Abstract
Agrobacterium radiobacter K84, used worldwide to biocontrol crown gall disease caused by Agrobacterium tumefaciens, produces an antiagrobacterial compound called agrocin 84. We report the nucleotide sequence of pAgK84, a 44.42-kb plasmid coding for production of this disubstituted adenine nucleotide antibiotic. pAgK84 encodes 36 ORFs, 17 of which (agn) code for synthesis of or immunity to agrocin 84. Two genes, agnB2 and agnA, encode aminoacyl tRNA synthetase homologues. We have shown that the toxic moiety of agrocin 84 inhibits cellular leucyl-tRNA synthetases and AgnB2, which confers immunity to the antibiotic, is a resistant form of this enzyme. AgnA, a truncated homologue of asparaginyl tRNA synthetase could catalyze the phosphoramidate bond between a precursor of the methyl pentanamide side group and the nucleotide. We propose previously undescribed chemistry, catalyzed by AgnB1, to generate the precursor necessary for this phosphoramidate linkage. AgnC7 is related to ribonucleotide reductases and could generate the 3′-deoxyarabinose moiety of the nucleoside. Bioinformatics suggest that agnC3, agnC4, and agnC6 contribute to maturation of the methyl pentanamide, whereas agnC2 may produce the glucofuranose side group bound to the adenine ring. AgnG is related to bacterial exporters. An agnG mutant accumulated agrocin 84 intracellularly but did not export the antibiotic. pAgK84 is transmissible and encodes genes for conjugative DNA processing but lacks a type IV secretion system, suggesting that pAgK84 transfers by mobilization. By sequence analysis, the deletion engineered into pAgK1026 removed the oriT and essential tra genes, confirming the enhanced environmental safety of this modified form of pAgK84.
Keywords: Agrobacterium radiobacter, biological control, pAgK84
Soil microbes produce exotic and potentially useful antimicrobial compounds. Among such agents is agrocin 84, a unique disubstituted analogue of adenosine (Fig. 1) produced by Agrobacterium radiobacter K84, a nonpathogenic member of the genus isolated in Australia. The nucleoside core, which is essential for toxicity, contains 3′-deoxyarabinose rather than ribose (1, 2). The methyl-substituted pentanamide at the C-5 position is also required for activity (3), whereas the glucofuranose at the N6 position does not confer toxicity but is required for transport by susceptible bacteria (3). Both side groups are linked to the nucleoside by phosphoramidate bonds.
Fig. 1.
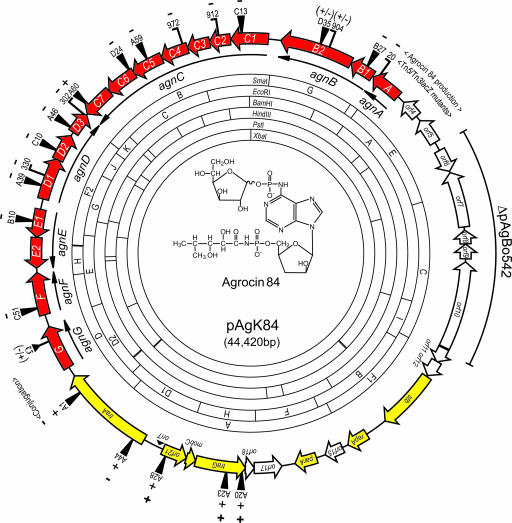
Gene structure and physical map of pAgK84. Restriction endonuclease sites were derived from the nucleotide sequence. Locations and organizations of identified ORFs are shown as arrows color-coded as genes involved in: synthesis, export, or immunity to agrocin 84 (red); replication and conjugative transfer (yellow); and unknown function (open). Sites of Tn5 (13) and Tn3HoHo1 (15) insertions are shown. Horizontal bars on Tn3HoHo1 symbols indicate orientation of the lacZ reporter with respect to the target gene. Effect of each insertion in agn or tra genes on production of agrocin 84 or conjugative transfer is indicated by (−), no detectable antibiotic or transfer; (+/−), detectable but reduced production; and (+), wild-type production or transfer. The region of the plasmid absent from pAgBo542 is indicated by the outer arc. The structure of agrocin 84 (2, 3) is shown in the middle of the map.
Agrocin 84 is inhibitory only against certain strains of the phytopathogenic agrobacteria, Agrobacterium tumefaciens and Agrobacterium rhizogenes. These two species induce neoplastic growths on host plants called crown galls and hairy roots, respectively. The neoplastic transformations result from the transfer from the pathogen to a plant cell of a small segment of DNA, called the T-DNA, that normally is part of large virulence elements called Ti or Ri plasmids (reviewed in ref. 4). The neoplasias result from expression of one set of T-DNA genes integrated into the nuclear DNA of the plant cell. The tumors characteristically produce novel carbon compounds, called opines, that are synthesized by enzymes coded for by additional genes on the integrated T-DNA. In turn, the inducing bacteria catabolize the released opines using metabolic systems also coded for by the resident Ti or Ri plasmids. Strains susceptible to agrocin 84 harbor Ti or Ri plasmids that code for biosynthesis by the plant and catabolism by the bacteria of one such class of opines, a set of sugar phosphodiesters called agrocinopines (5–7).
Agrocin 84, a mimic of the agrocinopines, is taken up by the pathogens via the opine transporter coded for by the Ti plasmid (5–8). Moreover, the agrocinopines produced by the tumors induce the expression of the Ti plasmid genes coding for their uptake and catabolism, thereby insuring that the tumorigenic strains are maximally susceptible to agrocin 84 produced by strain K84 (6, 7, 9). Clearly, evolution has tailored this antibiotic to a metabolic system in pathogenic isolates guaranteed to be expressed at high levels in the habitat provided by crown gall tumors.
The biology of the system has been harnessed in production agriculture; K84 is an extremely effective biocontrol agent and efficiently protects plants from infection by susceptible strains of Agrobacterium tumefaciens in the field (10). Production of agrocin 84 is important for biocontrol. Mutants of K84 that do not produce the antibiotic fail to control disease caused by susceptible pathogens. The bacterium and its improved derivative, K1026, are used in many parts of the world.
Biosynthesis of and immunity to agrocin 84 is coded for by an ≈45-kb plasmid pAgK84 (11, 12). pAgK84 can transfer by conjugation to other agrobacteria as well as to Rhizobium and Sinorhizobium and confers production of the antibiotic on these hosts (13). pAgBo542, a close relative of pAgK84 was identified in Bo542, a wild strain of Agrobacterium tumefaciens isolated in Germany (12). Its presence in isolates from different parts of the world suggests that pAgK84 is genetically mobile in nature and may be widely disseminated.
Despite its novel structure, virtually nothing is known about the pathway for the production of agrocin 84. Genes for synthesis of and immunity to the antibiotic map to five complementation groups covering a contiguous 15-kb region of the plasmid (13–15). An additional 8 kb is required for replication and transfer (13, 16). Here, we present the entire nucleotide sequence of pAgK84. Analysis of this sequence provides clues concerning the biosynthesis of agrocin 84 as well immunity to the antibiotic. The sequence also presents an interesting dilemma; although reported to be self-conjugative, pAgK84 does not code for a type IV secretion system.
Results and Discussion
Features of pAgK84.
The plasmid is a 44,420-bp covalently closed circle, 90% of which is predicted to encode proteins (Fig. 1) (GenBank accession no. AY442931). Annotation yielded 36 possible ORFs (see Table 2, which is published as supporting information on the PNAS web site). The G/C content of the plasmid is 53%, which is close to that of the Agrobacterium genome. The plasmid is divisible into three sections; regions coding for replication and transfer functions, for genes of unknown function, and for synthesis and export of, and immunity to, agrocin 84. We also located the precise sites of 22 transposon insertions previously mapped to the transfer and biosynthesis regions of the plasmid (13, 15) (Fig. 1; and see Table 3, which is published as supporting information on the PNAS web site).
Replication and Conjugative Transfer.
The 5.3-kb rep-tra region contains three putative replication genes stb, repA, and parA (Fig. 1). repA and parA are close homologues of replication genes of the Agrobacterium tartrate plasmid pTAR (17) (Table 2; and see Fig. 5, which is published as supporting information on the PNAS web site). The stb product contains a ParB domain and is related to a putative stabilization protein from Rhodopseudomonas palustris. The location of these genes is consistent with deletion analyses that yielded a minimum stable replicon containing this segment of the plasmid (16). Two small ORFs, orf11 and orf12, are located immediately upstream of stb, and could be part of the same transcriptional unit. The 42-bp intergenic region between orf12 and stb contains neither a good rho-independent terminator nor identifiable promoter sequences. However, transposon insertions mapping within the orf11-stb region have no effect on plasmid stability (16), suggesting that, if this gene set is expressed, neither stb nor orfs 11 and 12 are essential for replication or efficient partitioning (see below).
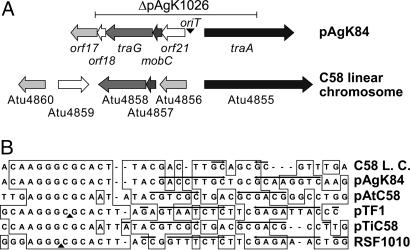
The 6.7-kb region required for conjugative transfer encodes four ORFs organized in two divergent gene sets (Fig. 1). traA, mobC, and traG are related to components of other conjugative transfer systems and most closely to a set of genes, organized in a similar fashion on the linear chromosome of Agrobacterium tumefaciens C58 (Figs. 2A and 5). The fourth gene, orf21, is not related to any known conjugative transfer gene and is not represented in the gene cluster on the linear chromosome of C58 (Table 2). A probable oriT, which contains a core nucleotide sequence closely related to the IncQ-type nic site of several transmissible plasmids (Fig. 2B), is located between the two divergently oriented gene sets.
Fig. 2.
The conjugative transfer determinants of pAgK84 and those on the linear chromosome of C58 are closely related. (A) The organization of the genes in the transfer regions of the two replicons are shown with identical fill patterns indicating significant amino acid sequence relatedness. Open arrows represent genes showing no significant relatedness. The region of pAgK84 deleted in the construction of pAgK1026 (24) is shown as a horizontal line above the maps. (B) Nucleotide sequences of oriT regions of RSF1010, pTF1, pTiC58, pAtC58, the linear chromosome (L.C.) of C58, and pAgK84 were aligned by using clustalx (38). Regions of sequence identity shared in the six segments are boxed. Arrows indicate inverted repeats. Filled triangles show experimentally determined nick sites.
Two Tn5 insertions, A1 and A44 (Fig. 1), are located in traA, which most probably codes for the oriT nicking enzyme (Tables 2 and 3). Mating experiments show that this gene is essential for conjugative transfer (Table 1) (13). The region also codes for an orthologue of traG, the normally essential coupling protein (Fig. 1 and Table 3). However, A20 and A23, two insertions located within traG (Fig. 1), do not affect conjugative transfer, even from UIA5, a donor that lacks any other plasmids (Table 1). TraG of pAgK84 is 80% identical to its orthologue (Atu4853) on the linear chromosome of C58. We suggest that this chromosomal gene, present in UIA5, complements the traG defect of the two transposon mutants. Interestingly, the traA homologue coded for by the linear chromosome of C58 apparently does not complement the traA mutation of pAgK84; the A1 plasmid does not transfer from either of the two donors tested (Table 1). With the exception of traG, pAgK84 does not code for any of the structural components of a recognizable Type IV secretion system (T4SS). Yet the plasmid transfers with wild-type frequency from strain UIA5 (Table 1), which lacks genes coding for a T4SS.
Table 1.
Conjugative transfer of pAgK84 and its tra-region mutants
| Donor strain* | tra mutation | Conjugation frequency† |
|
|---|---|---|---|
| Per donor | Per recipient | ||
| NT1-A46 | None | 3.40 × 10−5 | 3.81 × 10−5 |
| UIA5-A46 | None | 7.52 × 10−6 | 6.54 × 10−6 |
| NT1-A1 | traA | <10−8 | <10−8 |
| UIA5-A1 | traA | <10−8 | <10−8 |
| NT1-A20 | traG | 3.12 × 10−6 | 3.10 × 10−6 |
| UIA5-A20 | traG | 2.09 × 10−6 | 1.78 × 10−6 |
| NT1-A23 | traG | 1.36 × 10−6 | 1.34 × 10−6 |
| UIA5-A23 | traG | 6.45 × 10−7 | 4.91 × 10−7 |
*NT1 harbors pAtC58 but not pTiC58, whereas UIA5 lacks both plasmids.
†Expressed as transconjugants recovered per input donor or recipient. Frequencies represent the averages of three independent experiments.
An Unstable Region of Unknown Function.
An ≈8.7-kb region of pAgK84 codes for seven possible ORFs (Fig. 1 and Table 2), none of which are required for production of agrocin 84, for replication, or for conjugative transfer (13). Moreover, pAgBo542, an otherwise indistinguishable agrocinogenic plasmid from Agrobacterium tumefaciens strain Bo542, lacks a segment corresponding to this region (12). We cloned and sequenced a 7-kb BamHI fragment of pAgBo542 corresponding to the junctions of this deleted region (GenBank accession no. DQ460476). The 7,022-bp region of pAgK84 missing in pAgBo542 extends from orf6 to the 5′ half of orf11 (Fig. 1; and see Fig. 6, which is published as supporting information on the PNAS web site). orf5 and orf10, located adjacent to or within this region, could encode resolvases and transposases, suggesting a transposon origin for this segment of pAgK84 (Table 2). The 5′ end of the transposase ORF begins 30 bp upstream of the divergently oriented orf11 in the replication region of pAgK84. Because the orf11 product represents only the C-terminal segment of its closest homologues, we suggest that the transposon inserted into orf11 of a progenitor of pAgK84, thereby disrupting the protein product. Moreover, because orf11-orf12-stb may form a transcriptional unit, it is likely that the transposon insertion abolished expression of this gene set. This hypothesis is consistent with the fact that Tn5 insertions in the orf11-orf12-stb region have no effect on plasmid stability (16). We suspect that pAgBo542 is not ancestral but arose from an event in which this 7-kb region had been excised.
The Region Encoding Production of Agrocin 84.
Previous genetic analysis identified a large contiguous region of pAgK84 associated with production of and immunity to agrocin 84 (13–15). The locus contains 17 ORFs organized into seven transcriptional units, agnA, agnB1,2, agnC1–7, agnD1–3, agnE1,2, agnF, and agnG, within this 19.8-kb locus (Fig. 1 and Table 2). Of the 17 genes, only agnF codes for a product with no significant homologues in the databases (Table 2). However, agnF is essential for agrocin 84 biosynthesis; insertion C51, located within this monocistronic ORF, abolishes antibiotic production (Fig. 1 and Table 3).
Immunity to agrocin 84 is conferred independently by two loci, ImmA and ImmB (14). ImmA corresponds to agnB2, the gene product of which is strongly orthologous to bacterial leucyl-tRNA synthetases (leuRS) (Table 2). The aminoacyl tRNA synthetases (aaRSs) are essential enzymes that catalyze the ATP-dependent adenylylation and subsequent transfer of amino acids to their cognate tRNA. We recently reported that agrocin 84 strongly and specifically inhibits the activity of leuRS from Agrobacterium tumefaciens and that agnB2 codes for an active agrocin 84-resistant form of this enzyme (18).
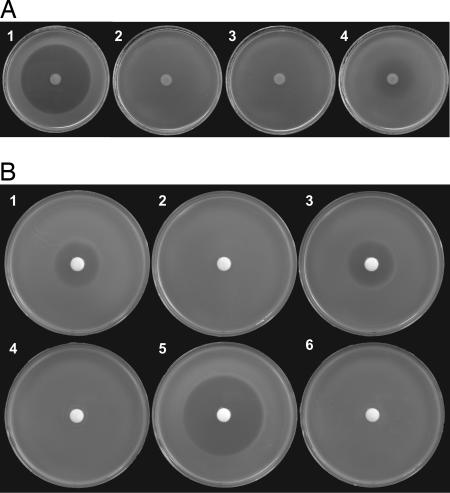
ImmB corresponds to agnG, which codes for a protein related to bacterial export pumps (Table 2). We tested the hypothesis that AgnG exports agrocin 84 in two ways. First, we examined the effect of expression levels of a clone of agnG on susceptibility to agrocin 84. When expressed from the low-copy-number clone pAgnG, the gene conferred partial resistance on the otherwise-susceptible strain C58 (Fig. 3A). However, expression of the gene from the high-copy-number clone pAgSmaD conferred complete resistance to the antibiotic (Fig. 3A). Resistance conferred by agnG, then, depends on level of expression. We then tested whether AgnG is required for export of agrocin 84 by measuring the relative amounts of antibiotic present in culture supernatants and in cell lysates of a strain harboring wild-type pAgK84 or the agnG mutant pAgK84-A1agnG::Sp. Although culture supernatants of a strain containing the agnG+ plasmid accumulated high levels of agrocin 84, those from the agnG mutant contained little or no detectable activity (Fig. 3B and Table 3). However, cell lysates of the strain harboring the agnG mutant contained high levels of agrocin 84 activity, whereas similar lysates from the strain harboring the wild-type plasmid failed to inhibit the indicator strain (Fig. 3B). Expression in trans of wild-type agnG in the agnG mutant restored accumulation of agrocin 84 in the culture supernatant concomitant with the disappearance of this activity in cell lysates (Fig. 3B). We conclude that AgnG is not involved in synthesis of or immunity to active agrocin 84 but functions in the export of the antibiotic.
Fig. 3.
agnG is required for agrocin 84 export. (A) Agrobacterium strains harboring pTiC58 were tested for influence of clones expressing agnG on susceptibility to agrocin 84 produced by strain NT1(pAgK84-A1) spotted in the center of the plate. Overlays of tested strains contain C58 (1), C58(pAgSmaG) (2), C58(pAgSmaD) (3), and C58(pAgAgnG) (4). (B) Agrobacterium strains harboring pAgK84-A1 (1 and 4) and the agnG mutant pAgK84-A1agnG::Sp without (2 and 5) or complemented with (3 and 6) pAgnG were grown in liquid medium and tested for accumulation of agrocin 84 in culture supernatants (1, 2, 3) and in cell-free lysates (4, 5, 6) with the supersensitive strain NT1(pTiC58ΔaccR) as the indicator, all as described in Materials and Methods.
The agn locus encodes a second aaRS homologue, agnA. This gene, which transposon mutagenesis indicates is essential for synthesis of agrocin 84 (Fig. 1), codes for a protein that is related to asparaginyl-tRNA synthetase (Table 2). AgnA lacks the tRNA anticodon-binding domain but retains the conserved catalytic domain responsible for the adenylylation of substrate amino acids (19). We suggest that this enzyme catalyzes a similar reaction, in which an amino acid-like precursor of the methyl pentanamide is added to the phosphate at the C-5 of the pentose moiety of the nucleotide (Fig. 1; and see below).
Two genes, agnB1 and agnC3, code for homologues of S-adenosyl-methionine-dependent methyl transferases (Table 2). AgnB1 is required for production of agrocin 84 (Fig. 1), but, because of polarity, we cannot conclude from previous genetic analysis whether agnC3 is an essential gene. Methylation reactions likely are required for synthesis of the antibiotic. The pentanamide at the C-5 position of the adenine-like nucleotide contains two methyl groups. Moreover, a methyl addition may be involved in synthesis of the pentanamide precursor preparatory to its condensation with the nucleotide (see below).
Other members of the agnC operon code for proteins showing significant relatedness to proteins or protein domains of known function. AgnC4, which contains an amidohydrolase motif, and AgnC6, which is weakly related to hydroxylases (Table 2), may be involved in maturation of the methyl pentanamide side group (see below). AgnC7, a weak homologue of iron–sulfur cluster dehydrogenases (Table 2) is related to members of the radical S-adenosyl-methionine family of oxidoreductases. As described below, we suggest that this gene product is involved in generating the 3′-deoxyarabinose moiety. AgnC2 contains extensive domain matches to many oxidoreductases, including activated-sugar epimerases, and may contribute a precursor of the N6-linked glucofuranosyl side group (see below).
Intriguingly, agnC5 is most closely related to mccF, a gene located in the microcin C7 operon of Escherichia coli (Table 2). Agrocin 84 and microcin C7 share common structural features; both contain an adenosine-like core with an N-acyl phosphoramidate-bound side group at the C-5 position of the nucleotide sugar (20). AgnC5 and MccF are members of a large family of bacterial proteins, all of which contain a conserved carboxypeptidase motif. The microcin C7 paralogue confers very weak immunity to the bacteriocin (20). However, the locus for synthesis of the closely related microcin, C51, lacks a homologue of mccF, suggesting that the product of this gene is not required for microcin synthesis or immunity (21).
AgnC1 and the five genes comprising the agnD1–3 and agnE1,2 operons code for proteins related to or containing motifs found in enzymes known or predicted to catalyze phosphorylation, dephosphorylation, and redox reactions using small substrates such as pyruvate and sugars (Table 2). Although we cannot predict functions for any of these products in the synthesis of agrocin 84, given their in silico-predicted activities, it is likely that synthesis of the antibiotic involves small phosphorylated intermediates.
The plasmid lacks genes with obvious relatedness to known transcriptional control factors. The absence of such regulatory proteins is consistent with our previous studies showing that expression of the agn genes is constitutive (14, 15).
Conclusions
The complete sequence of pAgK84 illuminates the genetics of a very successful biocontrol system, validates the development by site-directed deletion mutagenesis of an environmentally safer version of the plasmid, and provides clues concerning the pathway for synthesis of the novel nucleotide antibiotic which is, in large part, responsible for the biocontrol properties of strain K84.
The plasmid clearly is transmissible by conjugation (13). Moreover, such transfer has occurred under field conditions (22, 23), and otherwise-sensitive pathogens inheriting pAgK84 retain tumorigenicity but become resistant to agrocin 84 (13, 23). The likelihood of such transfer compromising the use of K84 as a biocontrol agent prompted the construction of pAgK1026, in which the genetically defined conjugative transfer region of pAgK84 was deleted (24). Our sequence analysis shows that the deletion removed all or large segments of traG and traA (Fig. 2) as well as the cis-acting oriT site. The two defects combined ensure that pAgK1026 cannot spread to recipients in the field by trans-mobilization.
Remarkably, the conjugative transfer genes and oriT of pAgK84 are closely related to a putative transfer system located on the linear chromosome of Agrobacterium tumefaciens C58 (Figs. 2 and 5). That the traG orthologue of the linear chromosome apparently complements traG mutants of pAgK84 (Table 1) lends significance to this relatedness. However, pAgK84 lacks genes coding for a type IV secretion system (T4SS), suggesting that the plasmid is not self-conjugative. pAgK84’s natural host, Agrobacterium radiobacter K84, codes for at least three T4SSs, one on the large circular chromosome (data not shown), one on the conjugative plasmid pAtK84b (25), and the third on the megaplasmid pAtK84c (data not shown). Any one of these systems could provide the mating bridge required for transfer of pAgK84 observed in the field.
Knowing the sequence of the ImmA gene agnB2 has led to an understanding of the mode of action of agrocin 84. Previous studies suggested that the antibiotic targets DNA synthesis (3, 26). However, that agnB2 codes for a functional leuRS led to the discovery that the toxic moiety of agrocin 84 specifically inhibits the aminoacylation reaction catalyzed by the agrobacterial orthologue of this tRNA synthetase (18). Moreover, the corresponding activity of the agnB2 product is not significantly inhibited by the antibiotic, which explains how this plasmid-encoded leuRS confers immunity to agrocin 84 (18). These observations are consistent with the structural similarity between the nucleotide with its 5′-pentanamide side group and leucyl-AMP, the product of the aminoacylation reaction catalyzed by leuRS (18). There can be little doubt that the active form of agrocin 84 targets protein synthesis and that the leuRS encoded by agnB2 confers immunity to this inhibitory activity.
Based on sequence, we also defined a function for ImmB; as predicted by our bioinformatics analysis, experimental studies clearly show that agnG, which maps to this locus, is required for the export of agrocin 84 (Fig. 3). Although not a true immunity function, these results explain why the gene, when expressed alone, confers resistance to agrocin 84 on a susceptible strain.
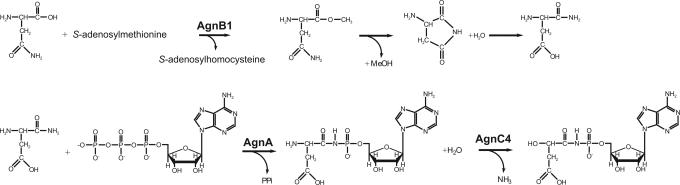
Sequence analysis of other agn genes also predicts biosynthetic function. That AgnA contains the catalytic domain of an aminoacyl tRNA synthetase strongly suggests that this protein is involved in the addition of the methyl pentanamide, or its precursor, to the C-5 of the nucleotide analogue. aaRSs normally catalyze phosphoanhydride bond formation between amino acids and ATP as the first step in the aminoacylation of tRNA. We propose a two-step mechanism by which AgnA could form the N-acyl phosphoramidate linkage. In the first, the hydroxyl group of the α-carbon carboxylate of an asparagine is methylated by AgnB1, a putative S-adenosyl-methionine-dependent O-methyl transferase (Fig. 4). The O-methyl addition, being a good leaving group, could drive an internal rearrangement resulting in migration of the side-chain amine group to the carbonyl carbon, thus displacing methanol and yielding an amidated aspartic acid (Fig. 4). Second, in a reaction that is analogous to adenylylation catalyzed by normal aaRSs, AgnA could catalyze an attack by the amine, a good nucleophile, on the α-phosphorous of the nucleotide, forming the N-acyl phosphoramidate bond (Fig. 4). No matter the mechanism, it is remarkable that a molecule that acts as an aaRS inhibitor is biosynthesized in part by an aaRS-like enzyme. Conscription of the adenylylation reaction catalyzed by these enzymes is not without precedent. Truncated paralogues of aaRS enzymes are involved in diverse noncanonical functions, including tRNA modifications (27), and biosynthesis of amino acids and other metabolites (28).
Fig. 4.
Proposed roles of AgnB1, AgnA, and AgnC4 in the formation of the phosphoramidate bond with the nucleotide analogue and subsequent deamination of the methyl pentanamide precursor. See text for details.
The condensation described above leaves an amine at the α-carbon of the amino acid substituent. AgnC4 contains a metal-dependent domain common to amidohydrolases, which catalyze reactions involving amino groups (29). We propose that AgnC4 removes this amine, yielding the hydroxyl substitution at the 2-carbon of the mature methyl pentanamide (Fig. 4).
AgnC6, which contains a hydroxylase domain (Table 2), also could be involved in modifying the amino acid precursor of the pentanamide by adding the hydroxyl group at the 3-carbon of this moiety.
The origin of the glucofuranose substitution at the N6 position is of some interest. The broad distribution among prokaryotes of aldohexofuranosyl moieties in complex carbohydrates has only recently become appreciated, and there is some interest in how these furanosyl forms are produced and incorporated into other compounds (30). Galactofuranose can be derived enzymatically from galactopyranose by UDP-galactopyranose mutase (epimerase) (31). AgnC2 contains a domain that exhibits significant relatedness to WcaG, a nucleoside-diphosphate sugar epimerase. We suggest that AgnC2 catalyzes the interconversion of an NDP-glucopyranose with NDP-glucofuranose, the putative side-chain precursor. Alternatively, because Agrobacterium encodes a UDP-galactose epimerase, it is conceivable that AgnC2 epimerizes UDP-galactofuranose to its glucofuranosyl form.
The origin of the 3′-deoxyarabinose-containing adenosine analogue, a nucleoside unique to agrocin 84, is problematic. However, AgnC7 is related to the radical S-adenosyl-methionine family, which includes the Class III ribonucleotide reductases. These enzymes convert ribonucleotides to 2′-deoxyribonucleotides (32), and it is conceivable that AgnC7 catalyzes such a reaction at the 3′ position of the adenosine pentose, generating the 3′-deoxy nucleoside analogue. However, insertion A60, which does not affect agrocin 84 activity, is located in agnC7 (Fig. 1). Because the insertion is at the far 3′ end of the gene and is distal to the active site, it is possible that the mutant protein retains activity. It also is possible that the modification at C-3 of the pentose is not essential for activity or that a redundant function in the bacterium substitutes for the mutant gene.
The predicted roles of agnA, agnB1, agnC2, agnC3, agnC4, agnC6, and agnC7 remain to be verified, but each is testable. We cannot speculate on the pathways that generate the deoxyarabinose nucleoside and the mature methyl pentanamide or the mechanism by which the glucofuranose is added to the adenine ring. It is clear, however, that the synthesis of agrocin 84 is genetically complex and represents novel chemistries and uniquely evolved enzymes. Knowing the sequence of the genes as well as their genetic and physical organization greatly facilitates approaches to understanding the pathways and their enzymes involved in synthesis of this unique antibiotic.
Materials and Methods
Bacterial Strains and Plasmids.
All Agrobacterium tumefaciens strains are C58 derivatives. NT1(pTiC58Δ accR) harbors a Ti plasmid that confers supersensitivity to agrocin 84 (9). Strain UIA5 lacks pTiC58 and pAtC58 and is resistant to rifampicin and spectinomycin (13). Media and culture conditions were described in ref. 13. pAgK84 was isolated from Agrobacterium radiobacter strain K84 as described in ref. 12. Broad host-range vectors pSa152 and pLAFR3 have been described (33, 34), as have the construction and properties of pAgSmaD, which encodes agnG, and pAgSmaG, which encodes agnB2 (14). pAgnG was constructed by cloning a 2.8-kb EcoRI–BamHI fragment that contains agnG driven by its own promoter into the low-copy-number vector pLAFR3.
Subcloning and Sequencing.
DNA fragments from partial digests of pAgK84 with BamHI or SmaI in pBR325 and pSa152 (14) were recloned into pBluescript SK(+) and sequenced by using BigDye terminator kits (Applied Biosystems) on ABI377-XL sequencers (Applied Biosystems) with universal and reverse primers. Synthetic primers were made for primer-walking when necessary. Sites of previously mapped Tn5 and Tn3HoHo1 insertions in pAgK84 (13–15) were determined by sequencing junction fragments by using appropriate primers homologous to transposon sequences.
Sequence Analysis and Annotation.
DNA sequences were assembled and ORFs identified by using the seqmanii subroutine of DNASTAR. All ORFs >249 bp were examined for possible ribosomal-binding sites and were annotated by using blastx and blastp protocols (35). Combining sequence annotations and genetic analyses (13–16), we assigned gene names to the predicted ORFs where possible.
Marker-Exchange Mutagenesis.
To generate the agnG::Ω mutant pAgK84-A1agnG::Sp, a 4.7-kb BamHI–HindIII fragment of pAgK84 that includes agnG, was cloned into pLAFR3 to give pAgnGF. pAgnGF was digested with PstI, which cuts within agnG, and blunt-ended with Klenow fragment (Takara Bio, Tokyo). The Ω fragment coding for resistance to streptomycin was recovered from pHP45Ω (36) by digestion with SmaI and inserted into the above blunt-ended site to give pAgnGSp. A 6.2-kb SmaI fragment including agnG::Ω was excised from pAgnGSp and inserted into the corresponding site in pBluescriptII SK(+), resulting in pBSagnGSp. pBSagnGSp was electroporated into NT1-A1 as described in ref. 15. Possible marker-exchanged mutants were isolated from nutrient agar (Difco) plates containing spectinomycin (100 μg/ml) and kanamycin (100 μg/ml). The marker-exchanged mutants were confirmed by restriction enzyme digestion patterns.
Agrocin 84 Bioassays.
Agrocin 84 was detected on Stonier’s agar plates by using C58 or NT1(pTiC58Δ accR) as indicator, as described in ref. 12. The role of AgnG in the accumulation of the antibiotic in culture supernatants and within cells was determined as follows: Strain NT1, harboring the wild-type or the agnG mutant of pAgK84 was grown for 24 h in AB minimal medium containing 0.2% glucose. The cells were removed by centrifugation, washed once with the same medium, and broken by sonication. One-hundred-microliter volumes of cell-free culture supernatant or cell lysate were loaded on an 8-mm-diameter paper disk that was placed onto a Stonier’s plate previously overlaid with NT1(pTiC58Δ accR).
Conjugation.
Conjugative transfer of pAgK84 from Agrobacterium donors to C58C1RS was assessed as described in ref. 37. Transfer was confirmed by analysis of plasmids in the transconjugants.
Supplementary Material
Acknowledgments
This work was supported by Crop Functional Genomics Center of the 21st Century Frontier R & D Program Grant CG1412, funded by the Ministry of Science and Technology of the Republic of Korea; by Korea Research Foundation Grant KRF-2004-005-F00013, funded by the Ministry of Education and Human Resource Development of the Korean Government (to I.H.); by National Institutes of Health Grant R01-GM52465 (to S.K.F.); and by National Science Foundation Grant EF-0333297 sub 763189 (to S.K.F.).
Abbreviations
- aaRS
amino acyl tRNA synthetase
- leuRS
leucyl tRNA synthetase.
Footnotes
References
- 1.Roberts W. P., Tate M. E., Kerr A. Nature. 1977;265:379–381. doi: 10.1038/265379a0. [DOI] [PubMed] [Google Scholar]
- 2.Tate M. E., Murphy P. J., Roberts W. P., Kerr A. Nature. 1979;280:697–699. doi: 10.1038/280697a0. [DOI] [PubMed] [Google Scholar]
- 3.Murphy P. J., Tate M. E., Kerr A. Eur. J. Biochem. 1981;11:539–543. doi: 10.1111/j.1432-1033.1981.tb06236.x. [DOI] [PubMed] [Google Scholar]
- 4.Gelvin S. B. Ann. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:223–256. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- 5.Ellis J. G., Murphy P. J. Mol. Gen. Genet. 1981;181:36–43. [Google Scholar]
- 6.Hayman G. T., Farrand S. K. J. Bacteriol. 1988;170:1759–1767. doi: 10.1128/jb.170.4.1759-1767.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H., Farrand S. K. J. Bacteriol. 1997;179:7559–7572. doi: 10.1128/jb.179.23.7559-7572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy P. J., Roberts W. P. J. Gen. Microbiol. 1979;114:207–213. [Google Scholar]
- 9.von Bodman S. B., Hayman G. T., Farrand S. K. Proc. Natl. Acad. Sci. USA. 1992;89:643–647. doi: 10.1073/pnas.89.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones D. A., Ryder M. H., Clare B. G., Farrand S. K., Kerr A. The Biological Control of Plant Disease. FFTC Book Series No. 42. Taipei, Taiwan: Food and Fertilizer Technology Center for the Asian and Pacific Region; 1991. pp. 161–170. [Google Scholar]
- 11.Ellis J. G., Kerr A., Van Montagu M., Schell J. Physiol. Plant Pathol. 1979;15:311–319. [Google Scholar]
- 12.Slota J. E., Farrand S. K. Plasmid. 1982;8:175–186. doi: 10.1016/0147-619x(82)90055-5. [DOI] [PubMed] [Google Scholar]
- 13.Farrand S. K., Slota J. E., Shim J.-S., Kerr A. Plasmid. 1985;13:106–117. doi: 10.1016/0147-619x(85)90063-0. [DOI] [PubMed] [Google Scholar]
- 14.Ryder M. H., Slota J. E., Scarim A., Farrand S. K. J. Bacteriol. 1987;169:4184–4189. doi: 10.1128/jb.169.9.4184-4189.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C.-L., Farrand S. K., Hwang I. Mol. Plant–Microbe Interact. 1994;7:472–481. [Google Scholar]
- 16.Farrand S. K., Wang C.-L., Hong S.-B., O’Morchoe S. B., Slota J. E. Plasmid. 1992;28:201–212. doi: 10.1016/0147-619x(92)90052-c. [DOI] [PubMed] [Google Scholar]
- 17.Leloup L., Lai E.-M., Kado C. I. Mol. Genet. Genomics. 2002;267:115–123. doi: 10.1007/s00438-002-0646-9. [DOI] [PubMed] [Google Scholar]
- 18.Reader J. S., Ordoukhanian P. T., Kim J.-G., de Crécy-Lagard V., Hwang I., Farrand S. K., Schimmel P. Science. 2005;309:1533. doi: 10.1126/science.1116841. [DOI] [PubMed] [Google Scholar]
- 19.Ibba M., Söll D. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 20.González-Pastor J. E., Millán J. L. S., Catilla M. Á., Moreno F. J. Bacteriol. 1995;177:7131–7140. doi: 10.1128/jb.177.24.7131-7140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fomenko D. E., Metlitskaya A. Z., Péduzzi J., Goulard C., Katrukha G. S., Gening L. V., Rebuffat S., Khmel I. A. Antimicrob. Agents Chemother. 2003;47:2868–2874. doi: 10.1128/AAC.47.9.2868-2874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panagopoulos C. G., Psallidas P. G., Alivizatos A. S. In: Soil-Borne Plant Pathogens. Schippers B., Gams W., editors. London: Academic; 1979. pp. 569–578. [Google Scholar]
- 23.Vicedo B., Penalver R., Asins M. J., Lopez M. M. Appl. Environ. Microbiol. 1993;59:309–315. doi: 10.1128/aem.59.1.309-315.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones D. A., Ryder M. H., Clare B. G., Farrand S. K., Kerr A. Mol. Gen. Genet. 1988;212:207–214. [Google Scholar]
- 25.Oger P., Farrand S. K. J. Bacteriol. 2002;184:1121–1131. doi: 10.1128/jb.184.4.1121-1131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das P. K., Basu M., Chatterjee G. C. J. Antibiotics. 1978;31:490–492. doi: 10.7164/antibiotics.31.490. [DOI] [PubMed] [Google Scholar]
- 27.Salazar J. C., Ambrogelly A., Crain P. F., McCloskey J. A., Söll D. Proc. Natl. Acad. Sci. USA. 2004;101:7536–7541. doi: 10.1073/pnas.0401982101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schimmel P., Ribas de Pouplana L. Trends Biochem. Sci. 2000;25:207–209. doi: 10.1016/s0968-0004(00)01553-x. [DOI] [PubMed] [Google Scholar]
- 29.Seibert C. M., Rauschel F. M. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 30.Soltero-Higgin M., Carlson E. E., Gruber T. D., Kiessling L. L. Nat. Struct. Mol. Biol. 2004;11:539–543. doi: 10.1038/nsmb772. [DOI] [PubMed] [Google Scholar]
- 31.Sanders D. A. R., Staines A. G., McMahon S. A., McNeil M. R., Whitfield C., Naismith J. H. Nat. Struct. Biol. 2001;8:858–863. doi: 10.1038/nsb1001-858. [DOI] [PubMed] [Google Scholar]
- 32.Kolberg M., Strand K. R., Graff P., Andersson K. K. Biochim. Biophys. Acta. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Tait R. C., Close T. J., Lundquist R. C., Hagiya M., Rodriguez R. L., Kado C. I. Bio/Technology. 1983;1:269–275. [Google Scholar]
- 34.Huynh T. V., Dahlbeck D., Staskawicz B. J. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 35.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Prentki P., Krisch H. M. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 37.von Bodman S. B., McCutchan J. E., Farrand S. K. J. Bacteriol. 1989;171:5281–5289. doi: 10.1128/jb.171.10.5281-5289.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgins D. G., Thompson J. D., Gibson T. J. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.