Abstract
Plant resistance proteins (R proteins) recognize corresponding pathogen avirulence (Avr) proteins either indirectly through detection of changes in their host protein targets or through direct R–Avr protein interaction. Although indirect recognition imposes selection against Avr effector function, pathogen effector molecules recognized through direct interaction may overcome resistance through sequence diversification rather than loss of function. Here we show that the flax rust fungus AvrL567 genes, whose products are recognized by the L5, L6, and L7 R proteins of flax, are highly diverse, with 12 sequence variants identified from six rust strains. Seven AvrL567 variants derived from Avr alleles induce necrotic responses when expressed in flax plants containing corresponding resistance genes (R genes), whereas five variants from avr alleles do not. Differences in recognition specificity between AvrL567 variants and evidence for diversifying selection acting on these genes suggest they have been involved in a gene-specific arms race with the corresponding flax R genes. Yeast two-hybrid assays indicate that recognition is based on direct R–Avr protein interaction and recapitulate the interaction specificity observed in planta. Biochemical analysis of Escherichia coli-produced AvrL567 proteins shows that variants that escape recognition nevertheless maintain a conserved structure and stability, suggesting that the amino acid sequence differences directly affect the R–Avr protein interaction. We suggest that direct recognition associated with high genetic diversity at corresponding R and Avr gene loci represents an alternative outcome of plant–pathogen coevolution to indirect recognition associated with simple balanced polymorphisms for functional and nonfunctional R and Avr genes.
Keywords: avirulence protein, resistance protein
Plants resist disease through a variety of preformed and induced barriers to infection. Among these is a genetically determined pathogen recognition system controlled by host resistance genes (R genes). In this gene-for-gene resistance system, plant R genes confer resistance to pathogen strains carrying corresponding avirulence (Avr) genes (so-called because their presence prevents growth on resistant plants). The recognition event involving the products of the R and Avr genes triggers host defense responses, including a localized host cell death or hypersensitive response (HR) that limits the spread of the pathogen from the infection site. Most known plant resistance proteins (R proteins) contain nucleotide binding site (NBS) and leucine-rich repeat (LRR) domains, with the latter implicated in pathogen recognition (1). In contrast, Avr proteins are diverse, and many have pathogenicity effector functions that play important roles in enhancing infection (2). The antagonistic relationship between R and Avr genes results in coevolutionary conflict as selection favors evolution of resistance in plants and virulence in their pathogens.
The Arabidopsis RPM1, RPS2, and RPS5 NBS-LRR-type R proteins confer resistance by detecting changes in host proteins that are modified by the effector function of their corresponding Avr proteins from the bacterial pathogen Pseudomonas syringae (3–6). P. syringae isolates virulent toward these R genes lack the corresponding Avr genes, and population studies of the R gene loci in Arabidopsis have found little genetic diversity and suggest that simple balanced polymorphisms for functional and nonfunctional alleles have been maintained over long evolutionary time scales (7, 8). This observation is consistent with predictions based on theoretical considerations (9), because mutations of the Avr protein that abolish recognition must also result in loss of its effector function, which can have a fitness penalty for the pathogen. Thus there is no selective pressure for diversification of the corresponding R and Avr genes. In contrast, recognition by direct R–Avr protein interaction raises the possibility of a gene-specific arms race leading to diversification of both R and Avr genes (10), because such an R protein in the host may be countered by alterations to the pathogen Avr protein that abolish recognition but retain effector function with little or no fitness penalty to the pathogen. However, there has been very little empirical evidence available to test this prediction. Although many plant R gene loci are highly polymorphic with diversifying selection playing an important role in the evolution of new recognition specificities (11), and some pathogen Avr gene families also show significant variation and diversifying selection (12), there are few examples where diversity in corresponding R and Avr genes has been studied. In one case, diversifying selection and high levels of polymorphism were observed in the corresponding ATR13 and RPP13 genes, from Hyaloperonospora parasitica and Arabidopsis, respectively (13, 14). However, it is not known whether the variation in these genes is associated with differences in recognition specificity as expected if diversification results from R–Avr counterselection. Conversely, although the Arabidopsis RRS1-R and rice Pita NBS-LRR class R proteins interact directly with their cognate Avr proteins PopP2 and Avr-Pita from Ralstonia solanacearum and Magnaporthe grisea, respectively (15, 16), the level of genetic diversity at the host and pathogen loci has not been reported to our knowledge.
In flax (Linum usitatissimum), diversifying selection has resulted in at least 12 allelic variants at the polymorphic L locus, which encode NBS–LRR proteins with specific recognition of different Avr proteins in various strains of the flax rust fungus (Melampsora lini; refs. 17 and 18). The polymorphic flax rust AvrL567 genes encode 150-aa proteins, predicted to release 127-aa proteins after secretion and cleavage of the signal peptide, that are recognized by the flax L5, L6, and L7 R proteins inside the plant cell (19). That study provided preliminary evidence for a possible arms race, with the observation of diversifying selection also driving amino acid variation between the three AvrL567 proteins (A, B, and C) encoded by rust strain CH5. Here we show that diversifying selection has led to extreme levels of polymorphism at the AvrL567 locus in different rust strains and that AvrL567 sequence divergence leads to qualitative differences in recognition specificity by the corresponding R genes. This recognition is based on direct R–Avr protein interactions, and AvrL567 variants that evade recognition nevertheless appear to have a conserved protein structure and stability. They may therefore retain an as-yet-unknown effector function with little or no fitness penalty to the pathogen. Thus AvrL567 diversity appears to be the result of R-gene-imposed selection in a gene-for-gene arms race.
Results and Discussion
Diversifying Selection Has Generated AvrL567 Variants that Are Differentially Recognized by Their Corresponding R Proteins.
Genetic diversity of AvrL567 genes was examined in six rust strains from geographically separated locations. In total, 12 members of the AvrL567 gene family have been identified, including the AvrL567-A, AvrL567-B, and AvrL567-C genes previously isolated (10), with one to four copies present at each haplotype (Table 1). Intralocus recombination events may have played a role in generating this haplotype diversity, because some variants are present in different combinations; for instance, AvrL567-F occurs in a haplotype with AvrL567-E in rust H, but with AvrL567-A and AvrL567-B in rust I. These genes are highly variable in sequence, with 35 polymorphic amino acid sites in the encoded 127-residue mature proteins (Fig. 1). There is a significant excess of nucleotide changes at nonsynonymous sites (average distance = 0.0484) over synonymous sites (average distance = 0.0067) in the coding sequence (ratio = 7.2; P < 10−6, t test; ref. 20), which indicates that diversifying selection has favored the accumulation of amino acid variation in these proteins. Analysis of shared polymorphisms provided no significant evidence for intragenic recombination contributing to AvrL567 sequence diversity.
Table 1.
Avirulence genotypes of flax rust strains
| Rust strain | Avr genotype* | AvrL567 genotype† |
|---|---|---|
| CH5‡ | Avr/avr | A, B/C |
| H | Avr/Avr | A, B/E, F |
| C | avr/avr | C/G, K |
| I | Avr/avr | A, B, F/H, I |
| Bsl | Avr/Avr | D, J/D, J |
| Fi | Avr/avr | A, B, F, L/C |
| 339 | Avr/? | A, B, L |
*Inferred rust avirulence (Avr) or virulence (avr) genotype from infection phenotypes of self or outcross progeny on plants containing L5 or L6 (31).
†AvrL567 gene variants (A to L) isolated from each rust strain by PCR were assigned to haplotypes based on segregation analysis in progeny derived from these rust strains, with the exception of rust strains Fi and 339 for which no progeny were available. The haplotype assignment in Fi is inferred from the phenotype observed in the in planta transient expression assays for these genes (Fig. 1) and the known heterozygous genotype for avirulence of this rust. No information on the genotype or haplotype arrangement of AvrL567 genes in rust 339 is available.
‡Data from ref. 19.
Fig. 1.
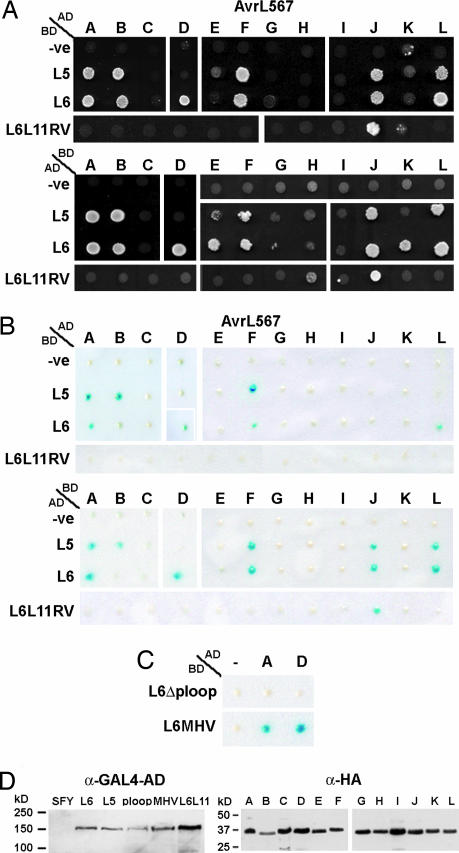
Amino acid variation between AvrL567 homologs is associated with differences in recognition specificity. (A) (Left) The consensus amino acid at polymorphic positions (numbered vertically above the consensus line) in the AvrL567 proteins is shown above the individual sequences with identical residues indicated by dots. (Right) The columns indicate whether a necrotic response was observed when these proteins were expressed in flax lines containing L5, L6, L7, or the recombinant L6L11RV gene. ++ indicates a very strong necrotic response observed within 4 days; + indicates necrosis observed within 10 days; +/− indicates a chlorotic response observed after 10 days; − indicates no response observed. ∗, A slight chlorotic response was observed for AvrL567-E on L6 in some but not all assays, suggesting very weak recognition. (B) Leaves of near-isogenic flax lines containing the L9, L5, L6, or L7 R genes or transgenic flax plants containing L6L11RV were infiltrated with Agrobacterium cultures containing T-DNA expression vectors encoding the predicted 127-aa mature AvrL567-D, AvrL567-F, AvrL567-J, or AvrL567–L proteins under the control of the cauliflower mosaic virus 35S promoter. Images were prepared 10 days after infiltration.
To assess which of the AvrL567 variants could act as Avr factors, we used Agrobacterium infiltration to transiently express these genes in leaves of flax plants containing the corresponding L5, L6, or L7 R genes. Transient expression of six of the AvrL567 variants (A, B, D, F, J, and L) induced an HR-like necrotic response dependent on the presence of L5, L6, or L7 (Fig. 1), indicating that these are functional Avr genes. A seventh gene, AvrL567-E, induced a very weak chlorotic response with L6 only. Likewise, AvrL567-D was L6-specific, whereas the others were recognized by both L5 and L6. Five AvrL567 variants (C, G, H, I, and K) failed to elicit a necrotic response. Genetic analysis of these rust strains indicated that all seven HR-inducing AvrL567 variants were derived from haplotypes conferring avirulence on L5, L6, and L7, whereas the five noninducing variants were derived from virulence alleles of this locus (Table 1). Thus, the transient assay results are completely consistent with the genetic descriptions of the infection phenotypes of each rust strain. In these assays, the difference between the L6 and L7 specificities seems to be largely one of degree, with L7 acting as a weak L6 allele. This finding is consistent with the weaker resistance phenotype conferred by L7, which allows limited urediospore production by avirulent rusts, relative to L6, which completely restricts urediospore production (21), and the high sequence similarity between the encoded proteins that differ by only 11 aa, all in the N-terminal Toll and IL-1 receptor-related domain (17).
We also tested recognition of the AvrL567 variants by L6L11RV, an in vitro-generated chimeric gene encoding a protein identical to L6 except for 11 amino acid differences in three C-terminal LRR units derived from L11. This chimeric gene confers resistance to rust strain Bs1, but not to any of the other rusts in Table 1, thus representing a recognition specificity distinct from other known L alleles (J.G.E., G.J.L., and P.N.D., unpublished results). Transient expression assays showed that L6L11RV represents a modified L6 recognition specificity, because it can induce an HR in response to AvrL567-J, derived from Bs1, but not to other AvrL567 genes (Fig. 1). Recognition of these AvrL567 proteins by L6 therefore depends on one or more of the 11 amino acid differences in the C-terminal region.
AvrL567 and L5/L6 Proteins Interact in Yeast.
We examined whether recognition of AvrL567 proteins involves direct R protein interaction. GAL4 DNA binding domain (BD) and transcriptional activation domain (AD) fusion constructs for the L5, L6, L6L11RV, and AvrL567 proteins were tested for activation of expression from the GAL4 upstream activating sequence in a yeast two-hybrid assay. Coexpression of some of these protein fusions activated expression of the HIS3 (Fig. 2A) and lacZ (Fig. 2B) reporter genes in transformed yeast cells, indicating that these corresponding R and Avr proteins can form physical interactions in yeast. There were some differences between assay systems, with the HIS3 auxotrophy assay more sensitive than the lacZ color assay, whereas fusion of the L proteins to GAL4-AD and AvrL567 proteins to GAL4-BD was more sensitive than the reverse fusions (Fig. 2 A and B). However, overall there was a very close correspondence between the detection of a protein interaction in yeast and the induction of HR during transient expression in planta (Fig. 1), including those combinations that distinguish different specificity classes. For instance, L6 but not L5 interacts with AvrL567-D, and the recombinant L6L11RV protein interacts only with AvrL567-J, in yeast as in planta. Protein interactions between R–Avr combinations that gave only weak responses in planta (Fig. 1) were detected only in the more sensitive yeast assays. For instance, AvrL567-B induced only a chlorotic response when expressed in L6 plants, and interaction between these proteins was detected by the HIS3 assay but not the lacZ assay. The L6 and AvrL567-E combination induced a very weak chlorotic response in planta, and interaction between these proteins was detected only by HIS3 assay with the more sensitive L6-AD/AvrL567-E-BD fusions. The only example where an interaction was detected in yeast but not in planta was between L6 and AvrL567-K, again only by the most sensitive HIS3 assay with the L6-AD/AvrL567-E-BD fusions. It is possible that the interaction between these proteins is too weak to lead to a detectable response in planta. The close correspondence between interaction in yeast and induction of HR in plants indicates that recognition specificity between these resistance and Avr proteins is based on direct interaction. The difference between the L6 and L6L11RV specificities confirms that the LRR plays a critical role in determining the specificity of these interactions.
Fig. 2.
AvrL567 proteins interact specifically with the corresponding R proteins in yeast. (A) Growth of yeast strain HF7c expressing both GAL4-AD and GAL4-BD fusion proteins on minimal media lacking histidine. (Upper) Growth of strains expressing AD::AvrL567 fusion proteins (A to L) with BD alone (-ve), or BD::L5, BD::L6, or BD::L6L11RV fusion proteins. (Lower) Growth of strains expressing the reversed fusions, i.e., BD::AvrL567 proteins (A to L) with AD alone (-ve), AD::L5, AD::L6, or AD::L6L11RV. Growth indicates expression of the HIS3 reporter gene as a result of interaction between the GAL4-AD and GAL4-BD fusion proteins. (B) Yeast strain SFY526 expressing the GAL4 fusion proteins was assayed for β-galactosidase activity. (Upper) Activity of strains expressing AD::AvrL567 proteins (A to L) with BD alone (-ve), BD::L5, BD::L6, or BD::L6L11RV. (Lower) Activity of strains expressing the reversed fusions. (C) β-Galactosidase activity of yeast strain SFY526 expressing GAL4-BD fused to modified L6 proteins containing a K271M (L6ΔP-loop) or a D541V (L6-MHV) mutation along with AD alone (-ve), AD::AvrL567-A, or AD::AvrL567-D. (D) Protein extracts from yeast strain SFY526, and SFY526 expressing AD::L5, AD::L6, AD::L6-D541V (MHV), AD::L6-K271M (P-loop) ADand::L6L11 (Left) or AD::AvrL567 (A to L; Right) were analyzed by immunoblotting with anti-hemagglutinin (HA) or anti-GAL4-AD mAbs. Positions and sizes of protein molecular mass standards are indicated.
AvrL567 Amino Acid Variation Is Associated with Specificity Differences.
RNA gel blot and RT-PCR analysis indicated that all 12 AvrL567 genes were expressed at similar levels in the transiently transformed leaves and rust-infected flax (data not shown), and immunoblot analysis indicated that all of the AvrL567 fusion proteins accumulated to similar levels in yeast (Fig. 2D and data not shown). This finding indicates that the differences in AvrL567 recognition in both flax and yeast result from amino acid sequence differences and several polymorphic sites are associated with specificity variation. For instance, AvrL567-D interacts with L6 but not L5, whereas AvrL567-A, AvrL567-B, AvrL567-F, AvrL567-J, and AvrL567–L interact with L5 and L6 (Figs. 1B and 2 A and B). Three polymorphic amino acid sites (sites 50, 90, and 96) distinguish these proteins (Fig. 1A) and may define residues that form critical contacts with the L5 protein. One of the AvrL567-D polymorphisms (Thr-50) is also present in AvrL567-E, which is weakly recognized by L6 but not L5 in planta and in yeast. The five noninteracting proteins derived from avr haplotypes (C, G, H, I, and K) also contain a polar (Thr) or charged (Lys) residue at this position, rather than the hydrophobic residues (Ile or Val) found in the L5 interacting proteins, which could explain their lack of recognition by L5. In addition, the five inactive proteins share an Asp-56 polymorphism, whereas the active proteins contain either Asn or Lys at this site, which may be critical for L6 interaction. However, the noninteracting proteins also each contain a number of unique polymorphisms that may collectively inhibit recognition. AvrL567-J contains a single distinguishing polymorphism, Ser-96, that may explain its recognition by the L6L11RV recombinant. AvrL567-E differs from AvrL567-J at only two amino acid positions and is in fact unique in combining the His-26 and Thr-50 polymorphisms, which may explain its lack of recognition by L5 and L6L11RV and very weak recognition by L6. Secondary structure predictions (22) suggest that residues 50 and 56 are located in a β-strand with their side chains likely to be solvent-exposed, whereas the polymorphic residues at positions 90 and 96 are predicted to be located in a loop region, so these residues are also well positioned for contacting an interacting protein.
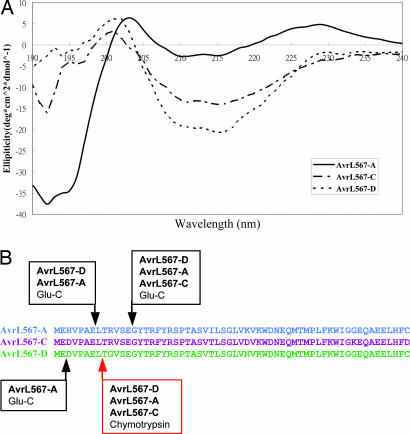
These amino acid differences could affect recognition through the presentation of an altered protein surface on a conserved structural backbone or through significant differences in the overall protein structure. To distinguish these possibilities we compared several structural characteristics of Escherichia coli-expressed AvrL567 proteins. These proteins show no significant sequence or structural similarities to known proteins or functional motifs by blast (23) or threading analysis (wurst, ref. 24; 3d-pssm, ref. 25), suggesting that they may have a novel 3D structure. AvrL567-A, AvrL567-C, and AvrL567-D proteins expressed in E. coli behave as monomeric species, as assessed by size exclusion chromatography (Fig. 4, which is published as supporting information on the PNAS web site) and dynamic light scattering (data not shown). To analyze the secondary structure content of the expressed proteins, we measured CD spectra (Fig. 3A). All three proteins showed very similar spectra, with minima and maxima occurring at similar wavelengths, and estimates of secondary structure content were consistent among the three proteins, indicating that they adopt a similar folded structure with mostly β-sheet and only a small proportion of helical structure as estimated by the programs k2d (24), selcon 3, continll, and cdsstr (26). Limited proteolysis with endopeptidase Glu-C or chymotrypsin indicated that the majority of the proteins correspond to a proteolytically resistant structure, suggesting that they contain a single structural domain with a flexible N-terminal region of ≈13 residues accessible to the protease active sites (Fig. 3B). Thus, these three proteins behave as monomeric, single-domain proteins consisting mainly of β-sheet secondary structure, and the specificity differences between them, including lack of recognition of AvrL567-C, are very likely caused by differences in surface-exposed residues rather than gross structural alterations.
Fig. 3.
AvrL567-A, AvrL567-C, and AvrL567–D proteins exhibit similar structural characteristics. (A) CD spectra of purified recombinant AvrL567-A, AvrL567-C, and AvrL567-D proteins. The spectra indicate that the proteins consist mainly of β-sheet secondary structure. (B) AvrL567 proteins were subjected to limited proteolysis with endopeptidase Glu-C (preferentially cleaves C terminal to glutamic acid residues) and chymotrypsin (cleaves C terminal to bulky hydrophobic residues). Treatment with either protease yielded a protected fragment of ≈14 kDa, which remained stable over a period of 24 h (data not shown). N-terminal sequencing and mass spectrometry were used to identify the protected fragments. The observed cleavage sites in the mature AvrL567 protein are indicated by arrows: red, chymotrypsin cleavage sites; black, Glu-C cleavage sites. No cleavage in the C-terminal region was observed.
Mutation of the L6 NBS Abolishes AvrL567 Protein Interaction.
We also examined the effects on AvrL567 interaction of mutations in the conserved P-loop and MHD (Met-His-Asp) motifs of the L6 NBS domain. The P-loop motif contains a conserved lysine residue (Lys-271 in L6) that interacts with the phosphate group of bound nucleotides in many nucleotide binding proteins, and mutations of this residue abolish ATP binding of the tomato I-2 NBS-LRR R protein (27). Furthermore, a Lys-271 to Met substitution in the L6 P-loop motif abolished L6 resistance function and the HR-inducing activity of an autoactive L6 mutant (28). L6-GAL4 fusion proteins carrying this mutation failed to interact with AvrL567 proteins (Fig. 2C) although the mutant protein was equally abundant in yeast as the WT version (Fig. 2D). This finding suggests that the presence of a bound nucleotide is required before the L6 protein can adopt a recognition-competent conformation. A mutation in the P-loop motif also disrupted intramolecular interactions between the N-terminal coiled-coil (CC) and C-terminal NBS-LRR domains of the potato Rx protein (29). Although L6 contains an N-terminal Toll and IL-1 receptor-related domain rather than a CC domain, similar disruption of intramolecular interactions may explain the inability of the L6 P-loop mutant to bind to the AvrL567 ligand. Plant R proteins also contain a conserved tri-peptide motif characterized by the sequence Met-His-Asp (the MHD motif). Substitution of Asp by Val in the MHD motif of Rx results in an autoactive protein that triggers defense signaling in the absence of the Avr product (30), which we have also observed for a similar mutation in the L6 protein (L6-MHV; ref. 28). However, when tested in yeast, the L6-MHV mutant protein maintains the same interaction with AvrL567 proteins as the WT L6 protein (Fig. 2C), indicating that adoption of an active signaling conformation by this protein does not preclude binding of the AvrL567 ligand. It is possible that continued association with the AvrL567 ligand may be required to maintain an active signaling conformation of WT L6 protein during a resistance response.
Conclusions
The flax rust AvrL567 locus is characterized by high levels of sequence variation with 12 variants identified from six rust strains showing up to 20% amino acid differences. This amino acid variation has resulted from diversifying selection and in turn confers clear differences in recognition specificity. Thus the ability to escape host resistance is the most likely source of the selection pressure driving evolution of AvrL567 genes. Similar diversifying selection has also occurred at the L locus of flax (18), although L alleles other than L5, L6, and L7 have evolved to recognize unrelated Avr genes (19) so changes in AvrL567 are clearly not the only influence on L gene evolution. These data are consistent with a coevolutionary arms race between these corresponding Avr and R genes (10). A prerequisite for such a coevolutionary outcome is that the pathogen Avr gene can accumulate mutations that affect recognition without imposing a significant fitness cost by impairing an important pathogenicity function. However, R proteins that confer resistance by detecting changes in host proteins modified by the effector function of their corresponding Avr proteins impose selection against this function. Theoretical modeling has suggested that indirect recognition can lead to stable long-term resistance, whereas direct recognition is likely to lead to relatively rapid evolution of new virulence phenotypes (9). Indeed the Arabidopsis Rpm1, Rps2, and Rps5 loci are characterized by low levels of genetic diversity and the presence of ancient polymorphisms, suggesting that simple balanced polymorphisms for functional and nonfunctional alleles have been maintained over long evolutionary time scales (7, 8). Recognition by a direct R–Avr protein interaction provides an alternative molecular basis that can explain a plant–parasite arms race leading to extensive diversification in corresponding R and Avr genes. In this situation Avr recognition is not related to effector function, so mutations that abolish recognition may retain function with little or no fitness penalty to the pathogen. Indeed we find that gene-for-gene specificity between the flax L and flax rust AvrL567 genes is correlated with the interaction between the encoded proteins in yeast (Fig. 2). Although other yeast- or plant-derived proteins may also be involved in this protein interaction, any ancillary proteins clearly are not critical to the specificity of the interaction, which is observed only when corresponding R and Avr proteins are present. Thus formation of such a recognition complex primarily depends on interaction between the R and Avr proteins, which provides the basis of gene-for-gene specificity in this system. Although the amino acid variation between AvrL567 proteins alters host recognition, it conserves the overall protein structure (Fig. 3), consistent with the maintenance of an as-yet-unknown pathogenicity function. It is noteworthy that all of the virulent rust strains retain and express intact copies of the AvrL567 gene, suggesting a positive fitness value of these genes to the pathogen.
The data presented here for direct recognition associated with high genetic diversity at R and Avr gene loci and previously published data for indirect recognition associated with simple balanced polymorphisms in Arabidopsis–Pseudomonas systems (3–8) represent two alternative outcomes in plant–pathogen coevolution. An important question raised by this observation is what factors affect the adoption of one or the other of these alternative paths? One possible influence is the different evolutionary histories of each plant–pathogen system. P. syringae isolates can infect a range of host plants, and many of the Avr genes detected by R genes in Arabidopsis are derived from P. syringae pathovars specialized to infect other plant species such as peas, beans, tomato, or soybean. On the other hand, flax rust is an obligate parasite that infects only flax. Similarly, H. parasitica is an obligate pathogen coevolved with Arabidopsis, and diversifying selection operates on the corresponding ATR13 and RPP13 loci in this pathosystem (15, 16). The narrow host range and obligate parasitism of these pathogens means that any R–Avr interactions coevolve in a closed system that may impose different constraints than the wider context in which P. syringae effectors have evolved. At a mechanistic level, differences in pathogenicity function of effector proteins may also influence the recognition mechanism adopted. The P. syringae effectors mentioned above generate modifications of their host protein targets involving phosphorylation or proteolytic cleavage, which are recognized by the corresponding resistance proteins. Other effectors may influence host target protein function without producing an easily distinguishable modified form of the target protein (for instance by acting as competitive inhibitors of enzyme function) in which case direct resistance–Avr recognition may be the only option for Avr detection. Insight into these issues will come from defining recognition mechanisms in a range of plant disease systems.
Materials and Methods
Rust and Plant Material.
Flax rust strains C, H, I, Fi, Bs1, and 339 and inoculation procedures have been described (17, 31). Near-isogenic flax lines containing the L5, L6, L7, or L9 alleles have been described by H. H. Flor (32).
Analysis of AvrL567 Sequences from Rust Strains.
Genomic DNA was isolated from germinated rust spores and amplified with the primers 10–1.15 (AAGCTTGAGAGCTCCGCTC) and 10–1.16 (TAATCCTCGTTGACATCAGTC) as described (19). Up to 50 independently cloned PCR products were sequenced from each rust strain to identify all sequence variants, and the number of variants was consistent with the number of AvrL567-hybridizing fragments observed in genomic DNA gel blots (data not shown). AvrL567 variants from rust strain I were assigned to haplotypes by comparing the sequence variants amplified from several diploid progeny of a cross between CH5 and I that were genotyped by restriction fragment length polymorphism analysis and pathotyping. Bs1 is an F2 rust derived from a cross between rust strain WA (contains AvrL567-D and AvrL567–J, data not shown) and Fi, so the presence of only AvrL567–D and AvrL567–J in Bs1 indicates that it is homozygous. AvrL567 coding sequences were aligned, and nucleotide sequence distances were calculated for nonsynonymous and synonymous sites by using the Jukes-Cantor algorithm of mega software, version 1.02 (33). The significance of differences between average pairwise nucleotide distances was assessed by a t test.
Gel Blot Analysis.
Restriction enzyme-digested genomic DNA was separated on 1.0% agarose gels and transferred to Hybond N+ nylon membranes (Amersham Pharmacia). RNA samples were separated on 1.5% agarose gels and transferred to Hybond N+. Prehybridization and hybridization with 32P-dCTP-labeled DNA probes were carried out in 7% (wt/vol) SDS, 1% (wt/vol) BSA, 0.5 M sodium phosphate (pH 7.2), and 1 mM EDTA at 65°C, and filters were washed in 1 × SSC and 0.1% SDS at 65°C.
Transcript Analysis by RT-PCR.
RNA was isolated from leaves of the rust-susceptible flax variety Hoshangabad 6 days after inoculation with rust. Total RNA was reverse-transcribed by using Superscript Reverse Transcriptase (GIBCO/BRL) with an oligo(dT)25 primer and then amplified by Taq polymerase with primers 10–1.15 and 10–1.16 (spanning two intron sites to distinguish cDNA or genomic DNA products). At least 20 independent RT-PCR clones were sequenced.
Transient Expression Assays.
DNA constructs encoding AvrL567 proteins lacking a signal peptide controlled by the cauliflower mosaic virus 35S promoter were generated in the binary vector pTNotTReg as described (19). Agrobacterium tumefaciens (GV3101-pMP90) strains containing these constructs were prepared at an OD600 of 1.0 in liquid MS medium containing 200 μM acetosyringone and infiltrated into flax leaves.
Expression and Purification of AvrL567 Proteins.
The AvrL567-A, AvrL567-C, and AvrL567-D proteins were expressed in E. coli strain BL21(DE3) as ubiquitin fusion proteins containing an N-terminal hexahistidine tag by using the vector pHUE (34). Cultures were grown at 37°C to a OD600 of ≈0.8–1.0 in LB broth and induced with 1 mM isopropyl thio-β-d-galactoside for 18–21 h at 15°C. Cells were harvested by centrifugation at 5,000 × g for 10 min at 4°C and resuspended in 1/10 culture volume of buffer A (20 mM Hepes, pH 7/300 mM NaCl/10 mM imidazole/1 mM PMSF/1 mg/ml aprotinin/1 mg/ml leupeptin/1 mg/ml pepstatin). Cell suspensions were lysed by repeated cycles of freezing and thawing and the addition of lysozyme (0.5 mg/ml), then clarified by centrifugation at 39,000 × g for 30 min at 4°C. The soluble fraction was incubated with Talon Co2+ resin (BD Biosciences) for 1 h, and the resin was washed with buffer A, then buffer B (as buffer A but with 20 mM imidazole), and finally incubated with buffer A containing deubiquitinating enzyme (34) at a 1:50 enzyme to substrate ratio and 10 mM β-mercaptoethanol for 18–20 h. The cleaved AvrL567 proteins were eluted with buffer A and further purified by size exclusion chromatography by using a Hi-Load Superdex 200 16/60 gel filtration column (GE Healthcare, Chalfont St. Giles, U.K.). Purified proteins were concentrated to 10–20 mg/ml by using Amicon Ultra centrifugal filter devices (Millipore), frozen in liquid nitrogen, and stored at −80°C.
Limited Proteolysis.
Purified AvrL567 proteins (1 mg/ml) were digested at 25°C with chymotrypsin or endopeptidase Glu-C in 100 mM Tris (pH 8), 300 mM NaCl, 10 mM CaCl2 or 25 mM NH4HCO3 (pH 7.8), and 300 mM NaCl. Different protein-to-protease ratios were used, aliquots were removed at regular time intervals, and reaction products were separated by electrophoresis in 20% polyacrylamide gels (SDS/PAGE) and visualized by Coomassie brilliant blue R250 (BioRad).
Protein Sequencing.
Samples for N-terminal sequencing were transferred onto Hybond-P poly(vinylidene difluoride) membranes (GE Healthcare) after SDS/PAGE and stained with Ponceau S (Sigma). Appropriate bands were excised and sequenced with an Applied Biosystems Procise 492 cLC protein sequencer as per instructions in the users' manual.
Mass Spectrometry.
Samples for mass analysis were desalted and concentrated by precipitation with chloroform/methanol as described (35) and resuspended in 50% (vol/vol) acetonitrile/0.1% acetic acid. Samples were either infused directly into an Applied Biosystems API QSTAR Pulsar1 ESI mass spectrometer or spotted onto the sample stage with sinapinic acid for analysis by using an Applied Biosystems Voyager-DE STR mass spectrometer. Proteolysis samples were purified by chromatography on a Zorbax reverse-phase 300SB C3 column (5 mm, 150 × 2.1 mm) with a linear 0–80% acetonitrile gradient in 0.1% formic acid at 200 ml/min over 40 min and subjected to mass analysis by using a QSTAR Pulsar QqTOF ESI mass spectrometer (Applied Biosystems).
CD.
CD spectra were recorded on a Jasco J810 spectropolarimeter at 25°C by using protein samples at a concentration of 6.79, 6.75, and 3.62 mg/ml in 20 mM sodium phosphate, pH 7 for AvrL567-A, AvrL567-C, and AvrL567-D, respectively. Measurements were recorded at 0.5-nm wavelength increments from 180 to 260 nm at 50 nm/min by using a 0.1-mm path length cell, 0.5-nm band width, 1-s response time, and five accumulations and corrected for buffer base line contribution.
Yeast Two-Hybrid Analysis.
GAL4 DNA BD and transcriptional AD fusions were prepared in the pGBT9 and pGADT7 vectors, respectively (Clontech). An L6 cDNA clone was amplified by Pfu polymerase with the primers L6PC1 (GGGAGATCTTGAACAAGTTTTGGAGACCA) and L6PC2 (GGGCTGCAGGTCATCTGTAG-GGCTGATCGGG), and a BglII/PstI fragment encoding amino acids 29–1291 of L6 was inserted into BamHI/PstI-digested vectors. The first 28 amino acids of L6 were omitted because this region has a potential signal anchor function and may have interfered with correct localization of the fusion protein. The L6-D541V and L6-K271M mutations (28) and L6L11RV recombinant were introduced into the L6-GAL4 fusion constructs by insertion of appropriate restriction fragments. A cDNA clone of L5 was generated by RT-PCR of RNA from flax line Wilden with L6PC1 and L6PC2, and then inserted into the fusion vectors as above. AvrL567 fusion constructs encode amino acids 21–150 fused to GAL4-BD or GAL4-AD through an in-frame BamHI site. Yeast transformation, His growth, and lacZ assays were performed as described in the Yeast Protocols Handbook (Clontech). Yeast proteins were extracted by the trichloro-acetic acid method, separated by SDS/PAGE, and transferred to nitrocellulose membranes (Pall) by electroblotting in a Bio-Rad Mini2D apparatus. Membranes were blocked with SuperBlock-TBST (Pierce,) probed with anti-GAL4-AD, anti-GAL4-DNA-BD (Clontech), or anti-hemagglutinin (Roche Molecular Systems) mAbs followed by a blocking step with normal goat serum (Pierce) and detection with goat anti-mouse/horseradish peroxidase (Pierce). Labeling was detected with the SuperSignal West Pico chemiluminescence kit (Pierce).
Supplementary Material
Acknowledgments
We thank Valerie Ryle, Kim Newell, and Patricia Atkinson for excellent technical assistance and Alun Jones and Chris Wood for assistance with mass spectrometry and N-terminal sequencing. This research was supported by grants from the Grains Research and Development Corporation and the Australian Research Council. A.-M.C. was supported by an Australian Postgraduate Award. B.K. is a National Health and Medical Research Council Research Fellow.
Abbreviations
- NBS
nucleotide binding site
- LRR
leucine-rich repeat
- Avr
avirulence
- R gene
resistance gene
- R protein
resistance protein
- HR
hypersensitive response
- BD
binding domain
- AD
activation domain.
See Commentary on page 8575.
References
- 1.Holt B. F., Hubert D. A., Dangl J. L. Curr. Opin. Immun. 2003;15:20–25. doi: 10.1016/s0952-7915(02)00014-6. [DOI] [PubMed] [Google Scholar]
- 2.Abramovitch R. B., Martin G. B. Curr. Opin. Plant Biol. 2004;7:356–364. doi: 10.1016/j.pbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Mackey D., Holt B. F., Wing A., Dangl J. L. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 4.Mackey D., Belkhadir Y., Alonsos J. M., Ecker J. R., Dangl J. L. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 5.Axtell M. J., Staskawicz B. J. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 6.Shao F., Golstein C., Ade J., Stoutemyer M., Dixon J. E., Innes R. W. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 7.Stahl E. A., Dwyer G., Mauricio R., Kreitman M., Bergelson J. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- 8.Mauricio R., Stahl E. A., Korves T., Tian D., Kreitman M., Bergelson J. Genetics. 2003;163:735–746. doi: 10.1093/genetics/163.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van der Hoorn R. A. L., De Wit P. J. G. M., Joosten M. H. A. J. Trends Plant Sci. 2002;7:67–71. doi: 10.1016/s1360-1385(01)02188-4. [DOI] [PubMed] [Google Scholar]
- 10.Stahl E. A., Bishop J. G. Curr. Opin. Plant Biol. 2000;3:299–304. doi: 10.1016/s1369-5266(00)00083-2. [DOI] [PubMed] [Google Scholar]
- 11.Michelmore R. W., Meyers B. C. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 12.Lahaye T., Bonas U. Trends Plant Sci. 2001;6:479–485. doi: 10.1016/s1360-1385(01)02083-0. [DOI] [PubMed] [Google Scholar]
- 13.Allen R. L., Bittner-Eddy P. D., Grenville-Briggs L. J., Meitz J. C., Rehmany A. P., Rose L. E., Beynon J. L. Science. 2004;306:1957–1960. doi: 10.1126/science.1104022. [DOI] [PubMed] [Google Scholar]
- 14.Rose L. E., Bittner-Eddy P. D., Langley C. H., Holub E. B., Michelmore R. W., Beynon J. L. Genetics. 2004;166:1517–1527. doi: 10.1534/genetics.166.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia Y., McAdams S. A., Bryan G. T., Hershey H. P., Valent B. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deslandes L., Olivier J., Peeters N., Feng D. X., Khounlotham M., Boucher C., Somssich I., Genin S., Marco Y. Proc. Natl. Acad. Sci. USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis J. G., Lawrence G. J., Luck J. E., Dodds P. N. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodds P. N., Lawrence G. J., Pryor A. J., Ellis J. G. In: Molecular Plant Pathology. Dickinson M., Beynon J., editors. Sheffield, U.K.: Sheffield Academic; 2000. pp. 88–107. [Google Scholar]
- 19.Dodds P. N., Lawrence G. J., Catanzariti A.-M., Ayliffe M. A., Ellis J. G. Plant Cell. 2004;16:755–768. doi: 10.1105/tpc.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes A. L., Nei M. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 21.Islam M. R., Mayo G. M. E. Plant Breed. 1990;104:89–100. [Google Scholar]
- 22.Andrade M. A., Chacon P., Merelo J. J., Moran F. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torda A. E., Procter J. B., Huber T. Nucleic Acids Res. 2004;32:W532–W535. doi: 10.1093/nar/gkh357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley L. A., MacCallum R. M., Sternberg M. J. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 26.Sreerama N., Woody R. W. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 27.Tameling W. I. L, Elzinga S. D., Darmin P. S., Vossen J. H., Takken F. L. W., Haring M. A., Cornelissen B. J. C. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howles P. H., Lawrence G., Finnegan J., McFadden H., Ayliffe M., Dodds P., Ellis J. Mol. Plant Microbe Interact. 2004;18:570–582. doi: 10.1094/MPMI-18-0570. [DOI] [PubMed] [Google Scholar]
- 29.Moffet P., Farnham G., Peart J., Baulcombe D. C. EMBO J. 2002;21:4511–4519. doi: 10.1093/emboj/cdf453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendahmane A., Farnham G., Moffett P., Baulcombe D. C. Plant J. 2002;32:195–204. doi: 10.1046/j.1365-313x.2002.01413.x. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence G. J., Mayo G. M. E., Shepherd K. W. Phytopathology. 1981;71:12–19. [Google Scholar]
- 32.Flor H. H. Adv. Agron. 1954;6:152–161. [Google Scholar]
- 33.Kumar S., Tamura K., Nei M. mega: Molecular Evolutionary Genetics Analysis. University Park: Pennsylvania State University; 1993. version 1.02. [Google Scholar]
- 34.Catanzariti A.-M., Soboleva T. A., Jans D. A., Board P. G., Baker R. T. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobe B., Jennings I. G., House C. M., Feil S. C., Michell B. J., Tiganis T., Parker M. W., Cotton R. G. H., Kemp B. E. Protein Sci. 1997;6:1352–1357. doi: 10.1002/pro.5560060626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.