Abstract
High NaCl causes DNA double-strand breaks and activates the transcription factor, TonEBP/OREBP, resulting in increased transcription of several protective genes, including those involved in accumulation of compatible organic osmolytes. Several kinases are known to contribute to signaling activation of TonEBP/OREBP, including ATM, which is a member of the phosphatidylinositol 3-kinase (PI3K)-like kinase family and is activated by DNA double-strand breaks. The purpose of the present studies was to investigate a possible role of PI3K Class IA (PI3K-IA). We found that high NaCl increases PI3K-IA lipid kinase activity. Inhibiting PI3K-IA either by expressing a dominant negative of its regulatory subunit, p85, or by small interfering RNA-mediated knockdown of its catalytic subunit, p110α, reduces high NaCl-induced increases in TonEBP/OREBP transcriptional activity and transactivation, but not nuclear translocation of TonEBP/OREBP, or increases in its abundance. Further, suppression of PI3K-IA inhibits the activation of ATM that is caused by either ionizing radiation or high NaCl. High NaCl-induced increase in TonEBP/OREBP activity is reduced equally by inhibition of ATM or PI3K-IA, and the effects are not additive. The conclusions are as follows: (i) PI3K-IA activity is necessary for both high NaCl- and ionizing radiation-induced activation of ATM and (ii) high NaCl activates PI3K-IA, which, in turn, contributes to full activation of TonEBP/OREBP via ATM.
Keywords: DNA damage, osmotic stress, TonEBP/OREBP
TonEBP/OREBP is a transcription factor that is activated by high NaCl and other forms of hypertonicity, leading to the to protective accumulation of compatible organic osmolytes and heat shock proteins (1). Regulation of TonEBP/OREBP transcriptional activity is complex. Within 30 min of hypertonicity, TonEBP/OREBP becomes phosphorylated and translocates into the nucleus. Some hours later, TonEBP/OREBP mRNA and protein abundance increase (2), mediated by stabilization of TonEBP/OREBP mRNA (3). Also, hypertonicity increases transactivation activity of TonEBP/OREBP, associated with phosphorylation of the C-terminal region containing its transactivation domain (4). Several protein kinases are known to contribute to a hypertonicity-induced increase of TonEBP/OREBP transcriptional activity and transactivation, including p38 (5, 6), Fyn (5), Ataxia telangiectasia mutated (ATM) (7), and protein kinase A (PKAc) (8), none of them, alone, being sufficient for full activation.
ATM is activated by autophosphorylation on S1981 early in the response to the DNA double-strand breaks that are caused by ionizing radiation (IR) (9). High NaCl causes DNA double-strand breaks (10, 11) and, like IR, activates ATM by phosphorylation of S1981 (7). This activation of ATM is necessary for full high NaCl-induced TonEBP/OREBP transcriptional activity (7). Wortmannin, which inhibits ATM kinase activity, reduces high NaCl-induced TonEBP/OREBP transcriptional activity in HEK293 cells (7). However, wortmannin also inhibits high NaCl-induced transcriptional activity in AT cells, which lack functional ATM, implying that other wortmannin-inhibitable kinases besides ATM are involved in activating TonEBP/OREBP (7). Other wortmannin-inhibitable kinases include phosphatidylinositol 3-kinase (PI3K), DNA-PK, and myosin light-chain kinase (12–14). Taken together, these findings suggest that other wortmannin-inhibitable kinases in addition to ATM contribute to high NaCl-induced activation of TonEBP/OREBP. PI3K seemed a likely candidate because high NaCl activates it, as reflected by the increase in phosphorylation (15, 16) and activity (15) of its downstream target, AKT/PKB. Further, high NaCl increases phosphorylation GSK-3β in renal medullary interstitial cells (17), which is significant because GSK-3 is a downstream target of PI3K (18) and phosphatidyl inositol 3-phosphate (PIP3) in Swiss 3T3 cells (19).
PI3Ks can be divided into three main classes on the basis of their in vitro lipid substrate specificity, structure, and mode of regulation. Class I PI3Ks can phosphorylate PI, PI (4)P, and PI(4,5)P2. Class I PI3Ks contain a p110 catalytic subunit of which there are four isoforms, α, β, γ, and δ. The class IA α, β, and δ subunits are tightly complexed to the p85 regulatory/adaptor subunit. In contrast, the class IB p110γ isoform utilizes a distinct p101 molecule as a regulatory partner. We used two strategies to study the specific effects of class IA PI3K (PI3K-IA) on TonEBP/OREBP activity. (i) Dominant negative (Δp85) down-regulation of p85 in Jurkat cells stably transfected with tetracycline-regulated constructs (20). Δp85 is a dominant negative of the p85α regulatory subunit. Because its inter-SH2 domain (amino acids 479–513, which constitute the p110 binding domain) is deleted, Δp85 is unable to bind to p110 catalytic domains. Overexpression of Δp85 prevents the recruitment and activation of the catalytic subunit by outcompeting endogenous p85 for binding sites. (ii) p110α small interfering RNA (siRNA) knocks down the protein expression of the catalytic subunit α (p110α) of PI3K-IA.
We also used those strategies to examine a possible role of PI3K-IA in activation of ATM by high NaCl and IR. ATM (9) is held inactive in unirradiated cells as a dimer or higher-order multimer, with the kinase domain bound to a region surrounding S1981. IR induces rapid intermolecular autophosphorylation of S1981 that causes dimer dissociation and initiates cellular ATM kinase activity (9). An interesting aspect of the response to IR is that most ATM molecules in the cell can be rapidly phosphorylated on this site after the introduction of only a few DNA double-strand breaks. This widespread activation of ATM at sites distant from the actual DNA breaks has been ascribed to changes in the structure of chromatin (9), but it is conceivable that PI3K-IA-mediated increase in PIP3 could contribute.
Results
Effect of High NaCl on PI3K-IA Activity.
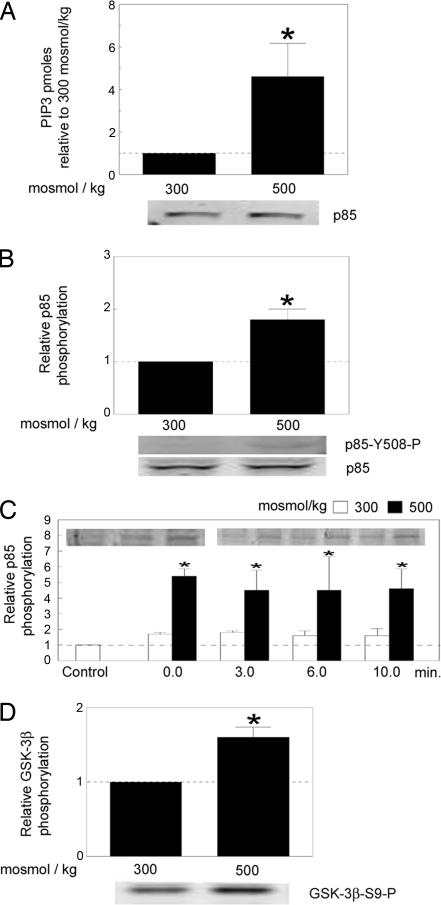
Increasing osmolality from 300 to 500 mosmol/kg for 1 hour by adding NaCl to the medium increases PI3K activity, as measured by in vitro ELISA of PIP3 generated by p85 immunoprecipitates from Jurkat cells (Fig. 1A). The Jurkat cells used in this and subsequent experiments contain a tetracycline-regulated dominant negative of the PI3K regulatory subunit, p85α (20). However, the dominant negative was not expressed in this experiment because of the presence of tetracycline.
Fig. 1.
PI3K-IA is activated by high NaCl. (A) PI3K-IA activity was measured in p85 immunoprecipitates 1 h after increasing osmolality to 500 mosmol/kg by adding NaCl to Jurkat cells expressing active PI3K-IA; i.e., tetracycline was present, which prevented expression of the c-Myc Δp85, PI3K-IA DN construct. (A Lower) Western blot showing that there were equal amounts of p85 protein in reactions from the cells at 300 and 500 mosmol/kg. (B and C) Osmolality bathing HEK293 was increased to 500 mosmol/kg for 1 h by adding NaCl. (B) p85 protein abundance and phosphorylation on Y508. (C) Time course of high NaCl-induced phosphorylation of p85 (Y508-P) in HEK293 cells. The cells were grown at 300 mosmol/kg. For the “control” at 300 mosmol/kg, that medium was not changed. For the points at 0 min, we changed the medium to a new one at 300 or 500 mosmol/kg, then immediately washed with ice-cold PBS, followed immediately by the protein extraction buffer. (D) GSK-3β phosphorylation on S9. All values are normalized to the activity in cells kept at 300 mosmol/kg. Mean ± SEM (∗, P < 0.05; n = 3).
p85 is phosphorylated on Y508 upon platelet-derived growth factor stimulation, which may regulate PI3K activity (21, 22). Because the high NaCl-induced increase in PI3K lipid-kinase activity could result from such a posttranslational modification, we measured phosphorylation of Y508 of p85 in HEK293 cells. Increasing osmolality from 300 to 500 mosmol/kg for 1 h by adding NaCl to the medium increases this phosphorylation (Fig. 1B). The increase in phosphorylation of p85 is very rapid. It is already apparent at the earliest time that we can measure “0 min,” which is immediately after the medium is changed to an identical one (300 mosmol/kg) or an otherwise identical one increased to 500 mosmol/kg by adding NaCl (Fig. 1C). High NaCl also increases phosphorylation of the PI3K target, GSK-3β (Fig. 1D). Thus, high NaCl increases PI3K activity, possibly by phosphorylation of p85-Y508.
Effect of PI3K-IA Activity on Transcriptional Activity of TonEBP/OREBP.
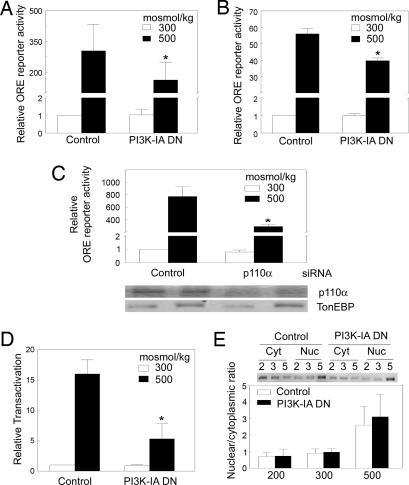
We used an ORE/TonE luciferase reporter system to measure TonEBP/OREBP transcriptional activity. We varied PI3K-IA activity by the presence or absence of tetracycline in the Jurkat cells or doxycycline in HEK293 Tet-on cells. p85α is the regulatory subunit of PI3K-IA, and p110 is the catalytic subunit (20). Δp85 lacks the p110 SH2 binding domain (amino acids 479–513), which makes it a dominant negative p85α (20). Tetracycline inhibits Δp85 expression in the Jurkat cells, and doxycycline induces Δp85 expression in the HEK-293 Tet-on cells. Raising osmolality to 500 mosmol/kg by adding NaCl increases TonEBP/OREBP transcriptional activity ≈250-fold in the Jurkat cells (Fig. 2A, control) and ≈56-fold in the HEK-293 (Fig. 2B, control) when endogenous PI3K-IA is active. Dominant negative inhibition of PI3K-IA has no significant effect on TonEBP/OREBP transcriptional activity at 300 mosmol/kg in either cell type (Fig. 2A and B). However, dominant negative inhibition decreases TonEBP/OREBP transcriptional activity at 500 mosmol/kg by 50% in the Jurkat cells (Fig. 2A) and by 30% in the HEK293 cells (Fig. 2B). As a control, we measured the effects of tetracycline and doxycycline on ORE/TonE reporter activity in Jurkat and HEK293 cells, respectively, in the absence of the dominant negative constructs. The effects are small, and the differences are not statistically significant (data not shown).
Fig. 2.
Inhibition of PI3K-IA decreases TonEBP/OREBP transcriptional activity but does not affect its subcellular localization. To inhibit PI3K-IA, the c-Myc Δp85 (PI3K-IA DN) construct was expressed in the Jurkat Tet-off cells by removing tetracycline for 48 h, or in the HEK293 Tet-on cells, by adding 2.0 μg/ml doxycycline for 24 h. (A) TonEBP/OREBP transcriptional activity in Tet-off Jurkat cells. The cells were transfected with ORE-X reporter plasmids. Twenty-four hours later, osmolality either was kept at 300 mosmol/kg or was increased to 500 mosmol/kg for 16 h by adding NaCl before measuring luciferase reporter activity. (B) TonEBP/OREBP transcriptional activity in HEK293 Tet-on cells. The cells were transiently cotransfected with ORE-X and c-Myc Δp85 constructs. Twenty-four hours later, osmolality either was kept at 300 mosmol/kg or was increased to 500 mosmol/kg for 16 h by adding NaCl before measuring luciferase activity. (C) Effect of siRNA targeting p110α on TonEBP/OREBP transcriptional activity. HEK293 cells stably transfected with ORE-X (HEK293 ORE-X cells) were transfected with 20 nM siRNA, either control (random, nontargeting) or targeting p110α for 48 h. After transfection, osmolality either was kept at 300 mosmol/kg or was increased to 500 mosmol/kg for 24 h by adding NaCl before measuring luciferase reporter activity. In the blots, abundance of TonEBP/OREBP and p110α was determined by Western analysis. (D) TonEBP/OREBP transactivation in Tet-off Jurkat cells. The cells were cotransfected with the GAL4 UAS reporter and GAL4dbd-548–1531. Twenty-four hours later, osmolality either was kept at 300 mosmol/kg or was increased to 500 mosmol/kg for 16 h by adding NaCl before measuring luciferase activity. In A–D, values are normalized to cells kept at 300 mosmol/kg. (E) TonEBP/OREBP intracellular location in Jurkat cells. Cells were exposed to 200, 300, or 500 mosmol/kg by varying NaCl concentration (“2,” “3,” or “5”) for 30 min before measuring TonEBP/OREBP protein by Western analysis in cytoplasmic and nuclear extracts and calculating TonEBP/OREBP nuclear/cytoplasmic ratio. A representative Western blot is shown. All values are mean ± SEM (∗, P < 0.05; n = 3).
In addition, we inhibited expression of p110α, a catalytic subunit of PI3K-IA, in ordinary HEK293 cells by using a specific siRNA. After 72 h of siRNA treatment, the expression of p110α is knocked down by 70% (Fig. 2C). This p110α siRNA reduces TonEBP/OREBP transcriptional activity by 60% at 500 mosmol/kg but has no significant effect at 300 mosmol/kg (Fig. 2C). We conclude that the increase in the catalytic activity of PI3K-IA contributes to high NaCl-induced increase in TonEBP/OREBP transcriptional activity.
Effect of PI3K-IA Activity on High NaCl-Induced Transactivation Activity of TonEBP/OREBP.
We used a binary GAL4 reporter system (4) to measure TonEBP/OREBP transactivation. Although PI3K-IA is active, high NaCl increases TonEBP/OREBP transactivation activity ≈15-fold (Fig. 2D, control). Expression of Δp85, the dominant negative of PI3K-IA, inhibits TonEBP/OREBP transactivation activity by 67% at 500 mosmol/kg but has no significant effect at 300 mosmol/kg (Fig. 2D). We conclude that increased PI3K-IA activity contributes to high NaCl-induced TonEBP/OREBP transactivtion.
Effect of PI3K-IA Activity on TonEBP/OREBP Protein Abundance.
Sixteen to twenty-four hours of high NaCl significantly increases TonEBP/OREBP protein abundance in tetracycline-regulated PI3K mutant Jurkat cells and HEK293 Tet-on cells (data not shown). In those cells, expression of Δp85, the dominant negative of PI3K-IA, does not significantly affect TonEBP/OREBP protein abundance at either 300 or 500 mosmol/kg (data not shown). Further, specific siRNA knockdown of p110α does not affect the protein abundance of TonEBP/OREBP in ordinary HEK293 cells at either 300 or 500 mosmol/kg (Fig. 2C). Thus, we find no evidence that PI3K-IA activity affects TonEBP/OREBP protein abundance.
Effect of PI3K-IA Expression on High NaCl-Induced Nuclear Translocation of TonEBP/OREBP.
We determined the nuclear to cytoplasmic ratio of TonEBP/OREBP by measuring the amounts of it in nuclear and cytoplasmic protein extracts by Western blot. Increasing osmolality from 300 to 500 mosmol/kg by adding NaCl increases TonEBP/OREBP nuclear to cytoplasmic ratio 3-fold in the tetracycline-regulated PI3K mutant Jurkat cells, whether they are expressing PI3K-IA (Fig. 2E, control) or not (Fig. 2E, PI3K-IA DN). We conclude that PI3K-IA activity does not contribute to high NaCl-induced nuclear localization of TonEBP/OREBP.
Effect of PI3K-IA Activity on High NaCl-Induced Activation of ATM.
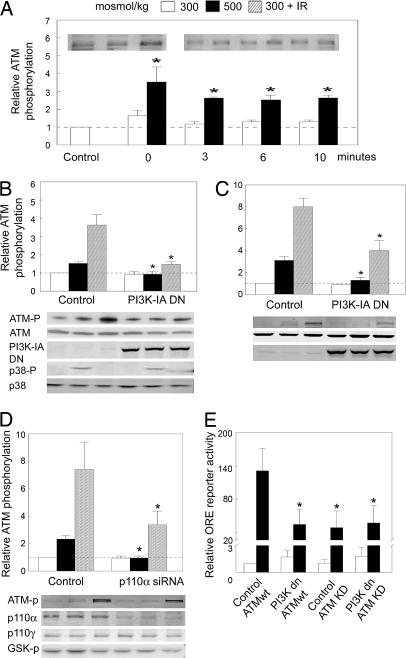
We used a phosphospecific antibody against ATMS1981-P to determine the effect of PI3K-IA activity on ATM activation in tetracycline-regulated PI3K mutant Jurkat cells and HEK293 cells. When PI3K-IA is active, elevating osmolality from 300 to 500 mosmol/kg by adding NaCl increases phosphorylation of ATM on S1981 (Fig. 3A). The increase in phosphorylation is very rapid. It is already apparent at the earliest time that we can measure, which is immediately after the medium is changed to an identical one (300 mosmol/kg) or an otherwise identical one increased to 500 mosmol/kg by adding NaCl (0 min in Fig. 3A). Dominant negative PI3K-IA (Fig. 3 B and C, PI3K-IA DN) or p110α siRNA (Fig. 3D, p110α siRNA) prevents the increase in ATM phosphorylation. The p110α siRNA does not affect expression of p110γ, the catalytic subunit of PI3K-IB, confirming its specificity (Fig. 3D). In the same experiment, we also measured phosphorylation of GSK-3β, a downstream target of PI3K that becomes phosphorylated after high NaCl (Fig. 1D). siRNA-mediated knockdown of p110α prevents this effect (Fig. 3D), which further supports the conclusion that high NaCl activates PI3K-IA. We conclude that PI3K-IA activity is necessary for high NaCl-induced activation of ATM. On the other hand, high NaCl-induced phosphorylation of p38, does not depend on PI3K-IA activity (Fig. 3B).
Fig. 3.
Effects of PI3K-IA DN (Δp85) or p110α siRNA on ATM activation (phosphorylation of S1981). The cells were exposed to 5 Gy of IR, then analyzed 1 h later or exposed to high NaCl for 2 h. (A) Time course of high NaCl-induced phosphorylation of ATM (S1981-P) in HEK293 cells. The cells were grown at 300 mosmol/kg. For the control at 300 mosmol/kg, that medium was not changed. For the points at 0 min, we changed the medium to a new one at 300 or 500 mosmol/kg, then immediately washed with ice-cold PBS, followed also immediately by the protein extraction buffer. (B) Jurkat Tet-off cells. The c-Myc Δp85 (PI3K-IA DN) construct was expressed by removing tetracycline for 48 h. (B Lower) Western blots analysis of Δp85 expression and of ATM and p38 phosphorylation. (C) HEK293 Tet-on cells. The c-Myc Δp85 construct was expressed by adding 2.0 μg/ml doxycycline for 24 h. (C Lower) Western blot analysis of Δp85 expression and ATM phosphorylation. (D) Effect of siRNA targeting p110α on ATM activation. HEK293 cells stably transfected with ORE-X (HEK293 ORE-X cells) were transfected with 20 nM siRNA for 48 h, either control (random, nontargeting) or targeting p110α. (D Lower) Western blot analysis of p110α and p110γ expression and the phosphorylation of ATM and GSK-3β. (E) Effects of expression of PI3K-IA and/or ATM on TonEBP/OREBP transcriptional activity. PI3K-IA activity in the tetracycline-regulated PI3K mutant Jurkat cells was modified by the presence or absence of tetracycline for 48 h, then the cells were cotransfected by electroporation with ORE-X reporter and with wild type (wt) or kinase dead (KD) ATM, which is a dominant negative. Twenty-four hours later, osmolality either was kept at 300 mosmol/kg or was increased to 500 mosmol/kg (NaCl added) for 2 h before measuring reporter activity. All values are relative to control kept at 300 mosmol/kg. Mean ± SEM (∗, P < 0.05; n = 3).
Effect of PI3K-IA Activity on Ionizing Radiation-Induced Activation of ATM.
IR activates ATM kinase through phosphorylation on S1981 (7, 9). Although PI3K-IA is being expressed, IR (5 Gy) increases phosphorylation of ATM on S1981 within 1 h (Fig. 3 B–D, control). Dominant negative PI3K-IA (Fig. 3 B and C, PI3K-IA DN) and p110α siRNA (Fig. 3D, p110α siRNA) greatly reduce the increase in phosphorylation. We conclude that PI3K-IA activity is necessary for full IR-induced activation of ATM.
Effect of Combined Inhibition of ATM and PI3K-IA Activities on High NaCl-Induced Increase in TonEBP/OREBP Transcriptional Activity.
To examine the relationship between PI3K-IA and ATM, we varied PI3K-IA activity by the presence or absence of tetracycline and ATM activity by transfection of wild-type or dominant negative (kinase dead) ATM (23). The various combinations had no significant effect at 300 mosmol/kg (Fig. 3E). However, the increase in transcriptional activity at 500 mosmol/kg was suppressed equally by inhibition of PI3K-IA activity (PI3K-IA DN, ATM wt), ATM activity (Control, ATM KD; kinase dead, KD), or both (PI3K IA DN, ATM KD) (Fig. 3E). We conclude that because PI3K-IA activity is necessary for high NaCl-induced activation of ATM (Fig. 3) and the effects of inhibiting PI3K-IA and ATM activity are not additive (Fig. 3E), the effect of PI3K-IA activity on high NaCl-induced TonEBP/OREBP transcriptional activity is mediated via ATM.
Discussion
Kinases Involved in High NaCl-Induced Activation of TonEBP/OREBP.
Our original motive for these studies was our finding that wortmannin inhibits high NaCl-induced TonEBP/OREBP transcriptional activity, even in AT cells, implicating participation of some other wortmannin-inhibitable kinase in addition to ATM (7). We now find that PI3K-IA activity, which wortmannin also inhibits, contributes to high NaCl-induced activation of TonEBP/OREBP. However, because PI3K-IA acts through ATM, we cannot explain the effect of wortmannin in AT cells as being due to its inhibition of PI3K-IA. There are other kinases that are inhibited by wortmannin including DNA-PK, myosin light chain kinase, and other PI3Ks. All of these kinases are additional candidates for involvement in regulating TonEBP/OREBP.
Activation of ATM by DNA Double-Strand Breaks.
IR and high NaCl induce DNA double-strand breaks and activate ATM (7, 10, 11). How DNA breaks activate ATM is not completely understood. It has been proposed that IR causes generalized changes in chromatin structure that initially activate ATM, even at a distance from DNA breaks (9). We now find that activation of PI3K-IA is necessary both for IR- and for high NaCl-induced activation of ATM. This finding raises the possibility that the changes in chromatin structure might activate nuclear PI3K, in turn leading to activation of ATM. Alternatively, the same perturbations that affect chromatin structure also might activate PI3K-IA independently. In addition, we do not yet know how PI3K-IA activates ATM, whether it is a direct effect or involves intermediates.
Relation Between Protein Kinases Involved in High NaCl-Dependent Activation of TonEBP/OREBP.
Several protein kinases have been shown to contribute to high NaCl-induced increase of TonEBP/OREBP transcriptional activity and transactivation, including p38 (5, 6), Fyn (5), ATM (7), and PKAc (8). Based on the present studies, PI3K-IA is another. Like the other kinases, inhibition of PI3K-IA reduces activation of TonEBP/OREBP but not completely so (8). Further, although the effect of PI3K-IA is mediated through ATM, the activation of p38 is not. Whether the other kinases interact with each other is not yet established.
Besides ATM, Akt/PKB is also a major target of PI3K (24). However, there is disagreement regarding the effect of hypertonicity on Akt/PKB activity. Some studies concluded that hypertonicity activates Akt/PKB, including direct measurements of increased Akt/PKB activity in NIH 3T3 and CHO cells (25). Also, phosphorylation of Akt/PKB at S174 increases in Madin-Darby canine kidney cells subjected to high NaCl and in inner medullas of rats subjected to dehydration (16). In contrast, other studies concluded that Akt/PKB actually is inactivated by hypertonicity, including that high sorbitol decreases Akt/PKB activity in HEK293 and COS cells (26) and that high sucrose decreases it in Swiss 3T3 cells, despite increased PIP3 abundance and increased PI3K activity (19). In the latter study, failure of the increased PIP3 to activate Akt/PKB was attributed to concomitant activation of an inhibitory pathway.
Mechanism of Activation of PI3K-IA by High NaCl.
We do not know how high NaCl activates PI3K. However, several modes of activation were described previously in response to other signals. The most generally recognized one is through phosphorylation of tyrosines in plasma membrane-bound proteins, creating docking sites for p85, which then attracts and activates p110 (27). However, other mechanisms also have been reported. Some PI3K occurs in the nucleus (28, 29). Stimulation of cells by nerve growth factor activates nuclear PI3K, resulting in accumulation of 3-phosphorylated phosphoinositide lipids in the nucleus (29, 30). That is mediated by PLCγ, which activates PI3K Enhancer, a nuclear GTPase that increases PI3K activity (31, 32). Finally, high NaCl also induces phosphorylation of p85 on Y508 (Fig. 1 B and C). This posttranslational modification could be involved in the activation of PI3K.
Materials and Methods
Cell Culture.
We used four kinds of cells. (i) Tetracycline regulated PI3K mutant Jurkat cells, previously described in ref. 20, that Tet-conditionally express c-Myc Δp85, a dominant negative of PI3K-IA. We grew them in RPMI 1640 medium (HyClone, Logan, UT) supplemented with 10% FBS (HyClone), 0.5 mg of geneticin (GIBCO Life Technologies, Carlsbad, CA), 0.3 mg/ml Hygromycin B (Invitrogen) and 2.5 μg/ml tetracycline (Sigma-Aldrich). Tetracycline suppresses expression of the c-Myc Δp85 construct, but expression occurs within 48 h after tetracycline is removed. (ii) HEK293 Tet-on cells (BD Biosciences) expressed the reverse tetracycline-controlled transactivator. We grew the Tet-on HEK293 cells in DMEM (American Type Culture Collection, Manassas, VA), supplemented with 10% Tet System Approved FBS (BD Biosciences), and 2.0 μg/ml doxycycline (Sigma-Aldrich). c-Myc Δp85 construct was transiently transfected into the cells, either omitting doxycycline (control) or continuing it (experimental, dominant negative expressed). (iii) HEK293 cells stably expressing an ORE-X reporter of TonEBP/OREBP transcriptional activity were grown in Eagle's minimal essential medium (ATCC) supplemented with 10% FBS (HyClone) and 5 μg/liter blasticidin (Sigma-Aldrich). Osmolality of the basal medium was 300 mosmol/kg. Osmolality was increased by adding NaCl to a total osmolality of 500 mosmol/kg for 0.5, 2, or 16 h. (iv) Unmodified HEK293 cells were grown in EMEM medium (ATCC) supplemented with 10% FBS (HyClone). Osmolality of the basal medium was 300 mosmol/kg. Osmolality was increased by adding NaCl to a total osmolality of 500 mosmol/kg for 0.5, 2, or 16 h.
Plasmids and siRNA.
The ORE-X luciferase reporter construct contains two copies of human ORE-X (33) within a minimal IL-2 promoter (34) (hTonE-GL3, a gift from S. N. Ho, University of California at San Diego, La Jolla, CA). Wild-type and kinase dead Flag-ATM (23) were gifts of M. B. Kastan (St. Jude Children's Research Hospital, Memphis, TN). The c-Myc Δp85 construct was described in ref. 20. The binary GAL4 reporter system for measuring transactivation was described in ref. 4. Plasmids were transfected by electroporation by using the Gene Pulser Xcell Electropration System (Bio-Rad), according to the manufacturer's instructions. We designed the siRNA against p110α, as a synthetic dsRNA Dicer substrate to enhance the RNA interference potency and efficacy (35). The control (nontargeting) siRNA (Integrated DNA Technologies, Coralville, IA) duplex sequences were as follows: sense, 5′-Phos-UGAACCUGACCCAGGGGAGGGAGdTdT-3′ and antisense sequence 5′-AACUCCCUCCCCUGGGUCAGGUUCAUU-3′. The p110α siRNA (Dharmacon Research, Chicago) sequences were as follows: sense, 5′-Phos-GCCAGUACCUCAUGGAUUAGAAGdAdT-3′ and, antisense, 5′-AUCUUCUAAUCCAUGAGGUACUGGCUU-3′. The siRNAs were transient transfected by using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Twenty-four hours after siRNA transfection, the osmolality was either kept at 300 mosmol/kg or was increased to 500 mosmol/kg for 24 h by adding NaCl.
Western Blot Analysis.
Cells were lysed with M-PER protein extraction reagent (Pierce Biotechnology) for whole-cell extracts or with NE-PER (Nuclear and Cytoplasmic Extraction Reagents; Pierce Biotechnology) for separate nuclear and cytoplasmic fractions, according to supplier's instructions. Protease inhibitor mixture (Roche Diagnostics, Alameda, CA) and phosphatase inhibitor cocktails I (P2850; Sigma-Aldrich) and II (P5726; Sigma-Aldrich) were included in the lysis buffers. Proteins were separated on 4–12% Novex Tris-Glycine gels and transferred to nitrocellulose membranes (Invitrogen). Western blot analysis was performed according to instructions for the Odyssey Infrared Imaging System (Li-Cor, Lincoln, NE). In brief, nonspecific binding was blocked by incubating membranes overnight at 4°C with Odyssey Blocking Buffer diluted 1:1 with PBS. Membranes then were incubated with anti-NFAT5 (TonEBP/OREBP) rabbit polyclonal antibody (Affinity BioReagents, Golden, CO), anti c-Myc (Santa Cruz Biotechology), anti-ATMS1981-P (Rockland Immunochemicals), anti-p110α (Cell Signaling, Beverly, MA), anti-p110γ (Santa Cruz Biotechology), anti-p85 (Santa Cruz Biotechology, Santa Cruz, CA), anti-p85Y508-P (Santa Cruz Biotechology), anti-GSK-3βS9-P (Cell Signaling Technology, Beverly, MA), anti-p38T180/Y182-P MAPK, anti-p38 MAPK (Cell Signaling Technology) or anti-ATM (MAT3; a gift from M. B. Kastan) for 2 h at room temperature. After washing with 0.1% Tween-20 in PBS, blots were incubated with Alexa Fluor 680 goat anti-rabbit IgG or Alexa Fluor 780 goat anti-mouse IgG (Molecular Probes) for 1 h in the dark. Blots were visualized and quantitated by using a Li-Cor Odyssey Infrared Imager.
Calculation of Nuclear/Cytoplamic Ratios.
The relative amounts of TonEBP/OREBP in the cytoplasmic and nuclear fractions and the nuclear/cytoplasmic ratio were calculated from the relative concentrations of TonEBP/OREBP in each cytoplasmic or nuclear extract and the relative volumes of the extracts.
Luciferase Reporter Assay.
Cells grown in six-well plates were transfected with 10 μg of ORE-X luciferase reporter plasmids. Then, the medium was changed to the same osmolality (300 mosmol/kg) or elevated to 500 mosmol/kg by adding NaCl. Luciferase activity was measured either 2 or 16 h later by using the Luciferase Assay System (Promega) in a Victor 3 1420 Multilabel counter (PerkinElmer).
PI3K Activity and Immunoprecipitation.
Tetracycline-regulated PI3K mutant Jurkat cells were incubated at 300 mosmol/kg in a serum-free medium supplemented with 5% BSA and 2.5 μg tetracycline for 48 h, then fresh medium at 300 or 500 mosmol/kg (NaCl added) was substituted. One hour later, cells were pelleted by centrifugation. Subsequent steps were at 4°C. The pellet from one 75-cm2 cell culture flask was extracted for 5 min with 1 ml of lysis buffer containing 50 mM Tris·HCl (pH 8.0), 150mM NaCl, 1% Nonidet P-40, 1 mM CaCl2, 1 mM MgCl2, protease inhibitor mixture (Roche Diagnostics, Alameda, CA), and phosphatase inhibitor mixtures (Sigma-Aldrich) and centrifuged at 15,000 × g for 10 min. Samples were precleared with 0.2 mg of protein A-agarose (Roche Diagnostics, Alameda, CA), followed by 12 μg of IgG-agarose (Santa Cruz Biotechnology) for 1 h each, then centrifuged. For immunoprecipitation of p85α, the precleared supernatants were incubated overnight with 4 μg of rabbit anti-p85α (Santa Cruz Biotechnology) then for 2 h with the addition of 0.2 mg of protein A-agarose beads. The beads were resuspended in kinase buffer (20 mM Tris·HCl, pH 7.4/10 mM sodium chloride/4 mM MgCl2/25 μM ATP). To determine the amount of immunoprecipitated PI3K-IA, an aliquot was mixed with an equal volume of Laemmli sample buffer, and, after centrifugation, supernatant proteins were separated on 4–12% Novex Tris-Glycine gels and transferred to nitrocellulose membranes (Invitrogen). Membranes were then incubated with anti-p85α rabbit polyclonal antibody (Santa Cruz Biotechnology). Western blot analysis was performed according to instructions for the Odyssey Infrared Imaging System.
PI3K activity was measured by using a competitive ELISA Assay (Echelon Research Lab, Salt Lake City) and by following the manufacturer's instructions. The assays were performed on p85α immunoprecipitated from Jurkat cells 1 h after the medium was substituted by one in which osmolality was elevated to 500 mosmol/kg by adding NaCl or was left at 300 mosmol/kg. The reaction products were incubated with PI(3,4,5)P3 detector protein, then added to the PI(3,4,5)P3-coated microplate for the competitive binding assay. A peroxidase-linked secondary detection reagent was used to measure PI(3,4,5)P3 detector protein bound to the plate. The colorimetric signal is inversely proportional to the amount of PI(3,4,5)P3 produced by PI3K activity.
Statistical Analysis.
Data were compared by a one-way ANOVA followed by Student-Newman-Keuls post hoc test. Logarithmic transformation was applied first to any data calculated as ratios. Results are expressed as mean ± SEM (n = number of independent experiments). Differences were considered significant for P < 0.05.
Acknowledgments
We thank Drs. Michael B. Kastan and Christopher J. Bakkenist (St. Jude Children's Research Hospital, Memphis, TN) for ATM reagents and advice and Dr. Steffan N. Ho for hTonE-GL3. This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health.
Abbreviations
- IR
ionizing radiation
- PIP3
phosphatidyl inositol 3-phosphate
- PI3K
phosphatidylinositol 3-kinase
- PI3K-IA
PI3K-Class IA
- siRNA
small interfering RNA.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Dmitrieva N. I., Burg M. B., Ferraris J. D. Am. J. Physiol. 2005;289:F2–F7. doi: 10.1152/ajprenal.00041.2005. [DOI] [PubMed] [Google Scholar]
- 2.Miyakawa H., Woo S. K., Dahl S. C., Handler J. S., Kwon H. M. Proc. Natl. Acad. Sci. USA. 1999;96:2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai Q., Ferraris J., Burg M. B. Am. J. Physiol. 2005;289:F803–F807. doi: 10.1152/ajprenal.00448.2004. [DOI] [PubMed] [Google Scholar]
- 4.Ferraris J. D., Williams C. K., Persaud P., Zhang Z., Chen Y., Burg M. B. Proc. Natl. Acad. Sci. USA. 2002;99:739–744. doi: 10.1073/pnas.241637298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ko B. C., Lam A. K., Kapus A., Fan L., Chung S. K., Chung S. S. J. Biol. Chem. 2002;277:46085–46092. doi: 10.1074/jbc.M208138200. [DOI] [PubMed] [Google Scholar]
- 6.Sheikh-Hamad D., Di Mari J., Suki W. N., Safirstein R., Watts B. A., III, Rouse D. J. Biol. Chem. 1998;273:1832–1837. doi: 10.1074/jbc.273.3.1832. [DOI] [PubMed] [Google Scholar]
- 7.Irarrazabal C. E., Liu J. C., Burg M. B., Ferraris J. D. Proc. Natl. Acad. Sci. USA. 2004;101:8809–8814. doi: 10.1073/pnas.0403062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferraris J. D., Persaud P., Williams C. K., Chen Y., Burg M. B. Proc. Natl. Acad. Sci. USA. 2002;99:16800–16805. doi: 10.1073/pnas.222659799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakkenist C. J., Kastan M. B. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 10.Dmitrieva N. I., Cai Q., Burg M. B. Proc. Natl. Acad. Sci. USA. 2004;101:2317–2322. doi: 10.1073/pnas.0308463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kultz D., Chakravarty D. Proc. Natl. Acad. Sci. USA. 2001;98:1999–2004. doi: 10.1073/pnas.98.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karnitz L. M., Burns L. A., Sutor S. L., Blenis J., Abraham R. T. Mol. Cell. Biol. 1995;15:3049–3057. doi: 10.1128/mcb.15.6.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarkaria J. N., Tibbetts R. S., Busby E. C., Kennedy A. P., Hill D. E., Abraham R. T. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- 14.Davies S. P., Reddy H., Caivano M., Cohen P. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z., Yang X. Y., Soltoff S. P., Cohen D. M. Am. J. Physiol. 2000;278:F155–F164. doi: 10.1152/ajprenal.2000.278.1.F155. [DOI] [PubMed] [Google Scholar]
- 16.Terada Y., Inoshita S., Hanada S., Shimamura H., Kuwahara M., Ogawa W., Kasuga M., Sasaki S., Marumo F. Kidney Int. 2001;60:553–567. doi: 10.1046/j.1523-1755.2001.060002553.x. [DOI] [PubMed] [Google Scholar]
- 17.Rao R., Hao C. M., Breyer M. D. J. Biol. Chem. 2004;279:3949–3955. doi: 10.1074/jbc.M309325200. [DOI] [PubMed] [Google Scholar]
- 18.Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 19.Van der, Kaay J., Beck M., Gray A., Downes C. P. J. Biol. Chem. 1999;274:35963–35968. doi: 10.1074/jbc.274.50.35963. [DOI] [PubMed] [Google Scholar]
- 20.Curnock A. P., Ward S. G. J. Immunol. Methods. 2003;273:29–41. doi: 10.1016/s0022-1759(02)00416-7. [DOI] [PubMed] [Google Scholar]
- 21.Kavanaugh W. M., Klippel A., Escobedo J. A., Williams L. T. Mol. Cell. Biol. 1992;12:3415–3424. doi: 10.1128/mcb.12.8.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kavanaugh W. M., Turck C. W., Klippel A., Williams L. T. Biochemistry. 1994;33:11046–11050. doi: 10.1021/bi00202a026. [DOI] [PubMed] [Google Scholar]
- 23.Canman C. E., Lim D. S., Cimprich K. A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M. B., Siliciano J. D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 24.Scheid M. P., Woodgett J. R. FEBS Lett. 2003;546:108–112. doi: 10.1016/s0014-5793(03)00562-3. [DOI] [PubMed] [Google Scholar]
- 25.Konishi H., Matsuzaki H., Tanaka M., Ono Y., Tokunaga C., Kuroda S., Kikkawa U. Proc. Natl. Acad. Sci. USA. 1996;93:7639–7643. doi: 10.1073/pnas.93.15.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier R., Thelen M., Hemmings B. A. EMBO J. 1998;17:7294–7303. doi: 10.1093/emboj/17.24.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanhaesebroeck B., Leevers S. J., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., Waterfield M. D. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. 535–602. [DOI] [PubMed] [Google Scholar]
- 28.Lu P. J., Hsu A. L., Wang D. S., Yan H. Y., Yin H. L., Chen C. S. Biochemistry. 1998;37:5738–5745. doi: 10.1021/bi972551g. [DOI] [PubMed] [Google Scholar]
- 29.Neri L. M., Milani D., Bertolaso L., Stroscio M., Bertagnolo V., Capitani S. Cell. Mol. Biol. (Noisy-le-grand) 1994;40:619–626. [PubMed] [Google Scholar]
- 30.Tanaka K., Horiguchi K., Yoshida T., Takeda M., Fujisawa H., Takeuchi K., Umeda M., Kato S., Ihara S., Nagata S., et al. J. Biol. Chem. 1999;274:3919–3922. doi: 10.1074/jbc.274.7.3919. [DOI] [PubMed] [Google Scholar]
- 31.Ye K., Aghdasi B., Luo H. R., Moriarity J. L., Wu F. Y., Hong J. J., Hurt K. J., Bae S. S., Suh P. G., Snyder S. H. Nature. 2002;415:541–544. doi: 10.1038/415541a. [DOI] [PubMed] [Google Scholar]
- 32.Ye K., Hurt K. J., Wu F. Y., Fang M., Luo H. R., Hong J. J., Blackshaw S., Ferris C. D., Snyder S. H. Cell. 2000;103:919–930. doi: 10.1016/s0092-8674(00)00195-1. [DOI] [PubMed] [Google Scholar]
- 33.Ferraris J. D., Williams C. K., Ohtaka A., Garcia-Perez A. Am. J. Physiol. 1999;276:C667–C673. doi: 10.1152/ajpcell.1999.276.3.C667. [DOI] [PubMed] [Google Scholar]
- 34.Trama J., Lu Q., Hawley R. G., Ho S. N. J. Immunol. 2000;165:4884–4894. doi: 10.4049/jimmunol.165.9.4884. [DOI] [PubMed] [Google Scholar]
- 35.Kim D. H., Behlke M. A., Rose S. D., Chang M. S., Choi S., Rossi J. J. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]