Abstract
The low rate of homologous recombination exhibited by wild-type strains of filamentous fungi has hindered development of high-throughput gene knockout procedures for this group of organisms. In this study, we describe a method for rapidly creating knockout mutants in which we make use of yeast recombinational cloning, Neurospora mutant strains deficient in nonhomologous end-joining DNA repair, custom-written software tools, and robotics. To illustrate our approach, we have created strains bearing deletions of 103 Neurospora genes encoding transcription factors. Characterization of strains during growth and both asexual and sexual development revealed phenotypes for 43% of the deletion mutants, with more than half of these strains possessing multiple defects. Overall, the methodology, which achieves high-throughput gene disruption at an efficiency >90% in this filamentous fungus, promises to be applicable to other eukaryotic organisms that have a low frequency of homologous recombination.
Keywords: filamentous fungi, functional genomics, knockouts, recombinational cloning
Historically, it has proven difficult to perform gene knockouts on a large scale in filamentous fungi. Unlike Saccharomyces cerevisiae, wild-type Neurospora strains exhibit a low frequency of homologous recombination after transformation, typically <10%, even when deletion cassettes contain long stretches of flanking sequence. The predominance of ectopic insertions dictates that a large number of transformants be screened. Of the many published procedures for creating knockout constructs (e.g., refs. 1–6), none lends itself well to a reliable high-throughput approach. To fill this need, we have developed a scheme that takes advantage of recombinational cloning in yeast (7, 8), a method that bypasses traditional restriction digestion and ligation. The components of the final construct are synthesized individually with short overlapping ends by PCR and cotransformed into yeast for assembly by the recombination machinery. An additional significant advance is the creation of Neurospora recipient strains in which ectopic insertions are virtually eliminated; Inoue and coworkers (9) recently demonstrated that mutation of either of two Neurospora genes required for nonhomologous end-joining DNA repair (mus-51 and mus-52) results in a dramatic increase in the frequency of homologous recombination. We have combined our method for cassette synthesis, the use of Δmus-51 and Δmus-52 strains, and additional procedures including custom-designed software tools, into a successful high-throughput gene deletion strategy for filamentous fungi. Here we report application of this technology to mutation of 103 genes encoding transcription factors in Neurospora.
The filamentous fungus Neurospora crassa is a model for many fungal species that are important animal and plant pathogens. Neurospora possesses a more complex life cycle than yeasts, with three different sporulation pathways and production of at least 28 different cell types (10, 11). Although Neurospora has been studied for >60 years by classical and molecular genetics, relatively little is known about transcriptional regulation in this organism. The availability of the complete Neurospora genome sequence (12) has allowed annotation of 182 transcription factor genes (not including general factors that regulate RNA polymerase; see ref. 11). The majority of factors belong to the Zn(II)2Cys6 fungal binuclear cluster family, whereas others are members of the C2H2 zinc finger, GATA, bHLH, B-ZIP, CBF CAAT-binding factor, homeobox, RING finger, PHD finger, MIZ zinc finger, and other families (11). Only 21 transcription factor genes have been characterized in Neurospora, and fewer yet have been studied in any other filamentous fungal system. The genes that have been studied in Neurospora play roles in metabolism and during development, including nutrient utilization or uptake (acu-15, cpc-1, nit-2, nit-4, pco-1, nuc-1, cys-3, qa-1F, and sre; see refs. 13–21), vegetative growth and macroconidiation (fl, rca-1, and rco-1; see refs. 22 and 23), light responses and the circadian rhythm (wc-1 and wc-2; see refs. and 25), sexual development (asd-4, asm-1, and pp-1; see refs. 26–28), DNA repair (uvs-2; see ref. 29), and mating type functions (mat A-1, mat A-3, and mat a-1; see refs. 30 and 31). There are many other processes for which basic information regarding transcriptional control mechanisms is totally lacking. Therefore, a large-scale functional study of transcription factors in Neurospora is timely and will provide a foundation for understanding complex modes of gene regulation in filamentous fungi.
Results
The overall strategy for high-throughput generation of gene deletions required a series of methodological advances for the construction of the deletion cassette DNA. In addition, recipient strains were created for transformation (mus-51::bar and mus-52::bar; see Materials and Methods). Because the marker for our deletion cassettes is hph (see below), we were unable to use the mus-51/52::hph deletion strains reported by Ninomiya et al. (9).
Primer Design, Optimization, and Production of Deletion Cassettes.
The cassettes contain (in order, 5′ to 3′) 1–1.3 kb of 5′ sequence flanking the ORF to be deleted (target gene ORF); the selectable marker hph [the gene for hygromycin B phosphotransferase (32), which confers hygromycin resistance] driven by the trpC promoter (33) and flanked by engineered MmeI restriction sites; and 1–1.3 kb of 3′ flanking sequence for the target gene ORF.
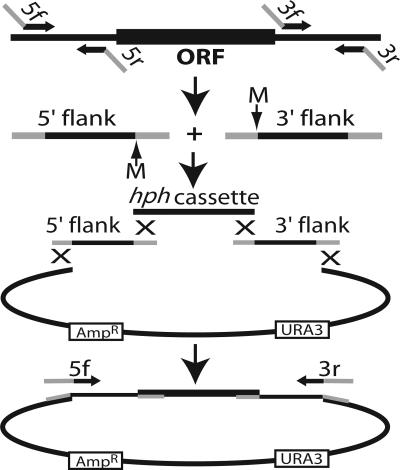
The method for construction of deletion cassettes is presented in Fig. 1. Briefly, four DNA fragments comprising the 5′ and 3′ DNA sequences flanking the target gene, the hph cassette, and a gapped yeast vector, respectively, were assembled in yeast using the endogenous homologous recombination system, and the final linear deletion cassette was amplified from the resulting construct by PCR (see Fig. 1 and Materials and Methods). To achieve this, four PCR primers, designated 5f, 5r, 3f, and 3r, were synthesized for each knockout cassette. The common 29-nt 5′ portions of the primers are shown in Supporting Text, which is published as supporting information on the PNAS web site; to select the gene-specific 20-nt portions of the primers, we used a custom-built software application (see Materials and Methods). This program successfully designed primers for 10,047 (96.8%) of the 10,620 ORFs predicted by Assembly 7 of the Neurospora genome sequence (www.broad.mit.edu/annotation/fungi/neurospora_crassa_7/index.html). Primer design failed for 16 genes because of their proximity to the ends of contigs. Modest alterations in the selection parameters enabled successful primer design for most of the other failures (data not shown).
Fig. 1.
Strategy for creating deletion constructs. 5′ and 3′ flank fragments are amplified separately from genomic DNA with primers 5f + 5r and 3f + 3r. Primers 5r and 3f incorporate MmeI sites (M) and have 5′ tails homologous to the hph cassette, whereas those for 5f and 3r are homologous to the vector. The two flanks are cotransformed into yeast along with the hph cassette and gapped yeast shuttle vector. Homologous recombination creates the circular construct and the final linear deletion cassette is amplified from the pooled yeast DNA with primers 5f and 3r. hph is transcribed in the antisense direction relative to the target gene.
MmeI sites were designed into the junctions between the flanks and the hph cassette (Fig. 1) to allow the adjacent gene-specific 20-bp sequences to serve as molecular bar codes. Digestion of genomic DNA by MmeI (which cuts 20 base pairs downstream of its recognition sequence) followed by ligation-mediated PCR will create products that can be amplified and then either sequenced or used in standard competition experiments (34–36).
In the development phase of this work, deletion cassettes were made with the hph gene driven by the Ashbya gossypii TEF promoter (pTEF); pTEF drives expression in both yeast and Neurospora, thereby allowing direct selection of yeast cells harboring the correctly assembled constructs on medium containing hygromycin B. Although sufficient for creation of deletion strains (e.g., C. A. Jones, S. E. Greer-Phillips, and K.A.B., unpublished work), the weak activity of pTEF in Neurospora led to slow growth and low numbers of transformants at the hygromycin concentration necessary for inhibiting growth of nontransformed cells. Therefore, to ensure dependable selection of true Neurospora transformants, we settled on the widely used and highly expressed Aspergillus nidulans trpC promoter-hph cassette (33) as our selectable marker for Neurospora deletion constructs. The yeast vector contains the URA3 gene, and selection is done on medium lacking uracil. DNA made from pooled yeast transformants serves as the template for specific amplification of the correct deletion cassette.
Before Inoue and coworkers published their findings regarding the mus-51 and mus-52 deletion strains (9), we explored methods for increasing the frequency of homologous recombination among transformants. Deletion cassette constructs were generated by yeast recombinational cloning as shown in Fig. 1, except that the flank fragments were 3 kb long. Split-marker fragments (37) were then amplified by PCR from the pooled yeast DNA (see Fig. 4, which is published as supporting information on the PNAS web site) and cotransformed into Neurospora. Combining our method for cassette construction with the published split-marker technique gave results that were a significant improvement over existing approaches. On average, 44% of the transformants obtained with this strategy had the proper gene replacement and, as determined by Southern analysis, 68% of those were free of ectopic insertions (results for 16 individual genes vary widely and are shown in Table 2, which is published as supporting information on the PNAS web site). Our subsequent adoption of Δmus-51 and Δmus-52, along with deletion cassettes with 1-kb flanks, rendered the split-marker approach unnecessary for the high-throughput deletion project. However, it remains a useful strategy for transforming strains not deficient in mus-51 or mus-52, especially when combined with our method for creating deletion cassettes.
Subsequent to the creation of the transcription factor deletion cassettes, this method was used to produce deletion cassettes for >9,600 Neurospora genes. The overall success rate for synthesis of flank fragments is ≈97%, whereas that for PCR amplification of the final deletion cassettes from the mixed yeast DNA pools is 90–95%. Electroporation of Neurospora with the final cassettes used for disrupting the transcription factors worked well over at least a 5-fold range of input DNA amounts, typically 200–1,000 ng.
Generation of Transcription Factor Deletion Mutants.
The 103 transcription factor genes chosen to illustrate the utility of this procedure were previously annotated (11) by blast searches (38) by using Assembly 3 of the Neurospora genome sequence (www.broad.mit.edu/annotation/fungi/neurospora_crassa_3/index.html). Genes with E values <10−5 were accepted as members of a particular class. For initial knockout experiments, genes representing all of the annotated groups (11) were chosen. Cassettes were made and used to transform Δmus strains as described above and in Materials and Methods, and the heterokaryotic primary transformants bearing putative gene knockouts were selected on the basis of hygromycin resistance.
Southern blot analysis of 627 primary transformants (six to seven transformants per gene) showed that homologous recombination without any ectopic insertion occurred in 98% of the cases. Ectopic insertion of knockout cassette DNA in addition to homologous recombination was observed in 0.8% of transformants, but stable homokaryons free of ectopic sequences were eventually obtained from these transformants after crossing (Table 1). Ectopic insertions without concurrent homologous recombination, as well as abnormal recombination events, were observed in 1.2% of the transformants (Table 1).
Table 1.
Results of Southern blot analysis of 627 primary transformants
| HR* with no ectopic† | HR with ectopic‡ | Ectopic with no HR§ | Unusual recombination¶ | |
|---|---|---|---|---|
| Number | 613 | 5 | 6 | 3 |
| Percent of total | 98 | 0.8 | 0.8 | 0.4 |
*HR, Homologous recombination.
†Transformants with bands indicative of gene replacement as well as wild type, but with no additional bands indicative of ectopic insertion.
‡Transformants with bands indicative of gene replacement and wild type, as well as additional bands.
§Transformants with no bands indicative of gene replacement but with both wild type and additional bands.
¶Transformants with no bands indicative of gene replacement or wild type but only extra bands.
Primary transformants were crossed to wild-type females to obtain homokaryotic knockout mutants free of the mus mutation. Although 99 of the transcription factor genes yielded homokaryotic deletion progeny, no hygromycin-resistant progeny were isolated for NCU00340 (pp-1), NCU01345 (asl-1), NCU01459 (asl-2), and NCU04050 (cpc-1).
Phenotypic Analysis of Transcription Factor Deletion Strains.
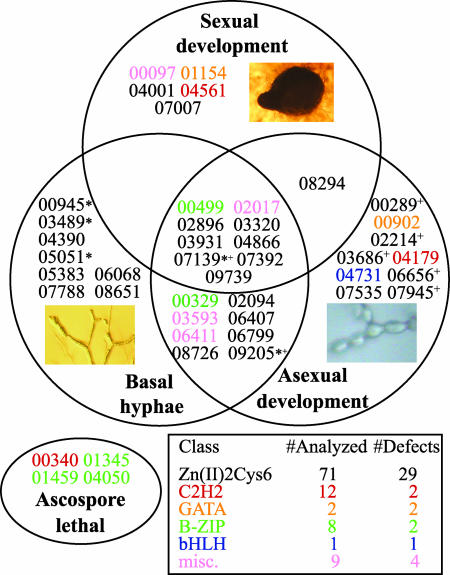
A substantial fraction of the viable mutants displayed visible phenotypes; the results indicate that 40 of the 99 genes studied are required for normal vegetative or sexual growth and development in Neurospora (Fig. 2). Table 3, which is published as supporting information on the PNAS web site, provides a complete list of genes deleted and the results of the phenotypic analyses, as well as names applied to mutants exhibiting novel phenotypes. Further details and photographs of the mutant strains are available at www.broad.mit.edu/annotation/genome/neurospora/Alleles.html.
Fig. 2.
Venn diagram showing the distribution of transcription factor knockout mutants with observed phenotypes. Mutants are represented by the NCU numbers of the deleted genes. Font color indicates gene family (see Inset table). Miscellaneous genes (pink) are from three classes: RING-type zinc finger (NCU06411), CBF CAAT-binding factor (NCU02017), and homeobox (NCU00097 and NCU03593). Genes showing ascospore lethality are shown in the oval. The numbers of transcription factor mutants analyzed and the numbers with phenotypes are listed in the Inset table. Knockout mutants indicated with * or + exhibited greater basal hyphal extension rates or aerial hyphae heights than wild type, respectively. For the wild-type controls, the basal hyphae extension rate was 65–80 mm per day, whereas the height of aerial hyphae achieved in 3 days was 30–45 mm. The three photographs show a perithecium (Upper), a conidiophore (Lower Right), and basal hyphae (Lower Left).
Basal hyphal extension and asexual development.
Neurospora grows by apical extension, branching, and fusion of basal hyphae to form a colony. Nutrient deprivation or desiccation leads to differentiation of aerial hyphae from these basal hyphae. The ends of aerial hyphae form budding structures called conidiophores that give rise to mature vegetative spores, the macroconidia (39).
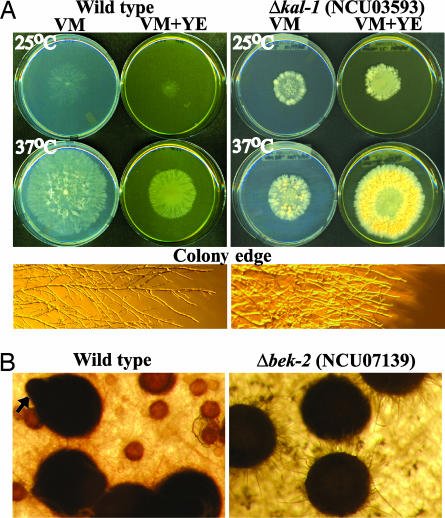
In our study, 20 mutants exhibited significantly reduced basal hyphal extension rates compared to wild type as measured in race tubes (see Supporting Text), whereas five mutants had faster growth rates (Fig. 2 and Table 3). Most mutants with defects in extension of basal hyphae also demonstrated abnormalities during macroconidiation (15 strains; Fig. 2). A positive correlation between production of normal levels of mature macroconidia and the height of aerial hyphae was observed for many mutants (data not shown). For example, the Δfluffy (fl):NCU08726 mutant exhibited short aerial hyphae and a block in macroconidiation as previously reported (40, 41). However, nine mutant strains with short aerial hyphae formed macroconidia normally (Table 3). Five mutants extended longer aerial hyphae than wild type (Fig. 2). Of particular interest, two mutants (Δvad-6:NCU09205 and Δbek-2:NCU07139; see below) exhibited both faster basal hyphal extension rates and longer aerial hyphae than wild type (Fig. 2).
The Δkaleidoscope-1 (kal-1:NCU03593) mutant was so named because of its radial and variable colony appearance under different growth conditions. The Δkal-1 mutant has a slower basal hyphal extension rate than wild type at 30°C and 37°C, but not at 25°C (Fig. 3A and data not shown). When 2% yeast extract was added to the medium, extension of basal hyphae was decreased in wild type but increased in the Δkal-1 strain (Fig. 3A). Δkal-1 mutants also exhibit increased branching of basal hyphae and shorter aerial hyphae (Fig. 3A and Table 3).
Fig. 3.
Colony morphology and asexual and sexual development of transcription factor knockout mutants. (A) Colony morphology of wild type and Δkal-1:NCU03593. Strains were grown for 24 h on VM or VM + yeast extract at 25°C and 37°C. The colony edge images show basal vegetative hyphae at ×11. (B) Transcription factor mutant exhibiting aberrant development of perithecia. Images at ×77 were taken 7 days after fertilization of protoperithecia with opposite mating type conidia. The arrow indicates a beak in wild type; this structure does not form in the mutant.
Sexual development.
Neurospora differentiates female reproductive structures (protoperithecia) in response to nitrogen starvation. Fertilization is accomplished by chemotropic growth of a specialized female hypha toward a male cell (typically a macroconidium) of opposite mating type, transport of the male nucleus into the protoperithecium, cell proliferation, and karyogamy. Meiosis ensues and the protoperithecium develops into the mature fruiting body (perithecium) containing ascospores (42).
All 99 viable knockout mutants exhibited normal male fertility (data not shown). In contrast, 14 of the transcription factors studied are required for various stages of female sexual development and ascospore production (Fig. 2 and Table 3). Nine of the mutants that do not produce perithecia also form few or no protoperithecia. The arrested development (Δadv-1:NCU07392) mutant is blocked in protoperithecial formation (as well as basal hyphal extension and asexual development; see below), in contrast to strains lacking the homologous gene pro1 in Sordaria macrospora (43). Submerged protoperithecia were observed in the Δsub1:NCU01154 mutant. Several mutants develop aberrant perithecia, including defective beaks (Δbek-1:NCU00097 and Δbek-2; see above; Fig. 3B) and less melanization (Δmel-1:NCU04561). Of these three mutants, only Δmel-1 is able to efficiently eject ascospores. Interestingly, the Δnit-4:NCU08294 mutant forms more protoperithecia and perithecia than wild type (data not shown).
Eight transcription factor knockout mutants possess significant defects in all three phenotypes: basal hyphal extension, asexual development, and sexual development (Fig. 2). In contrast, Δbek-2 has a defect in sexual development but demonstrates increased extension of both basal and aerial hyphae (Fig. 2). With the exception of all development altered (ada)-2:NCU02017, ada-4:NCU03320, and adv-1:NCU07392, these genes and their closest homologues have no reported functions in any filamentous fungal system.
Discussion
Methodological advances reported here significantly facilitate the high-throughput production of deletion mutants. We found recombinational cloning in yeast, a well characterized and widely used technique, to be the most suitable method for high-throughput synthesis of deletion constructs. Most steps are performed on a pipetting robot, allowing the creation of 400–600 constructs per week. Cassettes are electroporated into the mus deletion mutants in a 96-well format. Strains and processes are tracked by using a custom-written laboratory information management system (lims; see Supporting Text), and streamlined methods for preliminary phenotypic analysis have been developed. Both the general procedures developed for Neurospora (e.g., the use of mus-51/52 deletions to reduce ectopic insertions and a strong promoter to drive selectable marker expression) and the technical developments (primer selection and Southern analysis programs; high-throughput cassette construction, preparation, and transformation) should be readily adaptable to other organisms. In addition, this powerful methodology can be used to generate strains with tagged or mutant alleles or promoter replacements at the endogenous locus of any given gene.
The 99 viable transcription factor mutants were analyzed for a variety of characteristics during vegetative and sexual growth and development. A total of 40 mutants displayed at least one defining phenotype during our testing. Among the genes producing phenotypes, only four had been studied previously in Neurospora [wc-2:NCU00902, acu-15:NCU06656, fluffy (fl), and nit-4], and 14 others have characterized homologues in other fungi.
Our results revealed that highly conserved transcription factors can play different roles in various fungi. For example, Sordaria macrospora pro1 mutants are able to form protoperithecia, but not perithecia (43). In contrast, the Aspergillus nidulans Pro1 homologue RosA is required for sexual development at low carbon levels (44). Our analysis of the Neurospora homologue of pro1, adv-1, demonstrates that it is required for normal protoperithecial differentiation at both high and low sucrose concentrations. Thus, adv-1 is likely to be required during an earlier stage of female sexual development than pro1, and it does not possess a carbon concentration-dependent function, as observed for rosA. In addition, adv-1 is also essential for normal vegetative growth and development in Neurospora.
That no viable ascospore progeny were observed for four transcription factor mutants (Δpp-1:NCU00340, Δcpc-1:NCU04050, Δasl-1:NCU01345, and Δasl-2:NCU01459) suggests these factors are required for ascospore germination and/or perform some other essential function during the life cycle. Similar results were obtained during analysis of S. cerevisiae transcription factor genes, with 3.4% found to be essential for viability in rich medium (45). Interestingly, with the exception of pp-1, all of the ascospore lethal Neurospora genes are members of the B-ZIP family (11). The results for pp-1 are supported by a recent report that ascospores carrying a deletion of pp-1 are inviable, but that Δpp-1 strains can be maintained in the vegetative phase (26). Viable Δcpc-1 mutants have been reported (46), and we were eventually able to isolate Δcpc-1 strains by using the microconidiation procedure in the vegetative phase to bypass the need for a sexual cross (data not shown; see ref. 47).
Our work uncovered an important regulator of asexual growth and development in Neurospora, kaleidoscope-1 (kal-1), which encodes a homeobox-containing transcription factor. kal-1 is homologous to Podospora anserina pah1, a gene required for normal microconidiation and hyphal branching (48). The Δkal-1 mutation is highly pleiotropic, leading to substantial changes in colony morphology and conidial development (Fig. 3A). However, supplementation with yeast extract increases apical extension and conidiation of the kal-1 mutant, indicating a possible role for kal-1 in nutrient metabolism or sensing. Despite their extensive vegetative defects, kal-1 mutants are female-fertile. Two additional homeobox transcription factor genes, bek-1 and NCU03070, were also examined in our study; bek-1 was found to be required for normal development of perithecial beaks during sexual development. Homeobox transcription factors are well known as regulators of cell proliferation, differentiation, and pattern formation in mammals (49–51); our analysis supports conservation of developmental roles for homeobox factors in Neurospora.
It is notable that 59 of the transcription factor genes studied here do not possess obvious functions during growth and development, suggesting they may have roles under environmental conditions not included in our analyses. Some of the genes may also possess overlapping functions. Functional redundancy is observed for many proteins in eukaryotes, including transcription factors (52–54), and can be preserved over long periods of evolution because the compensatory activities of related genes can buffer the effects of deleterious mutations (55). Remarkably, numerous transcription factor genes in our study (33%) are similar to another Neurospora transcription factor (e−64 to e−5), and the majority of these proteins belong to the Zn(II)2Cys6 fungal binuclear cluster family (11). In addition, many (79%) did not yield obvious phenotypes when mutated. Mutation of multiple homologous transcription factor genes will be necessary to comprehend complex gene regulation in Neurospora.
Characterization of the null phenotype is a requisite step in determining the functions of a gene, and the accessibility of a complete collection of mutants would begin to unlock the secrets of any genome. We have reported here the development and implementation of a scheme for high-throughput production of targeted gene deletions in Neurospora that should be widely applicable to the filamentous fungi and beyond. We have used these methods to systematically delete genes encoding 103 transcription factors, and the resulting strains have provided a window into novel processes controlling vegetative growth, as well as sexual and asexual development.
Materials and Methods
Neurospora Strains.
Strains FGSC 4200 (mat a), FGSC 2489 (mat A), FGSC 6103 (his-3 mat A), FGSC 4317 (fl mat A), and FGSC 4347 (fl mat a) were obtained from the Fungal Genetics Stock Center, Kansas City, KS. Strains FGSC 9717 (mus-51::bar his-3 mat A), FGSC 9718 (mus-51::bar mat a), FGSC 9719 (mus-52::bar mat a), and FGSC 9720 (mus-52::bar his-3 mat A) were created in this study.
Primer Design and Synthesis of Deletion Cassettes.
A software application written by John Jones (John Jones Consulting, Moreno Valley, CA) was designed to retrieve regions adjacent to each ORF and pass them to primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), which would automatically select a list of candidate primers with designated parameters (see Supporting Text). For gene-specific flanks, the program searched a 1,500-bp region on either side of the ORF for primers that would synthesize a 1- to 1.3-kb fragment, thus allowing a gap between the ORF and either flank fragment of 0–500 bp. The resulting suggested primers were then tested for the absence of MmeI restriction sites (see above), uniqueness in the genome [by blast; (38)], and suitable GC content (to minimize primer–dimer formation), and the top-rated primer pair was selected. Primers were supplied in 96-well plates (Illumina, San Diego) and were diluted and mixed in three pairs for each gene by a Biomek NX robot (Beckman): 5f + 5r and 3f + 3r for PCR synthesis of the 5′ and 3′ flank fragments, respectively, and 5f + 3r for production of the final cassettes. The flanks were synthesized from genomic Neurospora DNA using LA Taq (TaKaRa; exact details of all procedures are in Supporting Text).
Yeast Transformation.
For the 103 transcription factor gene knockouts, yeast strain FY834 (56) was transformed with both flank fragments, the hph cassette, and gapped plasmid [pRS426 (57) digested with XbaI and EcoRI] in 96-well plates on the robot. A lithium chloride/polyethylene glycol procedure developed for a 96-well format was used essentially as described (58), except that the selection for transformants was done in liquid medium lacking uracil.
Preparation of Yeast DNA and Generation of Deletion Cassettes by PCR.
Yeast DNA was prepared from the liquid cultures on the robot with the Puregene Yeast DNA kit (Gentra Systems) with slight modifications: Zymolyase T-100 (Seikagaku America, Falmouth, MA) was used and, in place of the final precipitation step, CleanSEQ magnetic beads (Agencourt, Beverly, MA) were used to bind the DNA. For the synthesis of full-length cassettes, yeast DNA and the primer pair mixture 5f + 3r for each gene were added by the robot to a PCR mix containing LA Taq.
Construction of the mus-51 and mus-52 Deletion Mutant Strains.
Yeast recombinational cloning was performed to construct deletion cassettes with the bar cassette [which confers resistance to phosphinothricin (59, 60)] flanked by 3 kb upstream and 3 kb downstream of the ORF of either mus-51 or mus-52. The 7-kb deletion cassette fragment was generated by using the appropriate 5f and 3r primers with the yeast DNA as a template for PCR. The cassettes were transformed by electroporation into wild-type Neurospora strain FGSC 4200, as described (61, 62). Selection of transformants, Southern analysis, and sexual crosses to obtain homokaryons are described in Supporting Text. Four different genotypes of Δmus-51 and Δmus-52 were obtained (see above); all four showed normal vegetative growth and sexual fertility, consistent with the previous study (9).
High-Throughput Generation of 103 Transcription Factor Deletion Mutants.
Conidia from either Δmus-51 (FGSC 9718) or Δmus-52 (FGSC 9719) were transformed by electroporation with each knockout cassette, allowed to recover in Vogel’s minimal medium (VM; ref. 63) supplemented with yeast extract and histidine and plated in regeneration agar on sorbose plates (61) containing hygromycin B, yeast extract, and histidine. After incubation for 5 days at 30°C in the dark, colonies were picked onto VM slants containing hygromycin B.
Six or more transformants for each gene were subjected to Southern blot analysis, as described (64), with some modifications. A program was developed to allow automated identification of appropriate restriction enzymes to use for Southern blot analysis of gene replacement mutants (developed by John Jones; details in Supporting Text). The DIG High Prime labeling kit (Roche Applied Science, Indianapolis) was used for labeling full-length knockout cassette fragment probes.
Heterokaryotic transformants confirmed by Southern analysis to contain both wild-type and deletion mutant nuclei were crossed to wild-type (FGSC 2489) or his-3 (FGSC 6103) females. Ascospore progeny resistant to hygromycin were selected and tested for the mus deletion mutation by assessing sensitivity to phosphinothricin. Confirmation of homokaryons by Southern blot analysis, and determination of mating type and his-3 alleles were as described in Supporting Text.
Phenotypic Analysis.
Three major phenotypes were analyzed in verified transcription factor knockout mutants in an otherwise wild-type background: linear growth rates of basal hyphae, asexual development, and sexual development. Detailed procedures are presented in Supporting Text. Linear growth rates for basal hyphae were measured by using VM agar race tubes. Colony morphology and conidiation were assessed after inoculation of VM and VM + yeast extract plates and incubation at 25°C or 37°C. VM slant tubes inoculated with strains, incubated at 25°C for 3 days and then at room temperature for 3–5 days, were used to analyze aerial hyphae production, conidiation, and pigmentation. Conidiation was also assessed on plates. Extension of aerial hyphae in standing liquid cultures was measured after incubation for 72 h at 25°C.
Sexual development analysis was performed after inoculation of strains on synthetic crossing medium plates containing 0.1% or 1.5% sucrose and incubation at room temperature. Production of protoperithecia was scored after 7–8 days. At this time, plates were fertilized with opposite mating type wild-type strains and perithecial formation determined after an additional 7–8 days. Ascospore ejection was assessed 2 weeks after fertilization.
Supplementary Material
Acknowledgments
We thank John Jones for software design and lims implementation; Svetlana Krystofova and Suzanne Phillips for help with mus strain construction; Victoria Copeland for excellent technical assistance; and the following students who participated in the Neurospora Genetics and Genomics Summer Research Institute (NGGSRI) at the University of California, Los Angeles, for phenotypic analysis: Cynthia Aguirre, Eliana Alcantar, Andrea Cahuantzi, Natalie Cornejo, Zachary W. Cue, Evelyn De Los Santos, Anthony Dualo, Thomas J. Dunehew, Mina El-Masry, Jonathan Finley, Lizette C. Flores, Christopher Fonseca, Rukhsana A. Khan, Carolyn Kingsley, Juan Lupercio, Criseyda Martinez, Rosaura Ochoa, Olufisayo Oke, Cam M. Phu, Chloe Rivera, Michael Smith, Desiree L. Tejada, Tuan D. Tran, and Jackelyn Valladares. This work was supported by National Institutes of Health Grant P01 GM068087. The NGGSRI program was supported by National Institutes of Health Grants 5 R25 GM050067 and 5 R25 GM055052.
Abbreviation
- VM
Vogel’s minimal medium.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wendland J. Curr. Genet. 2003;44:115–123. doi: 10.1007/s00294-003-0436-x. [DOI] [PubMed] [Google Scholar]
- 2.Yang L., Ukil L., Osmani A., Nahm F., Davies J., De Souza C. P., Dou X., Perez-Balaguer A., Osmani S. A. Eukaryot. Cell. 2004;3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaveroche M. K., Ghigo J. M., d’Enfert C. Nucleic Acids Res. 2000;28:E97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu J. H., Hamari Z., Han K. H., Seo J. A., Reyes-Dominguez Y., Scazzocchio C. Fungal Genet. Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Kuwayama H., Obara S., Morio T., Katoh M., Urushihara H., Tanaka Y. Nucleic Acids Res. 2002;30:E2. doi: 10.1093/nar/30.2.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aronson B. D., Lindgren K. M., Dunlap J. C., Loros J. J. Mol. Gen. Genet. 1994;242:490–494. doi: 10.1007/BF00281802. [DOI] [PubMed] [Google Scholar]
- 7.Oldenburg K. R., Vo K. T., Michaelis S., Paddon C. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raymond C. K., Pownder T. A., Sexson S. L. BioTechniques. 1999;26:134–141. doi: 10.2144/99261rr02. [DOI] [PubMed] [Google Scholar]
- 9.Ninomiya Y., Suzuki K., Ishii C., Inoue H. Proc. Natl. Acad. Sci. USA. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bistis G. N., Perkins D. D., Read N. D. Fungal Genet. Newsl. 2003 [Google Scholar]
- 11.Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., Read N. D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., et al. Microbiol. Mol. Biol. Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galagan J. E., Calvo S. E., Borkovich K. A., Selker E. U., Read N. D., Jaffe D., FitzHugh W., Ma L. J., Smirnov S., Purcell S., et al. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 13.Zhou L., Marzluf G. A. Biochemistry. 1999;38:4335–4341. doi: 10.1021/bi982543f. [DOI] [PubMed] [Google Scholar]
- 14.Feng B., Marzluf V. Mol. Cell. Biol. 1998;18:3983–3990. doi: 10.1128/mcb.18.7.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan G. H., Fu Y. H., Marzluf G. A. Mol. Cell. Biol. 1991;11:5735–5745. doi: 10.1128/mcb.11.11.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang S., Metzenberg R. L. Mol. Cell. Biol. 1990;10:5839–5848. doi: 10.1128/mcb.10.11.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y. H., Kneesi J. Y., Marzluf G. A. J. Bacteriol. 1989;171:4067–4070. doi: 10.1128/jb.171.7.4067-4070.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baum J. A., Geever R., Giles N. H. Mol. Cell. Biol. 1987;7:1256–1266. doi: 10.1128/mcb.7.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T. D., Marzluf G. A. Curr. Genet. 2004;46:213–227. doi: 10.1007/s00294-004-0530-8. [DOI] [PubMed] [Google Scholar]
- 20.Barthelmess I. B. Genet. Res. 1982;39:169–185. doi: 10.1017/s0016672300020863. [DOI] [PubMed] [Google Scholar]
- 21.Bibbins M., Crepin V. F., Cummings N. J., Mizote T., Baker K., Mellits K. H., Connerton I. F. Mol. Genet. Genom. 2002;267:498–505. doi: 10.1007/s00438-002-0682-5. [DOI] [PubMed] [Google Scholar]
- 22.Bailey L. A., Ebbole D. J. Genetics. 1998;148:1813–1820. doi: 10.1093/genetics/148.4.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashiro C. T., Ebbole D. J., Lee B.-K., Brown R. E., Bourland C., Madi L., Yanofsky C. Mol. Cell. Biol. 1996;16:6218–6228. doi: 10.1128/mcb.16.11.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linden H., Ballario P., Arpaia G., Macino G. Adv. Genet. 1999;41:35–54. doi: 10.1016/s0065-2660(08)60150-9. [DOI] [PubMed] [Google Scholar]
- 25.Dunlap J. C. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 26.Li D., Bobrowicz P., Wilkinson H. H., Ebbole D. J. Genetics. 2005;170:1091–1104. doi: 10.1534/genetics.104.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng B., Haas H., Marzluf G. A. Biochemistry. 2000;39:11065–11073. doi: 10.1021/bi000886j. [DOI] [PubMed] [Google Scholar]
- 28.Aramayo R., Peleg Y., Addison R., Metzenberg R. Genetics. 1996;144:991–1003. doi: 10.1093/genetics/144.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomita H., Soshi T., Inoue H. Mol. Gen. Genet. 1993;238:225–233. doi: 10.1007/BF00279551. [DOI] [PubMed] [Google Scholar]
- 30.Ferreira A. V. B., An Z. Q., Metzenberg R. L., Glass N. L. Genetics. 1998;148:1069–1079. doi: 10.1093/genetics/148.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staben C., Yanofsky C. Proc. Natl. Acad. Sci. USA. 1990;87:4917–4921. doi: 10.1073/pnas.87.13.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gritz L., Davies J. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 33.Staben C., Jensen B., Singer M., Pollock J., Schechtman M., Kinsey J., Selker E. Fungal Genet. Newsl. 1989;36:79–81. [Google Scholar]
- 34.Giaever G., Chu A. M., Ni, Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 35.Shoemaker D. D., Lashkari D. A., Morris D., Mittmann M., Davis R. W. Nat. Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 36.Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 37.Catlett N. L., Lee B.-N., Yoder O. C., Turgeon B. G. Fungal Genet. Newsl. 2002;50:9–11. [Google Scholar]
- 38.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Springer M. L. BioEssays. 1993;15:365–374. doi: 10.1002/bies.950150602. [DOI] [PubMed] [Google Scholar]
- 40.Matsuyama S. S., Nelson R. E., Siegel R. W. Dev. Biol. 1974;41:278–287. doi: 10.1016/0012-1606(74)90306-6. [DOI] [PubMed] [Google Scholar]
- 41.Bailey-Shrode L., Ebbole D. J. Genetics. 2004;166:1741–1749. doi: 10.1534/genetics.166.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raju N. B., Leslie J. F. Genome. 1992;35:815–826. doi: 10.1139/g92-124. [DOI] [PubMed] [Google Scholar]
- 43.Masloff S., Poggeler S., Kuck U. Genetics. 1999;152:191–199. doi: 10.1093/genetics/152.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vienken K., Scherer M., Fischer R. Genetics. 2005;169:619–630. doi: 10.1534/genetics.104.030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chua G., Robinson M. D., Morris Q., Hughes T. R. Curr. Opin. Microbiol. 2004;7:638–646. doi: 10.1016/j.mib.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 46.Paluh J. L., Yanofsky C. Mol. Cell. Biol. 1991;11 doi: 10.1128/mcb.11.2.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ebbole D., Sachs M. S. Fungal Genet. Newsl. 1990:17–18. [Google Scholar]
- 48.Arnaise S., Zickler D., Poisier C., Debuchy R. Mol. Microbiol. 2001;39:54–64. doi: 10.1046/j.1365-2958.2001.02163.x. [DOI] [PubMed] [Google Scholar]
- 49.Abate-Shen C. Nat. Rev. Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 50.Ford H. L. Cell Biol. Int. 1998;22:397–400. doi: 10.1006/cbir.1998.0329. [DOI] [PubMed] [Google Scholar]
- 51.Mahaffey J. W. Curr. Opin. Genet. Dev. 2005;15:422–429. doi: 10.1016/j.gde.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Firulli A. B., Thattaliyath B. D. International Review of Cytology, A Survey of Cell Biology. 2002;Vol. 214:1–62. doi: 10.1016/s0074-7696(02)14002-2. [DOI] [PubMed] [Google Scholar]
- 53.Martinez E. Plant Mol. Biol. 2002;50:925–947. doi: 10.1023/a:1021258713850. [DOI] [PubMed] [Google Scholar]
- 54.Pickett F. B., Meekswagner D. R. Plant Cell. 1995;7:1347–1356. doi: 10.1105/tpc.7.9.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krakauer D. C., Nowak M. A. Semin. Cell Dev. Biol. 1999;10:555–559. doi: 10.1006/scdb.1999.0337. [DOI] [PubMed] [Google Scholar]
- 56.Winston F., Dollard C., Ricupero-Hovasse S. L. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 57.Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 58.Gera J. F., Hazbun T. R., Fields S. Methods Enzymol. 2002;350:499–512. doi: 10.1016/s0076-6879(02)50981-2. [DOI] [PubMed] [Google Scholar]
- 59.Pall M. L. Fungal Genet. Newsl. 1993;40:58. [Google Scholar]
- 60.Pall M. L., Brunelli J. P. Fungal Genet. Newsl. 1993;40:59–62. [Google Scholar]
- 61.Case M. E., Schweizer M., Kushner S. R., Giles N. H. Proc. Natl. Acad. Sci. USA. 1979;76:5259–5263. doi: 10.1073/pnas.76.10.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Q., Poole S. I., Borkovich K. A. Eukaryot. Cell. 2002;1:378–390. doi: 10.1128/EC.1.3.378-390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davis R. H., deSerres F. J. Methods Enzymol. 1970;71A:79–143. [Google Scholar]
- 64.Ivey F. D., Hodge P. N., Turner G. E., Borkovich K. A. Mol. Biol. Cell. 1996;7:1283–1297. doi: 10.1091/mbc.7.8.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.