosphorylation site (gray box). (B) Anti-p38 and anti-AID immunoblots of recombinant AID purified from E. coli (rAID) or AID purified from wild-type AID (wt) or AID−/− B cells (−/−) by immunoprecipitation with anti-AID or anti-p38 antibodies. (C) Anti-p38 and anti-AID immunoblots of rAID alone or after treatment with PKA in vitro. (D) Anti-p38 and anti-AID immunoblots of rAID, or anti-Flag immunoprecipitates from AID−/− B cells transduced with retroviruses encoding Flag-tagged AID (FlagAID), Flag-tagged AIDS38A (Flag AIDS38A), or empty vector (−/−). ∗, the expected product.
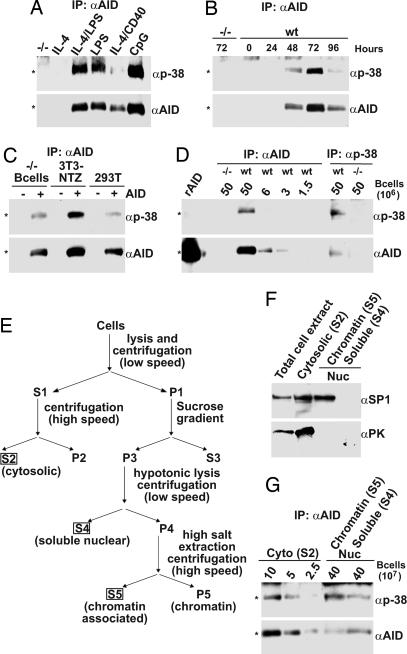
To determine which stimuli induce AID phosphorylation we activated spleen B cells with IL-4, LPS, LPS and IL-4, anti-CD40 and IL-4, or CpG (oligodeoxynucleotide containing unmethylated CpG dinucleotide motifs) and assayed for p38AID by immunoblotting. We found that IL-4 alone was not sufficient for AID induction but that all other stimuli tested induced AID and p38AID (Fig. 2A). The peak of p38AID production in B cells stimulated with LPS and IL-4 was 3 days after stimulation, and at this time there was less AID and p38AID in anti-CD40- and IL-4-stimulated cells than in those stimulated with CpG, LPS, or IL-4 and LPS (Fig. 2 A and B). Anti-CD40 treatment alone did not induce sufficient levels of AID expression to determine relative phosphorylation level (data not shown). Although AID phosphorylation at position 38 was found in B cells, it was not B cell-specific. This modification was also evident when AID was expressed in 293T and 3T3-NTZ cells at a level similar to or higher than in B cells (32, 33) (Fig. 2C).
Fig. 2.
p38AID expression in B cells and fibroblasts. (A) Expression of AID and p38AID in stimulated wild-type B cells or AID−/− controls (−/−). B cells were stimulated with IL-4, IL-4 and LPS, LPS, IL-4 and CD40, or CPG as indicated, and anti-AID immunoprecipitates were blotted with anti-AID or anti-p38. (B) Kinetics of AID versus p38AID expression. Anti-p38 and anti-AID immunoblots of anti-AID immunoprecipitates from wild-type B cells stimulated with LPS and IL-4. The numbers indicate the number of hours in culture. (C) Fibroblasts phosphorylate AID at position 38. Shown are anti-p38 and anti-AID immunoblots of anti-AID immunoprecipitates of AID−/− B cells, 3T3-NTZ cells, or 293T cells infected with AID encoding retrovirus or control (−). (D) Quantitation of AID and p38AID by dilution. Anti-p38 and anti-AID immunoblots of recombinant AID purified from E. coli (rAID), anti-AID, or anti-p38 immunoprecipitates from wild-type B cells or AID−/−. The numbers indicate the number of cells immunoprecipitated for each lane. Comparison of the signal intensities by densitometry reveal that 6% of AID was phosphorylated in this figure. (E) Biochemical fractionation scheme (see Materials and Methods for details). The fractions used for analysis, S2, S4, and S5, are boxed. (F) Anti-pyruvate kinase (a cytoplasmic marker) and anti-SP1 (nuclear marker) immunoblot of cytoplasmic (S2), soluble nuclear (S4), or chromatin (S5) fractions. (G) Anti-p38 and anti-AID immunoblots of AID immunoprecipitated from the cytoplasmic (S2), soluble nuclear (S4), or chromatin-associated (S5) fraction of B cells stimulated with LPS and IL-4. The numbers indicate the number of cells immunoprecipitated in each lane. ∗, the expected product.
To determine the relative amount of phosphorylated AID produced in B cells we compared anti-AID and anti-p38 immunoprecipitates by Western blotting. Whole-cell extracts were immunoprecipitated with anti-AID or anti-p38 and probed with either anti-AID or anti-p38 antibodies. Comparison of the signal intensities by densitometry revealed that only 5–15% of the AID expressed in activated B cells is phosphorylated (Fig. 2D). We conclude that a minor portion of the AID expressed in stimulated B cells is phosphorylated at position 38.
Under physiologic circumstances most of the AID expressed in B cells is in the cytoplasm, but translocation to the nucleus is required for AID function (29, 30, 35, 36). To investigate how phosphorylation might affect the subcellular localization of AID we probed fractioned cell extracts with anti-p38AID antibodies. B cells were stimulated with IL-4 and LPS and then separated into cytosolic (S2), soluble nuclear (S4), and chromatin-associated (S5) protein fractions (Fig. 2E). The cytosolic fraction (S2) was produced by hypotonic detergent lysis, and nuclei (P1) were purified by sucrose gradient centrifugation to remove contaminating cytosol. Nuclei were disrupted by hypotonic lysis to release soluble nuclear components (S4), and chromatin-associated proteins (S5) were released by salt extraction of the remaining pellet (37–39). The relative purity of the fractionated extracts was determined by immunoblotting against pyruvate kinase, a cytosolic marker, and specificity protein (Sp1), which is a chromatin-associated transcription factor (37, 40, 41). Our fractionation procedure allows leakage of nuclear material into the cytosolic fraction but yields soluble nuclear and chromatin-associated fractions that are relatively free of cytosolic contamination (Fig. 2F). AID was immunoprecipitated from each of the fractions, and the relative amounts of AID and p38AID were measured by Western blotting. In agreement with others (29, 30, 35), we found that most of the AID was in the cytoplasm and not in the nucleus (Fig. 2G). Furthermore, there was little enrichment of p38AID in the soluble nuclear fraction, suggesting that position 38 phosphorylation does not directly alter nuclear transport of AID (Fig. 2G). In contrast, chromatin-associated AID, which was 40% of the nuclear AID and <10% of the AID found in the cytoplasm, was highly enriched for p38AID (p38AID was 4.2-fold-enriched in chromatin versus cytoplasm; Fig. 2G). Thus, in activated B cells chromatin-associated AID is preferentially phosphorylated.
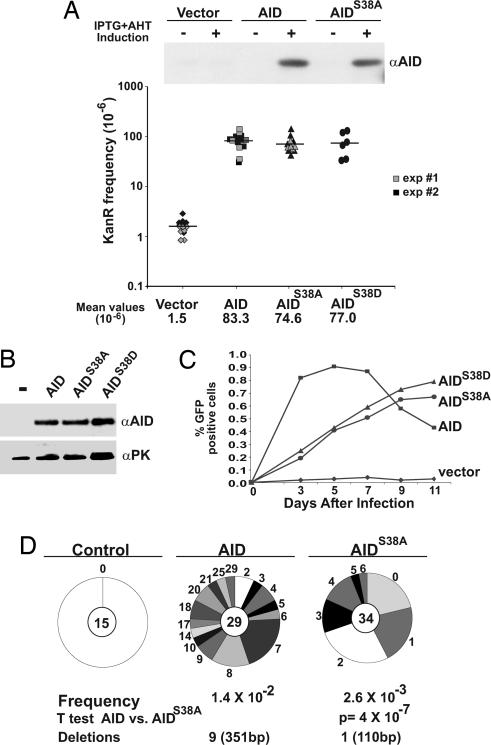
Serine-38 appears to be required for AID to interact with replication protein A and for initiation of CSR, but the role of AID position 38 phosphorylation in somatic mutation has not been examined (32, 33, 42). To examine the role of serine-38 in hypermutation we mutated AID serine-38 to alanine (AIDS38A) or aspartic acid (AIDS38D). We initially performed mutation assays in a uracil-N-glycosylase-deficient Escherichia coli that carry a plasmid encoding an inactivating point mutant kanamycin resistance gene to evaluate the activity of the mutants (12). In this assay, reversion of CCAP94 to CTAL94 confers kanamycin resistance and is a measure of AID cytidine deamination activity (12). We found that AID, AIDS38A, and AIDS38D displayed similar levels of activity (AID versus AIDS38A or AIDS38D; P = 0.48 and 0.5, respectively) (Fig. 3A). Because AID is not phosphorylated in E. coli (Fig. 1) we conclude that AIDS38A and AIDS38D mutation do not cause structural alterations that interfere with catalysis in E. coli.
Fig. 3.
AIDS38A is less active than AID in hypermutation in 3T3-NTZ fibroblasts. (A) Immunoblot of E. coli extracts from cells expressing AID, AIDS38A, or vector control before (−) or after (+) induction with IPTG. The graph shows a log plot of numbers of kanamycin-resistant (KanR) colonies after induction of AID, AIDS38A, and AIDS38D expression. (B) Anti-AID immunoblots of 3T3-NTZ whole-cell extracts from cells expressing AID, AIDS38A, or AIDS38D. Anti-pyruvate kinase (αPK) is shown as a loading control. (C) Accumulation of GFP-expressing 3T3-NTZ cells after transduction with AID-, AIDS38A-, or AIDS38D-expressing retroviruses. The x axis indicates the number of days after transduction, and the y axis indicates the percentage of GFP-positive cells measured by flow cytometry. (D) Number of mutations in the GFP gene cloned from 3T3-NTZ cells 11 days after transduction with retroviruses encoding AID, AIDS38A, or control. Segment sizes in the pie charts are proportional to the number of sequences carrying the number of mutations indicated in the periphery of the charts. The total number of independent sequences analyzed is indicated in the center of each chart. Statistical significance was determined by a two-tailed t test assuming unequal variance and comparing AID-expressing with AIDS38A-expressing cells. P values are indicated. The numbers of point mutations were as follows: 0 mutations per 14,115 bp for vector; 315 mutations per 22,401 bp for AID; and 64 mutations per 24,741 bp for AIDS38A.
To determine whether serine-38 phosphorylation regulates hypermutation in mammalian cells we used 3T3-NTZ indicator cells, which express a single integrated copy of an inactive form of GFP with a premature stop codon that can be reverted by mutation to produce active GFP (Fig. 5, which is published as supporting information on the PNAS web site) (43). These cells phosphorylate AID at position 38 at levels similar to or higher than B cells stimulated with LPS and IL-4 (Fig. 2C). Wild-type AID, AIDS38A, and AIDS38D were expressed at equivalent levels in 3T3-NTZ indicator cells, and both were able to induce GFP reversion mutations as measured by flow cytometry (Fig. 3 B and C). However, wild-type AID was more active than either of the mutants in this assay (Fig. 3C). At later time points GFP expression decreased because of continued accumulation of mutations and deletions. (Fig. 3 C and D) (43). The decreased activity of the AIDS38D mutant indicates that phosphorylation on serine-38 adds more than just a negative charge at that residue.
To measure mutations directly we sequenced the GFP genes cloned from cells expressing retrovirally encoded wild-type AID or AIDS38A 11 days after transduction (Fig. 3D). We found that there were fewer clones with mutations in cells expressing AIDS38A. All of the clones from AID-expressing cells showed at least two mutations, versus 21% with no mutations in AIDS38A (P = 0.008) (Fig. 3D). In addition, there were more mutations per mutated clone in cells expressing wild-type AID than in those expressing AIDS38A: 11 versus 1.8 mutations per clone, respectively (P = 5 × 10−7) (Fig. 3D), with a maximum of 29 mutations per clone in AID versus a maximum of 6 in AIDS38A. Finally, 28% of clones from AID-expressing cells displayed deletions versus 3% for AIDS38A (P = 0.006, Fig. 3D). Nevertheless, AIDS38A displayed the same hotspot preference and G/C bias as AID (Fig. 6, which is published as supporting information on the PNAS web site). We conclude that AID phosphorylation at position serine-38 is required for efficient hypermutation in 3T3-NTZ cells.
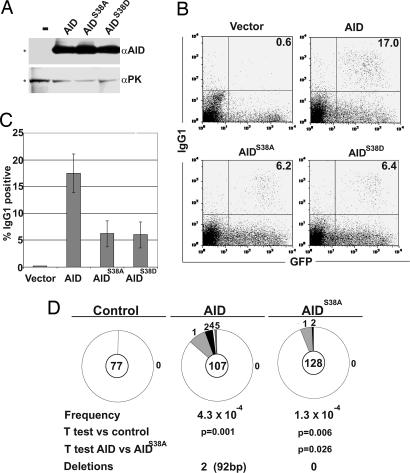
To determine whether phosphorylation also regulates hypermutation in B cells we measured the rate of hypermutation in the region 5′ of the Igμ switch in B cells stimulated with LPS and IL-4 (44–46). AID−/− B cells were infected with AID-, AIDS38A-, or AIDS38D-expressing retroviruses (Fig. 5) for 72 h and initially evaluated for cell-surface expression of IgG1 by flow cytometry (45). AID was produced at similar levels by all of the viruses (Fig. 4A) and at levels that were 10-fold higher than endogenous AID (47). Despite similar levels of AID protein expression in all samples (Fig. 4A), AIDS38A- and AIDS38D-expressing B cells showed a significantly lower rate of class switching to IgG1 than wild type: 6.2% and 6.4%, respectively, versus 17.0% with a background staining of 0.6% (Fig. 4B). Overall switching in AIDS38A-expressing cells was 35% of wild type in four experiments (P = 0.0013) (Fig. 4 B and C). The rate of 5′ of Igμ switch mutation was measured by cloning this region from purified, retrovirally infected IgM-expressing B cells that had not undergone CSR (45). B cells expressing AIDS38A displayed a lower rate of mutation than B cells expressing wild-type AID (P = 0.026) (Fig. 4D) because of a decrease in the number of mutated clones (5.2% versus 14%; P = 0.022) (Fig. 4D) and a slightly lower average number of mutations per mutated clone (1.1 mutations per clone in AIDS38A versus 1.7 mutations in AID; P = 0.09) (Fig. 4D), with similar distribution and hotspot preference (Fig. 4D and data not shown). We conclude that AID phosphorylation positively regulates hypermutation in B cells.
Fig. 4.
AIDS38A is less active than AID in CSR and hypermutation in B cells. (A) Anti-AID immunoblot of whole-cell extracts from AID−/− B cells infected with retroviruses encoding AID, AIDS38A, AIDS38D, or control (−). Anti-pyruvate kinase (αPK) is shown as a loading control. (A) Flow cytometric analysis of IgG1 expression in AID−/− B cells transduced with AID, AIDS38A, AIDS38D, or control vector and cultured in LPS plus IL-4 for 3 days after infection. The percentage of GFP+ cells that were IgG1+ is indicated on the right of each panel. (B) Percentage of CSR to IgG1 by AID−/− B cells transduced with vector control, AID, AIDS38A, or AIDS38D. Bars represent the mean and standard deviation from four independent experiments. (C) Number of mutations in the region 5′ of Sμ cloned from AID−/− B cells transduced with retroviruses encoding AID, AIDS38A, or control. Segment sizes in the pie charts are proportional to the number of sequences carrying the number of mutations indicated in the periphery of the charts. The total number of independent sequences analyzed is indicated in the center of each chart. Statistical significance was determined by a two-tailed t test assuming unequal variance and comparing AID-expressing and AIDS38A-expressing cells. (D) Anti-AID immunoblot of whole-cell extracts from AID−/− B cells infected with retroviruses encoding AID, AIDS38A, AIDS38D, or control (−). Anti-pyruvate kinase is shown as a loading control.
Discussion
Expression of AID in mouse or human B cells leads to programmed genomic damage and activation of signaling and repair pathways, which lead to antibody gene SHM and CSR under physiological circumstances (31). However, AID-induced lesions can also lead to permanent genomic damage by serving as substrates for chromosome translocations or by producing mutations in non-Ig genes, including oncogenes such as Bcl6 or Fas (26, 48–51). Translocations are normally prevented by activation of DNA damage signaling proteins such as p53, but loss of function of p53 is a common feature of B cell malignancies, and this may facilitate further transformation by allowing accumulation of AID-induced translocations (47). How AID is targeted to antibody genes and how untargeted mutations are prevented has not been determined, but the observation that AID expression can cause irreversible genomic damage suggests that regulation is likely to be essential in maintaining genomic integrity (31).
Overexpression of AID in B cells enhances hypermutation and CSR, suggesting that the amount of AID in the cell is rate-limiting and that controlling AID concentration in the nucleus might be critical. At least three mechanisms appear to regulate AID activity in B cells: transcription, cellular compartmentalization, and phosphorylation. AID expression is restricted to activated B cells undergoing SHM and CSR by a combination of specific transcription factors including PAX5 and E47 (52, 53), and the majority of AID protein is excluded from the nucleus by active nuclear export (29, 30). The importance of these forms of regulation was confirmed by the observations that transgenic expression of AID produced thymic lymphomas in mice and that interference of AID nuclear export in fibroblasts enhanced mutation (30, 54).
Our experiments confirm the assignment of serine-38 as a phosphorylation site on AID in activated B cells and show that phosphorylation regulates hypermutation, placing further limits on AID activity in vivo. In this study we used a retroviral overexpression system that we previously characterized to express AID at 10 times the native level in B cells (47). Despite the overexpression, we observed a significant difference between AID and AIDS38A activity in mammalian cells, suggesting that at lower, native levels of AIDS38A expression there may be even greater differences between mutant and wild-type forms.
The amino acid sequence in the region of AID serine-38 conforms to a PKA consensus site, and PKA has be shown to be physically associated with AID in activated B cells (33). PKA can also phosphorylate AID in vitro without affecting catalytic activity, suggesting that phosphorylation may impact AID function indirectly, possibly by facilitating interaction with other proteins (32). Consistent with this idea, phosphorylated AID showed enhanced deamination activity on DNA templates transcribed by T7 phage polymerase in the presence of replication protein A in vitro, and AIDS38A had severely impaired CSR activity compared with wild-type AID in retrovirally transduced AID−/− B cells (32, 42).
cAMP-dependent PKA exists as a holoenzyme tetramer consisting of two catalytic and two regulatory subunits (reviewed in refs. 55 and 56). PKA is a broad-spectrum serine/threonine kinase that phosphorylates a large number of sites in diverse protein substrates. Accumulation of intracellular cAMP causes dissociation of the holoenzyme tetramer and activation of PKA catalytic units. PKA substrate specificity is regulated by interaction with A kinase-anchoring proteins (AKAPs), a diverse family of proteins with at least 50 known members. AKAPs regulate PKA by anchoring the enzyme to specific subcellular locations including ion channels, actin, microtubules, mitochondria, centrosomes, nuclear matrix, and nuclear and cellular membranes (57, 58). Thus, AKAPs physically limit PKA activity by restricting the location of the active enzyme in the cell. In the nucleus, PKA regulates transcription factors and gene expression as well as chromosome condensation. A number of AKAPs are known to localize to the nucleus (e.g., AKAPs 7/95/150), but little is known about possible PKA/AKAP anchoring to active chromatin (55). PKA is expressed in all cells, and AID phosphorylation is not B cell-specific. Nevertheless, AKAPs are differentially expressed, and regulated phosphorylation of AID may be B cell-restricted, requiring physiologic levels of AID expression. In 3T3-NTZ indicator cells or B cells overexpressing AID, p38AID was not enriched in the chromatin fraction (Fig. 7, which is published as supporting information on the PNAS web site). However, overexpressed AID accumulates in the cytoplasm of those cells and may obscure p38AID enrichment in chromatin as assayed by cell fractionation. We speculate that nuclear AKAPs may regulate AID in B cells by producing the more active, phosphorylated form of the protein in the proximity of its DNA substrate. Consistent with this idea, p38AID was preferentially found in the chromatin-associated fraction in activated B cells.
p38AID represents a small fraction (5–15%) of the total AID expressed in B cells. Nevertheless, interfering with phosphorylation results in a >60% loss in both CSR and hypermutation. Therefore, the phosphorylated fraction of AID accounts for a disproportionate amount of its activity, and phosphorylation appears to be a key feature of AID regulation in vivo.
Materials and Methods
Cell Culture and Mutational Analysis.
Wild-type or AID−/− B cells were purified from mouse spleens by depletion with anti-CD43 beads (Miltenyi Biotec) and cultured in RPMI medium 1640 with 5 ng/ml IL-4 plus 25 μg/ml LPS (Sigma-Aldrich), 1 μg/ml anti-CD40 (Becton Dickinson), or 10 μg/ml CpG ODN 2395 (Coley Pharmaceutical) for 72 h. Retroviral supernatants for B cell infections were produced in BOSC 23 cells cotransfected with retroviral pMX-PIE and pCL-ECO plasmids. Supernatants were harvested 48 h after transfection and used to infect B cells that had been cultured in LPS plus IL-4 for 24 h. The cells were stained with biotinylated anti-mouse IgG1 antibodies and streptavidin phycoerythrin-Cy7 (Becton Dickinson) 72 h after infection with retrovirus and analyzed by flow cytometry. To perform switch μ-region mutational analysis, IgG−/GFP+ cells were sorted on a FACSVantage SE cell sorter (BD Immunocytometry Systems). PCR amplification and mutation analysis were performed as previously described (45). Retroviruses for infecting 3T3-NTZ cells (43) were produced by using pQCXIP and pCL-ECO plasmids (CLONTECH). The NTZ-3T3 assay and GFP gene mutational analysis were performed as previously described (30). Cells were cultured in the absence of tetracycline, and mutational analysis was performed on puromycin-selected cells regardless of GFP expression 11 days after retroviral infection. E. coli assays were performed exactly as previously described (12).
Protein Analysis.
To produce anti-p38 antibodies, rabbits were immunized with phosphopeptide CYVVKRRD(s-P)ATSCSLD (AID 30–45) coupled to keyhole limpet hemocyanin. Phosphospecific antibodies were purified by negative selection on unphosphorylated peptide AID 30–45 coupled to Sulfolink gel (Pierce) followed by positive selection on phosphopeptide AID 30–45 (59, 60). Cells were extracted in lysis buffer (20 mM Tris, pH 8/400 mM NaCl/1% Nonidet P-40/0.5 mM EDTA/25 mM NaF/1 mM DTT). To produce anti-AID antibodies, rabbits were immunized with AID residues 185–198 peptide-coupled to keyhole limpet hemocyanin (30). After seven rounds of immunization antibodies were affinity-purified (30). For immunoprecipitation, 2 mg of extracts were incubated with anti-AID antibody and protein A Sepharose (Amersham Pharmacia) for 2 h. For Flag immunoprecipitation, anti-Flag agarose beads (Sigma) were incubated with extracts for 2 h. Western blots were performed on the immunoprecipitated protein with anti-AID antibody or anti-p38 or on 50 μg of extracts with anti-pyruvate kinase (Polysciences) or anti-SP1 (Upstate Biotechnology). For retroviral AID-expressing B cells, Western blots were performed on samples with equal GFP expression. nih image was used for densitometry analysis.
PKA Phosphorylation.
A total of 100 ng of recombinant AID purified from E. coli purchased from Enzymax was incubated with 1,000 units of PKA (Calbiochem) at 30°C for 30 min in 50 mM Tris (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 200 μM ATP.
Cell Fractionation.
Activated B cells (4 × 107) were washed in PBS and resuspended in buffer A (10 mM Hepes, pH 7.9/0.1% Triton X-100/10 mM KCl/1.5 mM MgCl2/0.34 M sucrose/10% glycerol/1 mM DTT) for 8 min. Nuclei were collected by centrifugation (5 min at 1,300 × g). The supernatant (S1) was clarified by centrifugation (5 min at 20,000 × g) and collected (S2 cytoplasmic fraction). The nuclei (P1) were resuspended in buffer I (0.34 M sucrose/3 mM CaCl2/2 mM Mg acetate/10 mM Tris, pH 8/1 mM DTT/0.5% Nonidet P-40/10 mM NaF) and layered on a sucrose cushion buffer II (2 M sucrose/5 mM Mg acetate/0.1 mM EDTA/10 mM Tris, pH 8/1 mM DTT). The sucrose gradient was centrifuged (45 min at 30,000 × g). The pelleted nuclei (P3) were washed and lysed in buffer B (3 mM EDTA/0.2 mM EGTA/1 mM DTT) for 30 min and then centrifuged, and supernatant was collected (S4 soluble nuclear fraction) (39). The nuclei (P4) were resuspended in nuclear extraction buffer (20 mM Tris, pH 8/500 mM NaCl/0.5% Nonidet P-40/1 mM DTT/25 mM NaF) for 30 min to obtain the chromatin-associated fraction (S5). NaF was included to inhibit serine threonine phosphatase activity (38). RIPA buffer (50 mM Tris, pH 8/0.5% deoxycholate/0.1% SDS/1% Nonidet P-40/0.5 mM EDTA/25 mM NaF) was added to the cytoplasmic, soluble nuclear, and chromatin-associated fractions, and AID was immunoprecipitated from these fractions as above.
AID Purification and MS.
B cell extracts were made in lysis buffer, cell debris was removed by centrifugation, salt was adjusted to 200 mM, and extracts were precleared with protein A Sepharose (Amersham Pharmacia) and MonoQ Sepharose (Amersham Pharmacia) before immunoprecipitation with affinity-purified anti-AID antibodies coupled to Dynabeads (Dynal) (30). AID was eluted with elution buffer (50 mM Tris, pH 8.5/0.5% SDS/1 mM DTT), diluted into ion exchange buffer (20 mM Hepes, pH 7.8/7 M urea/1% Nonidet P-40/50 mM NaCl/1 mM DTT), and passed over SP Sepharose (Amersham Pharmacia) resin to retrieve AID and remove residual contaminants. AID was eluted with elution buffer run on an SDS/PAGE gel, and gel slices were collected from regions corresponding to AID detected by Western blot by using anti-AID antibodies. The slices were treated with 50 mM iodoacetamide to alkylate cysteine residues. After dehydration of gel slices with 100% acetonitrile, proteins were digested with 10 ng/μl modified sequencing-grade trypsin (Roche) in 50 mM ammonium bicarbonate for 7 h at 37°C. Tryptic peptides were collected with POROS R2 C18 reverse-phase resin, washed twice with 0.1% trifluoroacetic acid, and eluted with 50% methanol/20% acetonitrile/0.1% trifluoroacetic acid through C18 ziptips (Millipore). Peptides were crystallized in 2,5-dihydroxybenzoic acid for MALDI-MS. Phosphorylation of peptides was determined by using a hypothesis-driven MS approach as described previously (34). Putative AID peptides and phosphopeptides were systematically fragmented and analyzed by tandem MS by using MALDI–ion trap instrumentation (Finnegan). Peptide phosphorylation was identified in this experiment by the neutral loss of phosphoric acid (98 Da) upon fragmentation.
Supplementary Material
Acknowledgments
We thank the members of the M.C.N. laboratory, E. Besmer and F. Papavasiliou for discussion, K. Velizon for cell sorting, and T. Eisenreich for mice breeding. K.M.M. is supported by a fellowship from the National Institutes of Health. This work was supported in part by National Institutes of Health grants (to B.T.C. and M.C.N.). M.C.N. is a Howard Hughes Medical Institute Investigator.
Abbreviations
- AID
activation-induced cytidine deaminase
- CSR
class switch recombination
- SHM
somatic hypermutation
- AKAP
A kinase-anchoring protein.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Tonegawa S. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.McKean D., Huppi K., Bell M., Staudt L., Gerhard W., Weigert M. Proc. Natl. Acad. Sci. USA. 1984;81:3180–3184. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajewsky K. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M., Sankaranand V. S., Anant S., Sugai M., Kinoshita K., Davidson N. O., Honjo T. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 5.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 6.Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., et al. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 7.Bransteitter R., Pham P., Scharff M. D., Goodman M. F. Proc. Natl. Acad. Sci. USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F. W. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 9.Di Noia J., Neuberger M. S. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 10.Dickerson S. K., Market E., Besmer E., Papavasiliou F. N. J. Exp. Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen-Mahrt S. K., Harris R. S., Neuberger M. S. Nature. 2002;418:99–103. [PubMed] [Google Scholar]
- 12.Ramiro A. R., Stavropoulos P., Jankovic M., Nussenzweig M. C. Nat. Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- 13.Pham P., Bransteitter R., Petruska J., Goodman M. F. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 14.Rada C., Williams G. T., Nilsen H., Barnes D. E., Lindahl T., Neuberger M. S. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 15.Rada C., Di Noia J. M., Neuberger M. S. Mol. Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 16.Imai K., Slupphaug G., Lee W. I., Revy P., Nonoyama S., Catalan N., Yel L., Forveille M., Kavli B., Krokan H. E., et al. Nat. Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 17.Wiesendanger M., Kneitz B., Edelmann W., Scharff M. D. J. Exp. Med. 2000;191:579–584. doi: 10.1084/jem.191.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phung Q. H., Winter D. B., Alrefai R., Gearhart P. J. J. Immunol. 1999;162:3121–3124. [PubMed] [Google Scholar]
- 19.Frey S., Bertocci B., Delbos F., Quint L., Weill J. C., Reynaud C. A. Immunity. 1998;9:127–134. doi: 10.1016/s1074-7613(00)80594-4. [DOI] [PubMed] [Google Scholar]
- 20.Neuberger M. S., Di Noia J. M., Beale R. C., Williams G. T., Yang Z., Rada C. Nat. Rev. Immunol. 2005;5:171–178. doi: 10.1038/nri1553. [DOI] [PubMed] [Google Scholar]
- 21.Seki M., Gearhart P. J., Wood R. D. EMBO Rep. 2005;6:1143–1148. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poltoratsky V., Goodman M. F., Scharff M. D. J. Exp. Med. 2000;192:F27–F30. doi: 10.1084/jem.192.10.f27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrader C. E., Vardo J., Stavnezer J. J. Exp. Med. 2002;195:367–373. doi: 10.1084/jem.20011877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrader C. E., Edelmann W., Kucherlapati R., Stavnezer J. J. Exp. Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schrader C. E., Linehan E. K., Mochegova S. N., Woodland R. T., Stavnezer J. J. Exp. Med. 2005;202:561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramiro A. R., Jankovic M., Eisenreich T., Difilippantonio S., Chen-Kiang S., Muramatsu M., Honjo T., Nussenzweig A., Nussenzweig M. C. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Li Z., Scherer S. J., Ronai D., Iglesias-Ussel M. D., Peled J. U., Bardwell P. D., Zhuang M., Lee K., Martin A., Edelmann W., Scharff M. D. J. Exp. Med. 2004;200:47–59. doi: 10.1084/jem.20040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barreto V., Reina-San-Martin B., Ramiro A. R., McBride K. M., Nussenzweig M. C. Mol. Cell. 2003;12:501–508. doi: 10.1016/s1097-2765(03)00309-5. [DOI] [PubMed] [Google Scholar]
- 29.Ito S., Nagaoka H., Shinkura R., Begum N., Muramatsu M., Nakata M., Honjo T. Proc. Natl. Acad. Sci. USA. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBride K. M., Barreto V., Ramiro A. R., Stavropoulos P., Nussenzweig M. C. J. Exp. Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barreto V. M., Ramiro A. R., Nussenzweig M. C. Trends Immunol. 2005;26:90–96. doi: 10.1016/j.it.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Basu U., Chaudhuri J., Alpert C., Dutt S., Ranganath S., Li G., Schrum J. P., Manis J. P., Alt F. W. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 33.Pasqualucci L., Kitaura Y., Gu H., Dalla-Favera R. Proc. Natl. Acad. Sci. USA. 2006;103:395–400. doi: 10.1073/pnas.0509969103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang E. J., Archambault V., McLachlin D. T., Krutchinsky A. N., Chait B. T. Anal. Chem. 2004;76:4472–4483. doi: 10.1021/ac049637h. [DOI] [PubMed] [Google Scholar]
- 35.Rada C., Jarvis J. M., Milstein C. Proc. Natl. Acad. Sci. USA. 2002;99:7003–7008. doi: 10.1073/pnas.092160999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doi T., Kinoshita K., Ikegawa M., Muramatsu M., Honjo T. Proc. Natl. Acad. Sci. USA. 2003;100:2634–2638. doi: 10.1073/pnas.0437710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie S., Boyd F. M., Wong J., Bonham K. J. Biol. Chem. 2000;275:847–854. doi: 10.1074/jbc.275.2.847. [DOI] [PubMed] [Google Scholar]
- 38.Dignam J. D., Lebovitz R. M., Roeder R. G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wysocka J., Reilly P. T., Herr W. Mol. Cell. Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franca-Koh J., Yeo M., Fraser E., Young N., Dale T. C. J. Biol. Chem. 2002;277:43844–43848. doi: 10.1074/jbc.M207265200. [DOI] [PubMed] [Google Scholar]
- 41.Offterdinger M., Schofer C., Weipoltshammer K., Grunt T. W. J. Cell Biol. 2002;157:929–939. doi: 10.1083/jcb.200109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaudhuri J., Khuong C., Alt F. W. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 43.Yoshikawa K., Okazaki I. M., Eto T., Kinoshita K., Muramatsu M., Nagaoka H., Honjo T. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 44.Petersen S., Casellas R., Reina-San-Martin B., Chen H. T., Difilippantonio M. J., Wilson P. C., Hanitsch L., Celeste A., Muramatsu M., Pilch D. R., et al. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R. F., Nussenzweig A., Nussenzweig M. C. J. Exp. Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagaoka H., Muramatsu M., Yamamura N., Kinoshita K., Honjo T. J. Exp. Med. 2002;195:529–534. doi: 10.1084/jem.20012144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramiro A. R., Jankovic M., Callen E., Difilippantonio S., Chen H. T., McBride K. M., Eisenreich T. R., Chen J., Dickins R. A., Lowe S. W., et al. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gordon M. S., Kanegai C. M., Doerr J. R., Wall R. Proc. Natl. Acad. Sci. USA. 2003;100:4126–4131. doi: 10.1073/pnas.0735266100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muschen M., Re D., Jungnickel B., Diehl V., Rajewsky K., Kuppers R. J. Exp. Med. 2000;192:1833–1840. doi: 10.1084/jem.192.12.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pasqualucci L., Migliazza A., Fracchiolla N., William C., Neri A., Baldini L., Chaganti R. S., Klein U., Kuppers R., Rajewsky K., Dalla-Favera R. Proc. Natl. Acad. Sci. USA. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen H. M., Peters A., Baron B., Zhu X., Storb U. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 52.Sayegh C. E., Quong M. W., Agata Y., Murre C. Nat. Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 53.Gonda H., Sugai M., Nambu Y., Katakai T., Agata Y., Mori K. J., Yokota Y., Shimizu A. J. Exp. Med. 2003;198:1427–1437. doi: 10.1084/jem.20030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okazaki I. M., Hiai H., Kakazu N., Yamada S., Muramatsu M., Kinoshita K., Honjo T. J. Exp. Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor S. S., Kim C., Vigil D., Haste N. M., Yang J., Wu J., Anand G. S. Biochim. Biophys. Acta. 2005;1754:25–37. doi: 10.1016/j.bbapap.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 56.Baillie G. S., Scott J. D., Houslay M. D. FEBS Lett. 2005;579:3264–3270. doi: 10.1016/j.febslet.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 57.Wong W., Scott J. D. Nat. Rev. Mol. Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 58.Tasken K., Aandahl E. M. Physiol. Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- 59.Goueli S. A., Jarvis B. W. Methods Enzymol. 2001;332:337–343. doi: 10.1016/s0076-6879(01)32213-9. [DOI] [PubMed] [Google Scholar]
- 60.Lu M. J., Dadd C. A., Mizzen C. A., Perry C. A., McLachlan D. R., Annunziato A. T., Allis C. D. Chromosoma. 1994;103:111–121. doi: 10.1007/BF00352320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.