Abstract
Plants can perceive a wide range of biotic attackers and respond with targeted induced defenses. Specificity in plant non-self-recognition occurs either directly by perception of pest-derived elicitors or indirectly through resistance protein recognition of host targets that are inappropriately proteolyzed. Indirect plant perception can occur during interactions with pathogens, yet evidence for analogous events mediating the detection of insect herbivores remains elusive. Here we report indirect perception of herbivory in cowpea (Vigna unguiculata) plants attacked by fall armyworm (Spodoptera frugiperda) larvae. We isolated and identified a disulfide-bridged peptide (+ICDINGVCVDA−), termed inceptin, from S. frugiperda larval oral secretions that promotes cowpea ethylene production at 1 fmol leaf−1 and triggers increases in the defense-related phytohormones salicylic acid and jasmonic acid. Inceptins are proteolytic fragments of chloroplastic ATP synthase γ-subunit regulatory regions that mediate plant perception of herbivory through the induction of volatile, phenylpropanoid, and protease inhibitor defenses. Only S. frugiperda larvae that previously ingested chloroplastic ATP synthase γ-subunit proteins and produced inceptins significantly induced cowpea defenses after herbivory. Digestive fragments of an ancient and essential plant enzyme, inceptin functions as a potent indirect signal initiating specific plant responses to insect attack.
Keywords: elicitor, guard hypothesis, indirect perception, insect herbivory, plant defense
A mechanistic understanding and targeted improvement of plant resistance traits are recognized as essential in combating yield losses from crop pests. Plants can perceive and defensively respond to attack either directly by impeding pest growth or indirectly by promoting advantageous interactions with beneficial organisms (1–7). Great progress has been made in the identification of plant receptor-like kinase families mediating perception of biotic attack and the subsequent activation of signal transduction cascades spanning interactions of GTP binding proteins, mitogen-activated protein kinases, phytohormones, transcription factors, and ultimately induced biochemical defenses (2, 8). Despite these advances, relatively few candidate elicitors and ligands responsible for the initiation and specificity of induced plant defenses to pest attack have been identified (1, 2). This void is especially acute in the case of insect herbivore perception and is surprising given both the significance of plant–insect interactions in arthropod and angiosperm evolution and the role of insects in facilitating plant pathogen entry (9, 10).
Induced plant defenses are initiated in part by the direct perception of elicitors derived from offending organisms. For example, maize (Zea mays) and tobacco (Nicotiana attenuata) perceive insect attack through the direct detection of fatty acid amino acid conjugate (FAC) elicitors present in insect oral secretions (OS). Plants respond with indirect defenses in the form of induced volatile emissions that function as long-range attractants enabling predators and parasitoids to locate their respective prey and hosts (11–13). Receptor-like binding of FAC elicitors in maize has been reported; however, this plasma membrane protein remains unidentified (14). Alternatively, plants may also indirectly perceive biotic attack. For example, proteases released from phytopathogenic bacteria type-III secretion systems have been demonstrated to cleave specific host targets, resulting in biochemical defects that in turn are recognized by resistance proteins (R proteins) (3–5). Unlike plant pathogens, the analogous indirect perception of insect attack is unknown.
Efforts to understand plant perception of insect herbivory demonstrated that FAC elicitors possessed little or no volatile inducing activity in lima bean (Phaseolus lunatus) and cotton (Gossypium hirsutum) (15), suggesting the presence of unidentified elicitors. To explore additional mechanisms of plant perception of insects, we isolated a peptide elicitor from Spodoptera frugiperda OS that rapidly induces defenses in cowpea and beans (Phaseolus vulgaris). Larvae feeding studies on diets with the presence or absence of Escherichia coli-expressed proteins demonstrate that the elicitor is a proteolytic product of chloroplastic ATP synthase. Similar to “guard-based” perception systems for phytopathogenic bacteria, plants perceive insect herbivory through the indirect detection of perturbations in their proteome (3).
Results and Discussion
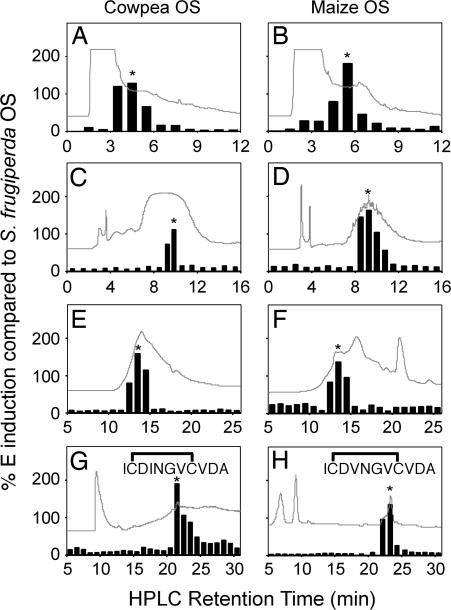
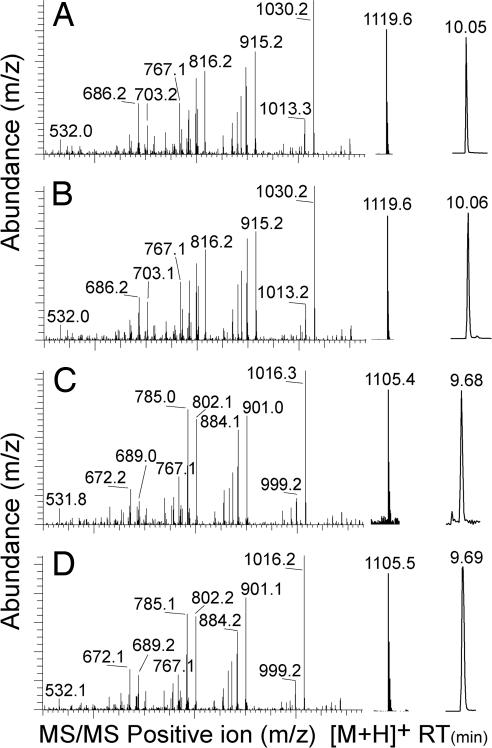
Outside of a few experimental systems, how many plants specifically recognize insect attack is largely unknown (1). Using cowpea plants, we confirmed the inactivity of established FAC elicitors yet consistently detected responses from S. frugiperda OS (see Fig. 6, which is published as supporting information on the PNAS web site). This result was unexpected given that FAC elicitors were first described in the genus Spodoptera (1, 12). We collected 100-ml samples of OS from S. frugiperda that had fed on either cowpea or maize (Z. mays), respectively, and used a cowpea leaf ethylene (E) induction assay to drive the fractionation of biological activity. Biotic attack often induces production of E and provides a useful marker for exploring plant non-self-perception (1, 16). Crude OS was acidified, partitioned with MeCl2, and centrifuged. The aqueous phase was sequentially fractionated on reverse-phase (RP) C18 and strong anion exchange solid-phase extraction (SPE) columns. Individual OS fractions containing the highest level of E-inducing activity were subjected to a series of HPLC separations including strong cation exchange (Fig. 1 A and B), RP-C18 (Fig. 1 C and D), gel filtration (Fig. 1 E and F), and normal-phase chromatography (Fig. 1 G and H). The final active fractions contained ≈2–3 μg of material. Positive ion electrospray LC-MS analysis of each active fraction revealed [M+H]+ ions with m/z ratios of 1,119.6 and 1,105.4 for the cowpea- and maize-derived S. frugiperda OS elicitors, respectively (Fig. 2 A and C). Chemical N-terminal sequencing demonstrated the peptide sequences +I-X-DINGV-X-VDA− and +I-X-DVNGV-X-VDA− for the cowpea- and maize-derived elicitors, respectively. Considering that cysteine produces a blank (X) cycle, we synthesized the two closely related 11-aa acidic disulfide-bridged peptides +ICDINGVCVDA− and +ICDVNGVCVDA− and confirmed their structure by chromatography and MS fragmentation with the isolated natural products (Fig. 2 B and D).
Fig. 1.
HPLC purification of inceptins from maize- and cowpea-derived S. frugiperda OS. Fractions inducing E production in cowpea leaves, denoted by an asterisk, were isolated by a series of strong cation exchange (A and B), RP-C18 (C and D), gel filtration (E and F), and normal-phase chromatography (G and H). UV traces (λ = 200 Å) are overlaid on an arbitrary scale. Fractions were sequentially collected, desalted, evaporated, and resolubilized in H2O for leaf bioassays. Final purification resulted in single fractions (∗) used for MS, Edman N-terminal sequencing, and confirmation with synthetic peptides.
Fig. 2.
LC-MS confirmation of natural and synthetic inceptins. From left to right, MS fragment ions, predominant positive m/z [M+H]+ ions, and LC retention time (RT) of parent ion peak of isolated natural product inceptin from cowpea-derived S. frugiperda OS (A), synthetic cowpea inceptin (B), isolated natural product inceptin from maize-derived S. frugiperda OS (C), and synthetic maize inceptin (D).
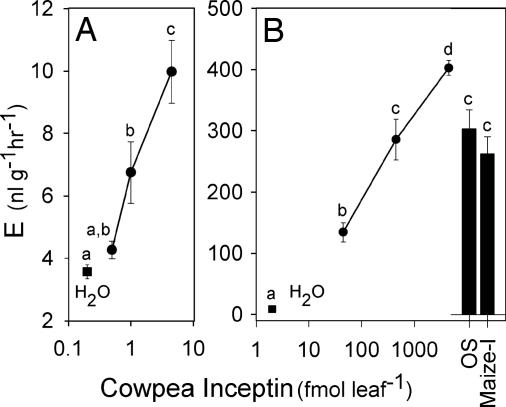
To confirm activity, wounded cowpea leaves were treated with synthetic inceptins. Plant responses to cowpea inceptin began at 1 fmol leaf−1 and proved highly linear up to 4,500 fmol leaf−1 (Fig. 3). Treatment of wounded leaves with 1 μl of cowpea-derived S. frugiperda OS produced an equivalent response to 450 fmol of either cowpea- or maize-derived inceptin (Fig. 3B). The independent isolation of nearly identical active OS peptides from S. frugiperda on diverse host plants demonstrates that this activity is highly specific to the inceptin sequence. Moreover, the exquisitely low level of inceptin required is consistent with the activity of established peptide signals with known ligand–receptor interactions (17–20).
Fig. 3.
Inceptins are potent inducers of E production in cowpea leaves. Average (n = 6; ±SEM) E production of damaged cowpea leaves treated with synthetic cowpea inceptin ranging from 0.45 to 4.50 fmol (A) and 45 to 4,500 fmol (B). Damaged leaves treated with H2O only (filled squares) were arbitrarily placed at 0.2 and 2.0 on the x axes of A and B, respectively. Filled bars represent leaf responses to 1 μl of cowpea-derived S. frugiperda OS and 450 fmol of maize inceptin (Maize-I). Different letters (a–d) represent significant differences. (All ANOVA P values were <0.0001. Tukey test corrections for multiple comparisons: P < 0.05.)
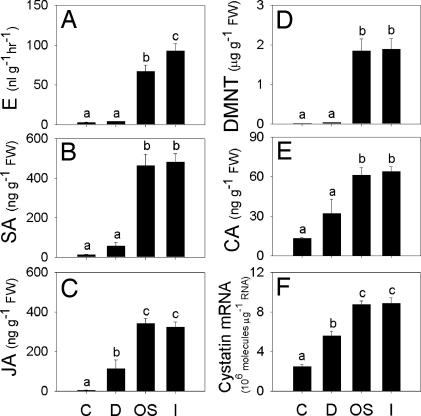
To test the hypothesis that inceptin is the primary elicitor of plant responses in this system, we compared the activity of S. frugiperda OS and inceptin on the induced accumulation of jasmonic acid (JA) and salicylic acid (SA), which are generally associated with wound and pathogen defense signaling, respectively (6, 7). Both 1 μl of cowpea-derived OS and inceptin (450 fmol) induced quantitatively similar levels of E, SA, and JA that were significantly greater than damage plus H2O alone (Fig. 4A–C). Through multiple synergistic and antagonistic crosstalk interactions E, JA, and SA are believed to mediate the specificity of induced plant defenses to biotic attack (21).
Fig. 4.
Inceptin and S. frugiperda OS induce similar phytohormone and defense responses. Leaves were either undamaged controls (C) or were damaged with the addition of H2O (D), 1 μl of cowpea-derived OS (OS), or 450 fmol of synthetic cowpea inceptin (I). Shown are average (n = 6, +SEM) E production (A) and leaf concentrations of SA (B), JA (C), DMNT (D), cinnamic acid (E), and cystatin protease inhibitor gene expression (F) 4 h after treatments. Different letters (a–c) represent significant differences. (All ANOVA P values were <0.0001. Tukey test corrections for multiple comparisons: P < 0.05.)
To estimate activation of both indirect and direct defenses, we quantified the volatile homoterpene (E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) and cinnamic acid and transcript levels of the protease inhibitor cystatin in leaves. S. frugiperda OS and inceptin induced equivalent leaf DMNT levels (Fig. 4D) and emission of volatiles including DMNT, (E)-β-ocimene, methyl salicylate, indole, (E)-β-farnesene, (E,E)-α-farnesene, and (3E,7E)-4,8,12-trimethyl-1,3,7,11-tridecatetraene (see Fig. 7, which is published as supporting information on the PNAS web site). These insect-inducible plant volatiles have established roles in the positive associative learning and attraction of natural enemies (22, 23). Inceptin and OS also increased cinnamic acid levels, a product of l-phenylalanine ammonia-lyase and precursor to phenylpropanoid defenses induced by biotic attack (Fig. 4E) (24). Transcripts for cowpea protease inhibitor cystatin were also equally induced in the leaves by S. frugiperda OS and inceptin (Fig. 4F). Wounding and insect herbivory often induce protease inhibitor production, which can function as an antinutritive defense by interfering with insect digestion (25).
To test the effect of inceptins on induced direct defenses, cowpea foliage-reared sixth-instar S. frugiperda were caged for 12 h on damaged leaves of intact plants treated 24 h previously with either H2O or 450 fmol inceptin. Larvae demonstrated a mass gain of 51.2 ± 2.6% and 42.2 ± 2.1%, respectively, demonstrating that inceptin treatment resulted in a significant (n = 35; paired Student’s t test, t = −2.572, P = 0.012) reduction in larvae biomass accumulation. Combined, these results demonstrate that inceptin activates plant defense in cowpea.
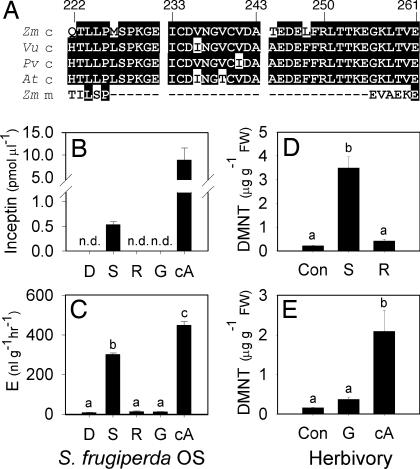
A blast (www.ncbi.nlm.nih.gov/blast) search using the amino acid sequences of cowpea and maize inceptin revealed high homology to chloroplastic ATP synthase γ-subunit (cATPC) 1 encoded by atpC1 from rice (GenBank accession no. XM_478377). A blast search using rice cATPC1 against all translated GenBank sequences revealed a nonannotated maize mRNA (GenBank accession no. AY108268) containing an identical match to the maize-derived inceptin sequence (+ICDVNGVCVDA−) (Fig. 5A). To obtain the cATPC sequence for cowpea, which was not present in the database, primers were designed to conserved regions of cATPC, and PCR product was amplified from cowpea leaf cDNA, cloned, and sequenced. The predicted amino acid sequence from the cowpea cATPC homologue (GenBank accession no. DQ312300) contained an exact match to the inceptin peptide isolated from cowpea-derived S. frugiperda OS (Fig. 5A). Alignments of the translated cATPC genes from multiple plants demonstrate a high degree of conservation in the amino acid sequence that corresponds to the predicted source of inceptins (see Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 5.
Inceptin is derived from cATPC and regulates perception of S. frugiperda attack. (A) Predicted amino acid sequences from chloroplastic (c) atpC mRNA sequences of Z. mays (Zm_c), V. unguiculata (Vu_c), P. vulgaris var. black bean (Pv_c), A. thaliana (At_c), and Z. mays mitochondrial (m) (Zm_m). Inceptin is located within amino acids 233–243 based on unprocessed full-length Zm_c. Shown are average (n = 4, +SEM) inceptin levels (B) and cowpea E production (C) from 1 μl of S. frugiperda OS from larvae fed artificial diet (D), cowpea shoots (S), or roots (R) containing H2O or E. coli-expressed proteins GST (G) and cATPC-GST (cA). n.d., not detectable (<10 fmol). (D and E) Average (n = 6, +SEM) cowpea DMNT concentrations 4 h after leaves were untreated (Con) or fed upon by larvae that had previously consumed shoots or roots (D) or artificial diet containing G or cA (E). Different letters (a–c) represent significant differences. (All ANOVA P values were <0.001. Tukey test corrections for multiple comparisons: P < 0.05.)
ATP synthase is a highly conserved enzyme catalyzing the synthesis of ATP from ADP and phosphate by means of a flux of protons over an electrochemical gradient. Chloroplastic ATP synthase consists of a CF1CF0 complex composed of nine different subunits encoded by the genes atpA through atpI (26). The cATPC protein resides in CF1 and contains an extra domain, absent from mitochondrial ATPC, spanning the inceptin peptide (Fig. 5A), which is susceptible to trypsin cleavage (27). ATPase is activated by reduction of the cATPC cysteine disulfide bridge, present in the inceptin fragment, which abolishes interactions with the inhibitory ε-subunit (27). Thus, inceptin fragments are derived from a regulatory domain unique to cATP synthases.
To test the hypothesis that inceptins are proteolytic fragments of cATPC that mediate plant defense during herbivore attack, we collected OS from S. frugiperda larvae fed an artificial diet, cowpea shoots, or roots mixed with either H2O or 60 μg of purified E. coli-expressed protein containing either GST or a partial cATPC-GST fusion protein. After 12 h of feeding, only OS from S. frugiperda on diets containing shoots or roots mixed with cATPC-GST contained inceptin (Fig. 5B). No inceptin was found in larval OS from artificial diet, roots, or roots plus GST protein. Similarly, only OS containing inceptin promoted E production (Fig. 5C). Purified cATPC-GST protein alone is not biologically active (see Fig. 9, which is published as supporting information on the PNAS web site). Importantly, cowpea leaf-derived OS contained 530 fmol μl−1 inceptin and accounts for the matching activity of synthetic inceptins (450 fmol μl−1) and crude OS (Fig. 3B). Cowpea responses to herbivory by sixth-instar S. frugiperda previously fed roots, shoots, or artificial diet with 9 μg of purified GST or cATPC-GST protein follow the same predicted patterns of DMNT (Fig. 5 D and E) and phytohormone (see Fig. 10, which is published as supporting information on the PNAS web site) production, demonstrating that inceptins regulate plant perception during insect attack. To examine inceptin perception in other species, we tested the synthetic predicted elicitor +ICDVNGVCIDA− from bean (P. vulgaris) on bean leaves at 1 pmol leaf−1 and found similar E- and volatile-inducing activity (see Fig. 11, which is published as supporting information on the PNAS web site). Preliminary experiments failed to detect similar biological activity of inceptins in maize and tobacco (data not shown).
S. frugiperda larvae assimilate the nitrogen in plant tissue through the proteolytic action of endopeptidases such as trypsin and exopeptidases like aminopeptidases and carboxypeptidases (28). A combination of these midgut enzyme activities is likely involved in the proteolysis of cATPC and production of inceptin. Curiously, the cATPC precursor to inceptin is predicted to be present only in chloroplasts and may enable pod-, stem-, and root-feeding insects to avoid elicitor generation. With processing external to plant tissues, inceptin signaling could offer a conceptual advantage in engineering plant resistance. Transgenic plants expressing recombinant proteins, each harboring multiple copies of inceptin, should preserve plants in an uninduced state yet increase perception and response during herbivore attack. Given the sequence conservation and similar response to corn- and cowpea-derived inceptins, S. frugiperda larvae that move onto cowpea plants are envisioned to be rapidly perceived through cATPC fragments originating from previously fed-on hosts.
Selected examples of defense-related peptide signaling in plants include systemin and bacterial-derived Flg22. Systemin is an 18-aa peptide produced upon wounding by plant proteolysis of prosystemin, which then binds the SR160/BRI1 receptor, a leucine-rich repeat receptor-like kinase, and is responsible for a broad array of jasmonate-mediated defense responses (19). Derived from the flagellin protein of phytopathogenic bacteria, the peptide fragment Flg22 interacts with the FLS2 receptor, also a leucine-rich repeat receptor-like kinase, and initiates disease-resistance responses (20). Flg22 is specific to bacterial invasion, highly conserved among classes of bacteria, and widely perceived by plants. Inceptin shares a number of similarities as well as differences with systemin and Flg22. Like systemin, inceptins are plant-derived yet are similar to Flg22 in their origination from highly conserved proteins with essential metabolic functions.
Unlike systemin and Flg22, inceptin perception is consistent with the guard hypothesis. In this model, R proteins function as a surveillance system to trigger resistance upon indirect detection of an intermediate plant protein that is modified by protease action from an offending organism (3). For example, in Arabidopsis, the Pseudomonas syringae type-III secretion systems releases the cysteine protease AvrRpt2 that cleaves the plant RPM1-interacting protein 4 (RIN4). Proteolysis of RIN4 is required for activation of the R protein RPS2 and resistance responses (4, 29). Similarly, the P. syringae type-III secretion system also releases the protease AvrPphB, which targets the plant serine/threonine kinase PBS1. Cleavage of PBS1 into two polypeptides is required for the activation of the R protein RPS5 and subsequent plant defense (5). Compared with the wealth of information on plant perception of bacterial attack, few R genes have been cloned and demonstrated to regulate insect resistance (30). Moreover, there are still no clear examples of defined ligand–R protein interactions that specifically regulate insect resistance. We hypothesize that cATPC is a target of specific insect proteases and, given the extremely low levels of inceptin required for activity, also a candidate ligand for plant receptors. In this work we demonstrate the indirect plant perception of attack by herbivores and validate the detection of inappropriate proteolytic fragments of “self” as a generalized strategy for plant non-self-recognition.
Materials and Methods
Plant and Insect Material.
S. frugiperda larvae were obtained from R. Meagher (Center of Medical, Agricultural, and Veterinary Entomology, Agricultural Research Service, U.S. Department of Agriculture) and reared on a pinto bean-based diet (31). Cowpea (Vigna unguiculata var. California Blackeye no. 5; The Wax Company, Amory, MS), maize (Z. mays var. Golden Queen), and black bean (P. vulgaris) were germinated in a professional grower’s soil mix (Piedmont Pacific, Statham, GA) supplemented with 14–14-14 Osmocote (Scotts, Marysville, OH). All plants were maintained in a greenhouse with a 12-h photoperiod, a minimum of 300 μmol−2 s−1 of photosynthetically active radiation supplied by supplemental lighting, 70% relative humidity, and a temperature cycle of 24°C/28°C (night/day). Hydroponic plants, for harvesting of clean root material, were grow as previously described (32).
Cowpea Leaf Bioassays.
All experiments used 2- to 3-week-old plants containing two fully expanded pairs of trifoliate leaves. For all induction assays, the adaxial sides of new fully expanded leaves were superficially scratched with a razor in three areas, removing ≈5% of the total waxy cuticle. The damage sites (2 cm2 each) included the central leaf tip spanning both sides of the midrib and two midbasal sections on opposite sides of the midrib. Test solutions in 5 μl of H2O were immediately applied and dispersed over the damage sites. Leaves remained on the intact plants for 1, 4, and 5 h before E, leaf metabolite/mRNA, and volatile emission sampling, respectively.
Elicitor Isolation.
Two 100-ml samples of OS from sixth-instar S. frugiperda larvae, fed separately on either cowpea or maize for at least 12 h, were collected (33). Crude OS was acidified with HCl to pH 1, partitioned with an equal volume of CH2Cl2, and centrifuged at 12,000 × g for 15 min. The aqueous phase was fractionated on a 5-g RP-C18 SPE column (Supelco, Bellefonte, PA), and activity was eluted with 10 ml of 1:1 CH3CN:H2O. Strong anion exchange was then performed on a 5-g SPE column by loading the sample in 1:4 ACN:H2O and 10 mM NH4CH3COOH (pH 5) and eluting activity with the same buffer containing 1.0 M NaCl. Activity was then HPLC-fractionated by using a P4000 pump, an AS3000 autosampler, and a UV6000LP detector (Thermo Separation Products, San Jose, CA). All HPLC samples were dissolved in the initial mobile phase (MP), and 1-min fractions were collected, desalted by using RP-C18 SPE columns, bioassayed, and stored for further purification at −70°C. Strong cation exchange HPLC used a Polysulfoethyl A column (250 × 9.4 mm, 5 μm, 300 Å; The Nest Group, Southboro, MA), a flow rate of 5 ml min−1, and MP A and B, both containing 1:4 CH3CN:H2O (pH 3.0) and 25 mM KH2PO4 with the addition of 0.5 M KCl to MP B. Activity, between 3 and 6 min, was eluted with binary gradient of 100% A to 100% B over 20 min. Multiple 10-mg injections were pooled, with each repeated 1-min fraction collection combined for each shared time interval. Fractions with maximal activity (4–5 min) were subjected to RP-C18 by using a YMC ODS-AQ column (250 × 4.6 mm, S-5 μm, 20 nm; Waters, Milford, MA) heated to 60°C with a flow rate of 1 ml min−1, with MP A and B containing 95:5 H2O:CH3CN and 9:1 CH3CN:H2O, respectively. Both solutions were buffered with 10 mM NH4COOH. Activity was eluted with MP A isocratic for 2 min followed by a binary gradient of 100% A to 100% B over 18 min. Active factions, eluting at 9–10 min, were separated by gel filtration by using a Tricorn Superdex Peptide 10/300 GL column (Amersham Pharmacia Biosciences) and an isocratic 1-ml min−1 flow of H2O containing 100 mM NH4CH3COOH. Activity eluting at 13 min was further fractionated by using a normal-phase carbamoyl-bonded TSKgel Amide-80 column (250 × 4.6 mm; Tosoh), a 1-ml min−1 flow rate, with MP A and B containing 95:5 CH3CN:H2O and H2O, respectively. Both solutions were buffered with 25 mM NH4COOH. MP A was held isocratic for 2 min and followed by a linear binary gradient reaching 1:1 A:B over 28 min. E-inducing activity eluted between 21 and 24 min.
Phytohormone and Biochemical Analysis.
GC-based quantification of elicitor-induced E production followed from Schmelz et al. (31) with modification. One hour after treatment, experimental leaves were excised and sealed in 13-ml tubes for an additional hour before headspace sampling. To estimate additional metabolites, cowpea leaves were left as undamaged controls or were damaged and treated with 5 μl of an aqueous solution containing H2O only, 1 μl of cowpea-derived S. frugiperda OS, or 450 fmol of inceptin. At time 0, half of this application was applied on one-half of the midrib, with the second half of the leaf treated 2 h later. All leaves were harvested at 4 h in liquid N2 for metabolite and transcript analyses. Isobutane chemical ionization GC-MS-based leaf tissue quantification of JA, SA, cinnamic acid, and DMNT was performed as described (34). Leaf pools of DMNT were quantified based on an external stand curve of synthetic DMNT and monitoring the [M+H]+ m/z ion 151 at retention time 7.15 min. Collection, quantification, and confirmation of leaf volatile emission followed established protocols (32). Five hours after treatments, additional individual leaves were excised, weighed, and placed in volatile collection chambers for 30 min. To normalize differences in leaf mass, volatile emission was reported as ng g−1 30 min−1.
Inceptin Characterization and Synthesis.
Selected samples were analyzed with an LC, identical to the purification system, coupled to an LCQ Deca XPMAX (Thermo Electron, San Jose, CA) ion trap MS. RP-C18 columns, MP, and gradients were as described in the HPLC isolation. The 1-ml min−1 flow was split, allowing 0.1 ml min−1 to enter the ion source. N-terminal sequencing was performed at the Institute of Biological Chemistry (Washington State University, Pullman) with Edmund chemistry degradation on a Model 475 sequencer (Applied Biosystems). Inceptin sequence and activity were confirmed by solid-phase peptide synthesis at the Protein Core Chemistry Facility (University of Florida, Gainesville) by using N-(9-fluorenylmethoxycarbonyl)-protected amino acids on an 432A Peptide Synthesizer (Applied Biosystems). Cysteine side chains and N-terminal amino acids were protected by acetamidomethyl and t-butyloxycarbonyl groups, respectively. The disulfide bond formation was performed on p-methyl benzyhydrylamine resin by iodine oxidation, and the peptides were cleaved from the resin with modified reagent K. All peptides were HPLC-purified.
Inceptin Quantification.
Internal standard-based quantification of inceptins from S. frugiperda OS was achieved by using ubiquitously labeled 13C and 15N valine-N-(9-fluorenylmethoxycarbonyl) (V*; Cambridge Isotope Laboratories, Andover, MA) incorporated into the synthetic peptide +ICDING-V*-CVDA−. Aliquots of crude OS, typically 50 μl, were sequentially spiked with 50 ng of the internal standard-based peptide, 5 μl of HCl, vortexed, and centrifuged 12,000 × g for 5 min. The aqueous phase was mixed with an equal volume of EtOH, stored at −70°C for 30 min, and centrifuged at 12,000 × g for 2 min. Samples were diluted to 5% EtOH, loaded on 100-mg RP-C18 SPE columns, washed with 2 ml of H2O, and eluted with 9:1 CH3CN:H2O. Samples were then concentrated to dryness under vacuum and brought up in 50 μl of 5:95 CH3CN:H2O containing 10 mM NH4COOH, and 10 μl was analyzed by LC-MS as described. Quantification was based on peak retention times (10.0–10.1 min) and monitoring [M+H]+ ions with a m/z of 1,119.5 and 1,125.5 for the natural and isotope-labeled inceptins, respectively. The identity of each sample was confirmed with MS daughter ion spectra.
RNA Isolation and Quantitative PCR of Cystatin.
Total RNA was isolated from control and treated cowpea leaves (500 ng) and used to synthesize cDNA by using the SuperScript II First-Strand Synthesis Kit (Invitrogen) (35). First-strand reactions, run in triplicate, were subjected to quantitative PCR with primers TTGAGATCGATAGTTTAGCTCGC and TAAGTACACTATGCAGGTGCATC designed from cowpea cystatin (36). All reactions were done in triplicate. Molecule numbers per microgram of total RNA were calculated by using a standard curve technique (37).
Identification of cATPC Sequences.
Sequences of atpC homologs were identified by blast analysis of the purified inceptin sequences against all translated GenBank sequences. Significant homology was first found with the rice (Oryza sativa) atpC mRNA, accession number XM_478377. Searches in maize returned mRNA sequences for chloroplastic and mitochondrial atpC with the GenBank accession nos. AY108268 and AY108441, respectively. Additional sequences located in the blast and The Institute for Genomic Research databases included Arabidopsis thaliana, pea (Pisum sativum), tomato (Lycopersicon esculentum), tobacco (Nicotiana tabacum), potato (Solanum tuberosum), medicago (Medicago truncatula), wheat (Triticum aestivum), and spinach (Spinacia oleracea), with accession nos. NM_116702, X63604, BT012794, X63606, BQ045978, TC94286, TC232362, and CAA68727, respectively. Primers ATPC-F (GGATCCGCTCTACACCAAGTTCGTGTC) and ATPC-R (GCGGCCGCATTGCTCATGGCACTCAT) were designed against conserved regions of the DNA sequence and contain BamHI and NotI sites for subcloning, respectively.
Cloning of Other Legume ATPC Sequences.
RNA and cDNA were made from leaf tissues as described above. Primers ATPC-F and ATPC-R were used to amplify sequences from cowpea; P. vulgaris varieties navy bean, kidney bean, and black bean; lima bean (P. lunatus); and peanut (Arachis hypogaea). Primers ATPC-F2 (GGCCATTGCTGATGATGT) and ATPC-R2 (GCATCAAGGATCTGAAC) amplified sequences for chickpea (Cicer arietinum), lentil (Lens culinaris), and sugar snap pea (P. sativum). All PCR products were subcloned into TOPO 2.1 vector (Invitrogen) and sequenced by using the vector primer M13-F. Sequences were translated by using editseq (DNASTAR, Madison, WI), and protein sequences were aligned in megalign (DNASTAR) with other ATPC proteins. Partial sequences of chloroplastic atpC from V. unguiculata, P. vulgaris (cultivars navy, kidney, and black bean), A. hypogaea, C. arietinum, L. culinaris, and P. lunatus are deposited in the GenBank database under accession nos. DQ312300, DQ317395, DQ317396, DQ317397, DQ317402, DQ317401, DQ317399, and DQ317400, respectively.
Bacterial Expression of cATPC Fragment.
The cowpea cATPC PCR fragment, corresponding to bases 716-1053 compared with the maize mRNA, was excised from the TOPO 2.1 vector with BamHI and NotI and was ligated into pET41b+ (Novagen, Darmstadt, Germany) cut with the same enzymes. Empty-vector GST controls and cATPC-GST fusion constructs were expressed in BL21(DE3)pLysS E. coli (Novagen) by induction with 1 mM IPTG for 16 h at 25°C and were purified by using GST·Bind Resin (Novagen).
S. frugiperda Feeding Studies.
Sixth-instar S. frugiperda larvae were isolated and allowed to feed for 12 h on 1 g of artificial diet, cowpea shoots, or roots spiked with 50 μl of either H2O only or 60 μg of bacterially expressed GST protein or cATPC-GST fusion protein. The OS was collected and pooled from four groups of four larvae (n = 4) for inceptin quantification and cowpea leaf E bioassays. Similarly, for S. frugiperda herbivory experiments larvae (n = 12) were allowed to feed on cowpea shoots or roots overnight. To avoid the presence of plant enzymes and moderate inceptin production, additional larvae (n = 12) were fed 0.1 g of artificial diet overnight containing either 9 μg of GST or cATPC-GST fusion protein. The OS was collected and pooled from a subset of six larvae (n = 1) for inceptin quantification. In these two experiments, larvae were carefully placed on paired cowpea leaves and covered with ultra-lightweight clear plastic domes (2.5-cm diameter) to partially restrict movement. Within 15 min, larvae typically initiated a single feeding bout, exited to the leaf underside through a hole (≈25 mm2) created by feeding, and were removed. Only leaf pairs with comparable physical damage were analyzed (n = 6). Purified GST and cATPC-GST proteins were estimated by using Coomassie blue staining of SDS/PAGE gels with BSA standards of 0.1, 0.5, 1.0, 2.5, and 5.0 μg and previously purified GST and cATPC-GST.
Supplementary Material
Acknowledgments
We thank R. L. Meagher, N. Lowman, and C. Dillard for supplying S. frugiperda larvae; A. Y. Chung, S. Stevens, and S. H. McClung for assistance in peptide analysis and synthesis; C. A. Ryan, G. Pearce, and G. Munske for facilitating amino acid sequencing; and H. J. Klee and A. R. Zangerl for useful editorial feedback. E.A.S. thanks previous mentors I. T. Baldwin, W. S. Bowers, and J. H. Tumlinson for years of guidance that enabled this work. This work was supported by U.S. Department of Agriculture–Agricultural Research Service base funds and U.S. Department of Agriculture–National Research Initiative Competitive Grants Program Grant 2002-35302-12375 subaward 2394-USDA-USDA-2375.
Abbreviations
- cATPC
chloroplastic ATP synthase γ-subunit
- OS
oral secretion
- E
ethylene
- RP
reverse-phase
- DMNT
(E)-4,8-dimethyl-1,3,7-nonatriene
- SPE
solid-phase extraction
- JA
jasmonic acid
- SA
salicylic acid
- R protein
resistance protein
- FAC
fatty acid amino acid conjugate
- MP
mobile phase.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kessler A., Baldwin I. T. Annu. Rev. Plant Biol. 2002;53:299–328. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J., Davis L. C., Verpoorte R. Biotechnol. Adv. 2005;23:283–333. doi: 10.1016/j.biotechadv.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Dangl J. L., Jones J. D. G. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 4.Mackey D., Belkhadir Y., Alonso J. M., Ecker J. R., Dangl J. L. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 5.Shao F., Golstein C., Ade J., Stoutemyer M., Dixon J. E., Innes R. W. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 6.Durrant W. E., Dong X. Annu. Rev. Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 7.Howe G. A. J. Plant Growth Regul. 2004;23:223–237. [Google Scholar]
- 8.Shiu S. H., Bleecker A. B. Proc. Natl. Acad. Sci. USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrlich P. R., Raven P. H. Evolution. 1964;18:586–608. [Google Scholar]
- 10.Nault L. R. Ann. Entomol. Soc. Am. 1997;90:521–541. [Google Scholar]
- 11.Turlings T. C. J., Tumlinson J. H., Lewis W. J. Science. 1990;250:1251–1253. doi: 10.1126/science.250.4985.1251. [DOI] [PubMed] [Google Scholar]
- 12.Alborn H. T., Turlings T. C. J., Jones T. H., Stenhagen G., Loughrin J. H., Tumlinson J. H. Science. 1997;276:945–949. [Google Scholar]
- 13.Kessler A., Baldwin I. T. Science. 2001;291:2141–2144. doi: 10.1126/science.291.5511.2141. [DOI] [PubMed] [Google Scholar]
- 14.Truitt C. L., Wei H. X., Pare P. W. Plant Cell. 2004;16:523–532. doi: 10.1105/tpc.017723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spiteller D., Pohnert G., Boland W. Tetrahedron Lett. 2001;42:1483–1485. [Google Scholar]
- 16.Felix G., Grosskopf D. G., Regenass M., Basse C. W., Boller T. Plant Physiol. 1991;97:19–25. doi: 10.1104/pp.97.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearce G., Strydom D., Johnson S., Ryan C. A. Science. 1991;253:895–898. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- 18.Felix G., Duran J. D., Volko S., Boller T. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 19.Ryan C. A., Pearce G. Proc. Natl. Acad. Sci. USA. 2003;100:14577–14580. doi: 10.1073/pnas.1934788100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boller T. Curr. Opin. Cell Biol. 2005;17:116–122. doi: 10.1016/j.ceb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Reymond P., Farmer E. E. Curr. Opin. Plant Biol. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- 22.Gouinguene S., Pickett J. A., Wadhams L. J., Birkett M. A., Turlings T. C. J. J. Chem. Ecol. 2005;31:1023–1038. doi: 10.1007/s10886-005-4245-1. [DOI] [PubMed] [Google Scholar]
- 23.Kappers I. F., Aharoni A., van Herpen T., Luckerhoff L. L. P., Dicke M., Bouwmeester H. J. Science. 2005;309:2070–2072. doi: 10.1126/science.1116232. [DOI] [PubMed] [Google Scholar]
- 24.Dixon R. A., Paiva N. L. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryan C. A. Annu. Rev. Phytopathol. 1990;28:425–449. [Google Scholar]
- 26.Malkin R., Niyogi K. In: Biochemistry and Molecular Biology of Plants. Buchanan B. B., Gruissen W., Jones R. L., editors. Rockville, MD: Am. Soc. Plant Physiol; 2000. pp. 568–628. [Google Scholar]
- 27.Hightower K. E., McCarty R. E. Biochemistry. 1996;35:4846–4851. doi: 10.1021/bi952913p. [DOI] [PubMed] [Google Scholar]
- 28.Ferreira C., Capella A. N., Sitnik R., Terra W. R. Arch. Insect Biochem. Physiol. 1994;26:299–313. [Google Scholar]
- 29.Axtell M. J., Staskawicz B. J. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 30.Rossi M., Goggin F. L., Milligan S. B., Kaloshian I., Ullman D. E., Williamson V. M. Proc. Natl. Acad. Sci. USA. 1998;95:9750–9754. doi: 10.1073/pnas.95.17.9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmelz E. A., Alborn H. T., Banchio E., Tumlinson J. H. Planta. 2003;216:665–673. doi: 10.1007/s00425-002-0898-y. [DOI] [PubMed] [Google Scholar]
- 32.Schmelz E. A., Alborn H. T., Tumlinson J. H. Planta. 2001;214:171–179. doi: 10.1007/s004250100603. [DOI] [PubMed] [Google Scholar]
- 33.Turlings T. C. J., McCall P. J., Alborn H. T., Tumlinson J. H. J. Chem. Ecol. 1993;19:411–425. doi: 10.1007/BF00994314. [DOI] [PubMed] [Google Scholar]
- 34.Schmelz E. A., Engelberth J., Tumlinson J. H., Block A., Alborn H. T. Plant J. 2004;39:790–808. doi: 10.1111/j.1365-313X.2004.02168.x. [DOI] [PubMed] [Google Scholar]
- 35.Nagy F., Kay S. A., Chua N. H. Analysis of Gene Expression in Transgenic Plants: Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic; 1988. [Google Scholar]
- 36.Fernandes K. V. S., Sabelli P. A., Barratt D. H. P., Richardson M., Xavierfilho J., Shewry P. R. Plant Mol. Biol. 1993;23:215–219. doi: 10.1007/BF00021433. [DOI] [PubMed] [Google Scholar]
- 37.Goncalves S., Cairney J., Maroco J., Oliveira M. M., Miguel C. Planta. 2005;222:556–563. doi: 10.1007/s00425-005-1562-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.