Abstract
Endocytosis is crucial for various aspects of cell homeostasis. Here, we show that proapoptotic death receptors (DRs) trigger selective destruction of the clathrin-dependent endocytosis machinery. DR stimulation induced rapid, caspase-mediated cleavage of key clathrin-pathway components, halting cellular uptake of the classic cargo protein transferrin. DR-proximal initiator caspases cleaved the clathrin adaptor subunit AP2α between functionally distinct domains, whereas effector caspases processed clathrin’s heavy chain. DR5 underwent ligand-induced, clathrin-mediated endocytosis, suggesting that internalization of DR signaling complexes facilitates clathrin-pathway targeting by caspases. An endocytosis-blocking, temperature-sensitive dynamin-1 mutant attenuated DR internalization, enhanced caspase stimulation downstream of DRs, and increased apoptosis. Thus, DR-triggered caspase activity disrupts clathrin-dependent endocytosis, leading to amplification of programmed cell death.
Keywords: caspase cleavage, TNF-related apoptosis-inducing ligand
Apoptosis plays crucial roles in the development and homeostasis of multicellular organisms (1). Two major signaling mechanisms, the cell-intrinsic and -extrinsic pathways, control apoptosis induction (2). These pathways activate cysteine proteases, called caspases, which cleave various cellular proteins that are essential for cell integrity. Caspases recognize specific tetrapeptide sequences containing aspartate and cleave the adjacent peptide bond (3).
Activation of the cell-extrinsic pathway occurs in response to ligands such as Fas ligand (FasL) and Apo2 ligand/TNF-related apoptosis-inducing ligand (Apo2L/TRAIL), through their respective cell-surface “death receptors” Fas (Apo1/CD95) (4) and death receptor (DR) 4 or DR5 (5). Ligand binding triggers recruitment of the adaptor FADD (Fas-associated “death” domain) to a death domain within the receptor’s cytoplasmic tail. FADD recruits the initiator protease caspase-8 to form the “death-inducing signaling complex” (DISC) (6). Proximity of caspase-8 molecules in the DISC stimulates enzymatic activity, resulting in self-processing (7). Cleaved caspase-8 then releases from the DISC to the cytoplasm and proteolytically activates effector caspases such as caspase-3 and -7. In certain cell types (type I) DR stimulation generates strong caspase-8 activity, which robustly activates effector caspases and commits the cell to apoptosis (8). Type II cells require signal amplification by the intrinsic pathway: caspase-8 cleaves the Bcl-2 homology domain 3 (BH3)-only protein Bid, which engages the intrinsic pathway through the multi-BH domain proteins Bax and Bak, enhancing effector-caspase activation and apoptosis (1, 2).
Endocytosis internalizes plasma membrane (PM)-associated proteins through membrane-bound vesicles, supporting various cellular functions including nutrient uptake, growth-factor signaling, and membrane homeostasis (9). One of the best-characterized endocytosis pathways relies on the protein clathrin (10). Clathrin adaptors, such as adaptor protein 2 (AP2), link clathrin to cytoplasmic determinants of endocytic cargo during the formation of PM invaginations known as clathrin-coated pits. Adaptors also perform scaffolding functions in endocytosis by recruiting accessory or regulatory proteins (11). GTP hydrolysis by dynamin drives the scission of deeply invaginated coated pits to release endocytic transport vesicles from the PM. After uncoating, the vesicles dock and fuse with early endosomes, where cargo sorts to different fates, e.g., tubulovesicular endosomal membranes for recycling to the PM or internal membranes of multivesicular late-endosomes for lysosomal degradation.
We discovered that DR activation leads to caspase-dependent cleavage of several proteins involved in clathrin-mediated endocytosis, halting cargo uptake. Experimental inhibition of endocytosis augmented caspase activation downstream of DRs, suggesting that DR-triggered destruction of the clathrin endocytic pathway reinforces caspase activation, thus amplifying cell death.
Results
DR Activation Induces Caspase-Mediated Cleavage of the Clathrin-Dependent Endocytosis Machinery.
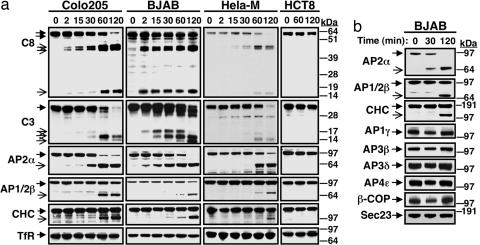
In studies on DR5 endocytosis, we observed that the DR5 ligand Apo2L/TRAIL triggered proteolytic cleavage of the α subunit of AP2 (AP2α). Analysis of several cancer cell lines showed that Apo2L/TRAIL promoted not only the expected processing of caspase-8 and -3, but also cleavage of AP2α, AP1/2β, and clathrin heavy chain (CHC) (Fig. 1a). These events occurred within 2 h and were restricted to cell lines susceptible to Apo2L/TRAIL-induced apoptosis, such as Colo205, BJAB, and HeLa-M, compared with resistant cell lines, such as HCT8 (Fig. 1a). Additional cell lines confirmed this observation (Fig. 7a, which is published as supporting information on the PNAS web site). FasL stimulated cleavage of AP2α and CHC in BJAB cells comparably to Apo2L/TRAIL (Fig. 7b). Apo2L/TRAIL also induced cleavage of dynamin, leading to depletion of the full-length protein that started within 1 h of stimulation (Fig. 7c). In a similar time frame, Apo2L/TRAIL did not induce proteolysis of structurally analogous adaptors that mediate other types of clathrin-dependent vesicular transport events (Fig. 1b). These included AP1γ, AP3β, AP3δ, and AP4ε, which support transport between the trans-Golgi network and endosomes (10), and the COP-I subunit β-COP or the COP-II subunit Sec23, which mediate transport between endoplasmic reticulum and Golgi (12). Hence, DR activation promotes rapid and specific cleavage of proteins involved in clathrin-dependent endocytosis.
Fig. 1.
Apo2L/TRAIL induces selective cleavage of the clathrin-dependent endocytosis machinery. (a) Cells were treated at 37°C with either trimeric Apo2L/TRAIL (Colo205 and HCT8) or antibody-crosslinked, tagged Apo2L/TRAIL (BJAB and HeLa-M), and cell lysates were analyzed by immunoblot for cleavage of caspase-8 (C8), caspase-3 (C3), adaptin (AP) 2α, AP1/2β (antibody does not distinguish the AP1 and 2 isoforms), CHC, or Tf receptor (TfR). (b) Colo205 cells were treated as in a and analyzed by immunoblot for processing of specific components of various types of clathrin-associated endocytic trafficking.
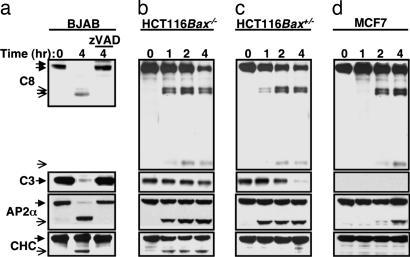
Pretreatment with the pan-caspase inhibitor N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone (zVAD-fmk) at a dose that inhibited ligand-induced processing of caspase-8 and -3 prevented cleavage of AP2α and CHC (Fig. 2a), as well as of AP2α in A549 cells (Fig. 8a, which is published as supporting information on the PNAS web site), indicating a requirement for caspase activity. Furthermore, although Apo2L/TRAIL and FasL induced cleavage of AP2α and CHC in WT Jurkat T cells, neither ligand stimulated processing of these targets in caspase-8-deficient or FADD-deficient mutant Jurkat cell lines (Fig. 8b). AP2α cleavage was fast and correlated in time with caspase-8 processing but preceded caspase-3 processing (Fig. 1a). By contrast, CHC cleavage was relatively slower and correlated better in timing with caspase-3 processing. In HCT116 cells, caspase-3 activation and apoptosis induction by Apo2L/TRAIL require Bax, whereas caspase-8 stimulation does not (5). As expected, Apo2L/TRAIL stimulated processing of both caspase-8 and -3 in Bax+/− HCT116 cells but only of caspase-8 in Bax−/− HCT116 cells (Fig. 2 b and c). AP2α cleavage was independent of Bax, whereas CHC cleavage required Bax (Fig. 2 b and c). In MCF7 cells, which are deficient in caspase-3 (13), Apo2L/TRAIL induced relatively weak and slow processing of caspase-8 and AP2α but did not stimulate CHC cleavage (Fig. 2d). Furthermore, small interfering RNA knockdown of caspase-3 in HT1080 cells blocked ligand-induced processing of CHC but not of AP2α (Fig. 8c). These findings suggest that initiator caspases in the DR pathway cleave AP2α, whereas effector caspases process CHC.
Fig. 2.
Involvement of different caspases in cleavage of AP2α and CHC. (a) BJAB cells were treated with the pan-caspase inhibitor zVAD-fmk (20 μM, 30 min) followed by treatment with crosslinked Apo2L/TRAIL (1 μg/ml) and analyzed by immunoblot for processing of caspase-8, caspase-3, AP2α, and CHC. Arrows with open heads indicate cleavage products, and arrows with filled heads indicate full-length proteins. (b–d) Bax−/− or Bax+/− HCT116 cells or caspase-3-defecient MCF-7 cells were treated with Apo2L/TRAIL and analyzed as in a.
AP2α contains two major functional parts, linked by a “hinge” region: the C-terminal “ear” and N-terminal “trunk” domains (11). To define the primary site of AP2α cleavage, we immunopurified the C-terminal 33-kDa cleavage product from BJAB cells and confirmed the identity of its tryptic peptides by mass spectrometry (Fig. 9a, which is published as supporting information on the PNAS web site). N-terminal sequencing revealed GPAAQPSLGPTPEEAFLS, a sequence immediately downstream from DVFD (Fig. 9a), a tetrapeptide sequence that resembles well characterized caspase recognition sites (14). The cleavage site has Asp and Gly at respective P1 and P1′ positions flanking the scissile bond and an Asp at the P4 position, which caspase-8 tolerates well (15). This tetrapeptide cleavage site sequence is present in isoform A of AP2α yet absent in isoform C. Consistent with this difference, Apo2L/TRAIL did not induce cleavage of isoform C, which was much less abundant than isoform A, in BJAB, Colo205, LS1034, and SW948 cells (data not shown). This AP2α cleavage site resides within the hinge (Fig. 9b), suggesting that its hydrolysis may disrupt AP2 function. Longer ligand stimulation of certain cell lines led to further processing of AP2α at a secondary site, which appeared to be within the initial 33-kDa fragment (Fig. 7 a and b).
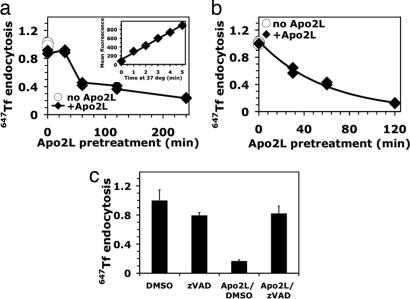
To assess the functional consequence of these caspase-mediated cleavage events, we studied uptake of transferrin (Tf). Internalization of receptor-bound Tf occurs via clathrin-coated pits and strictly requires AP2 (10). To determine the endocytosis rate of fluorescently labeled Tf, we confined measurements to the initial, linear phase of uptake, before internalized protein undergoes endocytic recycling to the PM (Fig. 3a Inset). Apo2L/TRAIL inhibited the rate of Tf endocytosis in BJAB and Colo205 cells by 75–85% (Fig. 3 a and b); pretreatment with zVAD-fmk substantially reversed this inhibition (Fig. 3c). Hence, DR activation leads to caspase-dependent disruption of clathrin-mediated endocytosis.
Fig. 3.
Pretreatment with Apo2L/TRAIL inhibits Tf endocytosis. (a and b) BJAB (a) or Colo205 (b) cells were pretreated at 37°C with or without crosslinked (a) or noncrosslinked (b) Apo2L/TRAIL for the indicated times and chilled on ice. The cells were then equilibrated on ice for 30 min with Alexa-647-conjugated Tf (647TF), and uptake at 37°C was measured by flow cytometry as described in Methods. Each endocytosis rate was derived from the slope of the initial linear phase of a 4-min uptake kinetics plot (a Inset). Rates were normalized to that observed in the absence of Apo2L/TRAIL (open circles), which was comparable to that observed when ligand was excluded from the preincubation step but present during the Tf incubation phase of the assay (filled diamonds; 0 min). (c) BJAB cells were preexposed to DMSO vehicle or zVAD-fmk for 30 min, treated for 4 h with crosslinked Apo2L/TRAIL, chilled on ice, and analyzed for 647Tf endocytosis rates as in a and b. Rates were normalized to the DMSO-treated sample (±SEM).
Apo2L/TRAIL Induces Clathrin-Mediated DR5 Endocytosis.
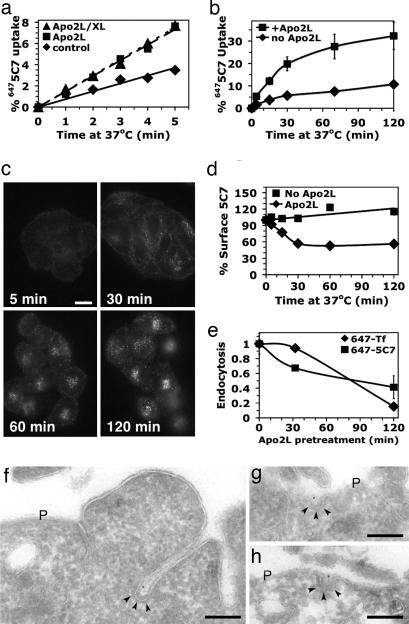
The rapid and selective cleavage of clathrin-pathway components suggested that physical proximity of DRs to clathrin coat proteins might facilitate the proteolytic events. Therefore, we asked whether DR5 undergoes clathrin-mediated endocytosis upon ligand stimulation. We used a monoclonal antibody (mAb 5C7) that recognizes the receptor’s extracellular domain without competing for ligand binding (13). Incubation with saturating amounts of 5C7 at 37°C did not affect surface DR5 levels as detected by another, nonoverlapping mAb (data not shown), verifying that 5C7 itself does not alter DR5 endocytosis. We prepared a fluorescent conjugate of mAb 5C7 (6475C7), bound it to Colo205 cells at 0°C, and followed it to measure the kinetics of DR5 endocytosis at 37°C (Fig. 4a). Exposure to trimeric or multimeric (antibody-crosslinked) forms of Apo2L/TRAIL similarly accelerated the rate of DR5 endocytosis by ≈2-fold (Fig. 4a), leading to ≈1/3 of the receptor internalizing over 2 h, mostly within the initial 30 min (Fig. 4b). Correspondingly, cell-surface DR5 declined during the first 30 min of stimulation, after which this DR5 pool stabilized (Fig. 4c). We observed similar results in BJAB cells (data not shown). In contrast to data obtained upon brief exposure to ligand at 37°C (Fig. 4a), extended exposure for 2 h inhibited DR5 endocytosis concomitantly with that of Tf, reducing the rate by 2.5- or 5-fold, respectively (Fig. 4d). These results raise the possibility that the clathrin pathway supports the endocytosis of DR5. Indeed in Colo205 cells briefly treated with Apo2L/TRAIL, a small fraction of the cell-surface 5C7 localized to clathrin-coated pits by electron microscopy, with no detectable localization to noncoated surface invaginations or caveolae (Fig. 4 f–h).
Fig. 4.
Characterization of DR5 endocytosis. (a and b) Colo205 cells with surface-bound 6475C7 mAb were incubated on ice for 30 min in the absence (diamonds) or presence of 10 μg/ml trimeric (squares) or crosslinked (triangles) Apo2L/TRAIL, then shifted to 37°C for the indicated time and rapidly chilled on ice. Surface fluorescence was removed by acid stripping, and DR5 uptake was quantified by flow cytometry. Mean values were plotted (±SEM in b). (c) HeLa-M cells were incubated at 37°C with 5 μg/ml 6475C7 and 5 μg/ml crosslinked Apo2L/TRAIL, then processed for immunofluorescence microscopy. (Scale bar: 20 μM.) (d) Colo205 cells were preequilibrated at 37°C with unlabeled mAb 5C7 for 30 min to bind surface and recycling DR5 pools. Incubations were continued with or without 10 μg/ml Apo2L/TRAIL, and the cells were rapidly chilled on ice. Cell-surface-exposed mAb 5C7 was probed with CY5 anti-mouse IgG and quantified by flow cytometry, and means were plotted (±SEM). (e) Colo205 cells were pretreated at 37°C with 10 μg/ml Apo2L/TRAIL, then rapidly chilled on ice and assayed for endocytosis as in a (6475C7) and Fig. 3b (647Tf). Endocytosis rates were normalized to the value without Apo2L/TRAIL pretreatment (±SEM). Similar results were observed when cells were prepared as in c, and endocytosis was assayed with surface-bound CY5 anti-mouse Fab (data not shown). (f–h) Colo205 cells with surface-bound mAb 5C7 were incubated on ice with Apo2L/TRAIL, shifted to 37°C for 5 min, and fixed. Ultrathin cryosections were labeled with rabbit anti-mouse IgG antibodies and Protein A gold (10 nm). The typical electron-dense clathrin coat is indicated by arrowheads. P, plasma membrane. (Scale bars in f–h: 200 nm.)
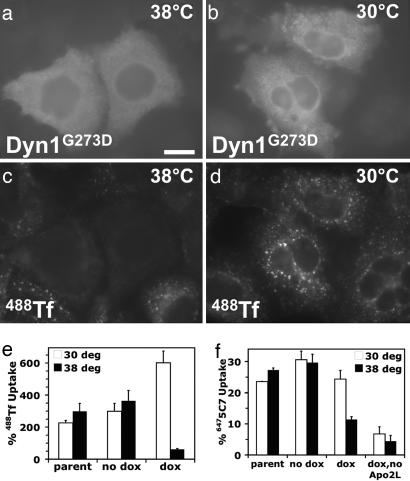
Next, we turned to an established intervention strategy based on doxycycline-regulated expression of a temperature-sensitive mutant of the GTPase dynamin-1 (dynG273D) in retrovirus-infected cells. This mutant rapidly and reversibly inhibits clathrin-dependent endocytosis upon shifting from a permissive (30°C) to a nonpermissive (38°C) temperature (16). Initial experiments in HeLa-M cells confirmed that doxycycline induction blocked Tf uptake at 38°C in cells with dynG273D expression but not in neighboring, nonexpressing cells or at 30°C (Fig. 5a–d; and see Fig. 10a, which is published as supporting information on the PNAS web site). Although inducible expression of dynG273D in HeLa-M or Colo205 cells was not sufficiently stable (data not shown), it was stable in a transduced BJAB cell line, with a strong block in Tf uptake at 38°C (Fig. 5e). Doxycycline-stimulated expression of dynG273D also decreased ligand-induced DR5 uptake from ≈25% at 30°C to ≈10% at 38°C. By contrast, endocytosis was similar at both temperatures in the absence of doxycycline (Fig. 5f). In HeLa-M cells dynG273D induction followed by a shift to 38°C also attenuated ligand-induced DR5 uptake; this effect was weaker than in BJAB cells, probably because of diminished dynG273D expression (Fig. 10b and data not shown). To exclude the unlikely possibility that decreased DR5 uptake in these 20-min uptake experiments was attributed to increased endocytic recycling rather than reduced endocytosis rates best measured over a 4-min uptake interval, we used a dominant-negative dynamin mutant (dynamin-1K44A) that unconditionally blocks clathrin-dependent endocytosis (17). Dynamin-1K44A inhibited both Tf endocytosis and ligand-induced DR5 endocytosis in HeLa-M cells (Fig. 10c).
Fig. 5.
Dynamin inactivation inhibits DR5 endocytosis. (a–d) DynG273D-transduced HeLa-M cells were puromycin-selected, doxycycline-induced, preincubated for 20 min at the indicated temperature, and incubated another 20 min in the presence of Alexa-488-conjugated Tf (488Tf). Cells were then processed for immunofluorescence microscopy by using a dynamin-1-specific antibody. (e and f) Nontransduced (parental) or clonal Dyn1G273D-transduced BJAB cells with (dox) or without (no dox) doxycycline induction were assayed for 488Tf or 6475C7 uptake over a 20-min period at 30°C (open bars) or 38°C (filled bars) by flow cytometry as described in Methods, and means were plotted (±SD).
These results indicate that Apo2L/TRAIL-induced DR5 uptake occurs primarily through clathrin-dependent endocytosis. Thus, physical proximity of the DISC within coated pits to clathrin-coat components may facilitate their cleavage. If true, DISC assembly and initiator-caspase activation might not require actual internalization of the coated pits. To test this, we chilled BJAB cells on ice to block Tf endocytosis (Fig. 11a, which is published as supporting information on the PNAS web site). Despite the block, Apo2L/TRAIL and FasL still recruited FADD and caspase-8 to their respective receptors and processed caspase-8 (Fig. 11b and data not shown). Furthermore, Apo2L/TRAIL induced comparable DISC formation, caspase-8 processing, and caspase-8 activity in the DISC at 38°C despite endocytosis inhibition by dynG273D (Fig. 11 c and d).
Inhibition of Clathrin-Mediated Endocytosis Augments DR-Induced Caspase Activation and Apoptosis.
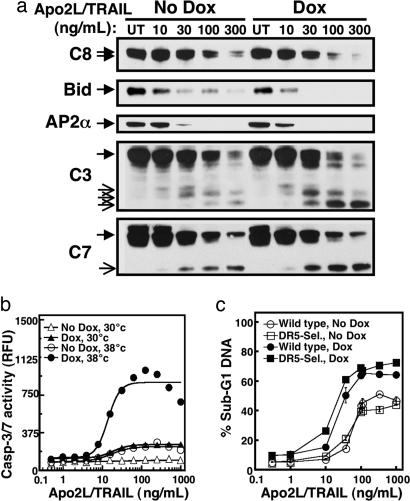
The rapid cleavage of clathrin-pathway proteins suggested that disruption of endocytosis might promote ligand-induced caspase activation and apoptosis. Indeed, dynG273D-mediated blockade of endocytosis markedly augmented ligand-induced processing of the total cellular caspase-8 pool, Bid, AP2α, and effector caspases-3 and -7 from whole-cell lysates (Fig. 6a), without significantly altering the formation or activation of the DISC itself (Fig. 11 c and d). Quantitative analysis showed a substantial increase in ligand-stimulated caspase-3/7 enzymatic activity after induction of dynG273D and temperature-shift to 38°C as compared with 30°C (Fig. 6b). Moreover, this condition also resulted in augmented apoptosis (Fig. 6c and data not shown), an effect observed after stimulation with either the WT ligand or a DR5-selective variant (18). Sensitization was evident not only by a left-shift of the dosage-response curve but also by greater maximal percentage of cells with fragmented DNA (Fig. 6c). These results indicate that caspase-mediated disruption of clathrin-dependent endocytosis amplifies caspase activation downstream of the DISC, leading to stronger apoptosis stimulation.
Fig. 6.
Dynamin inactivation augments DR-mediated caspase activation and apoptosis. (a) DynG273D-transduced BJAB cells with or without doxycycline induction were incubated at 38°C for 20 min to inactivate dynamin as in Fig. 5, incubated for an additional 4 h with or without crosslinked Apo2L/TRAIL, and analyzed by immunoblot for processing of the indicated proteins. (b) DynG273D-transduced BJAB cells with or without doxycycline induction were incubated at 30°C or 38°C for 20 min, incubated an additional 2 h with or without crosslinked Apo2L/TRAIL, and assayed for caspase-3/7 activity as described in Methods. (c) DynG273D-transduced BJAB cells with or without doxycycline induction were preincubated at 38°C for 20 min and incubated for an additional 4 h with or without crosslinked Apo2L/TRAIL or a DR5-selective Apo2L/TRAIL mutant (DR5-Sel.), and DNA fragmentation was assayed (±SEM) as described in Methods.
Discussion
Our studies uncover an unexpected interaction between the cell-extrinsic apoptosis pathway and the clathrin-dependent endocytosis machinery. Within minutes of proapoptotic DR engagement, activated caspases begin to cleave proteins that mediate clathrin-dependent endocytosis. The relatively rapid kinetics of AP2α cleavage, the requirement of this event for FADD and caspase-8, and the independence from Bax and caspase-3 suggest that DR-proximal caspases, most likely caspase-8 and/or caspase-10, carry out the initial processing of AP2α. Indeed, the caspase cleavage site in the AP2α hinge fits with sequences that caspase-8 is capable of recognizing (19).
The inhibition of clathrin-dependent endocytosis by DR activation occurred in the same time frame as caspase stimulation and was reversed by zVAD-fmk, demonstrating its dependence on caspase activity. The primary AP2α processing site mapped to the hinge region. The hinge links the ear domain, which supports accessory protein recruitment, to the trunk domain, which mediates PM association and cargo binding in conjunction with other AP2 subunits (11). Overexpression of the AP2α ear domain in COS7 cells inhibits clathrin-dependent endocytosis, an effect abrogated by mutations that prevent accessory-protein binding (20). Conversely, studies in Drosophila in which some cells express only earless AP2α suggest that the ear domain is not required in general endocytosis (21). Thus, it is unclear whether caspase cleavage of AP2α alone is sufficient to perturb endocytosis or whether the processing of other clathrin-pathway components also contributes. Regardless, DR-mediated caspase activation rapidly disrupts clathrin-dependent endocytosis.
How do activated caspases access clathrin-coat substrates? Our data suggest that after Apo2L/TRAIL stimulation, a substantial portion of surface DR5 moves into the cell through clathrin-dependent endocytosis. Our preliminary electron microscopy studies confirm that DR5 associates with clathrin-coated pits. Recent work indicates that RNA interference-mediated knockdown of clathrin and AP2 inhibits apoptotic signaling through Fas, a result interpreted as revealing an endocytosis requirement for receptor signaling (22). In light of our observations that Apo2L/TRAIL DISC assembly, activation, and apoptosis can still occur when endocytosis is inhibited by incubation on ice or through dynamin inactivation, we surmise that DR5 can bind FADD and caspase-8 while still at the cell surface, possibly within clathrin-coated pits. Indeed, blocking endocytosis with incubation on ice or temperature-sensitive dynamin does not interfere with coated-pit formation and cargo recruitment (16). DR activation caused nearly complete destruction of AP2α. In contrast, CHC and AP1/2β processing involved only a fraction of the total cellular proteins, perhaps the fraction that participated directly in DR endocytosis. Thus, caspase activation in the vicinity of the internalizing DR-associated DISC may lead to local destruction of specific clathrin-pathway components.
What is the biological significance of the finding that caspases disrupt clathrin-mediated endocytosis? One straightforward implication is that the elimination of this mechanism may represent a previously unrecognized part of the apoptotic program; a cell that is committed to apoptosis no longer needs to take up nutrients, respond to growth factors, or maintain membrane homeostasis. It is unlikely, however, that these cleavages on their own trigger apoptosis, because both clathrin and AP2 have been efficiently knocked down using small interfering RNAs by a number of different labs, with none reporting an increase in apoptosis. Notably, the cytotoxic, DNA-damaging drugs vinblastine and adriamycin also induced cleavage of components of the clathrin machinery, albeit with much slower kinetics than DR ligands (Fig. 12, which is published as supporting information on the PNAS web site). On the other hand, caspase-mediated processing of clathrin adaptors may play a role also in nonapoptotic cell modulation: in immature dendritic cells, basal caspase activity leads to cleavage of several adaptin subunits, whereas zVAD-inhibition of this effect promotes dendritic cell maturation (23).
A second, perhaps more surprising, implication of the interplay between DRs and the clathrin endocytic machinery is that this interaction provides a positive feedback loop that reinforces caspase activation through the extrinsic pathway. According to this model, endocytosis of activated DRs opposes apoptotic signaling to preserve cell survival if the clathrin machinery remains intact, for instance in response to subthreshold ligand levels. However, if DR stimulation generates enough caspase activity to disrupt endocytosis, then the signal persists, amplifying further caspase activation and promoting apoptosis. Conceivably, DR endocytosis could oppose DR signaling through lysosomal degradation, some other form of signal termination, or induction of competing antiapoptotic signals. Endocytosis did not regulate caspase activation at the level of initial DISC association. Nonetheless, dynamin inactivation enhanced the processing of total cellular caspase-8, and progressively of caspase-3 and -7, and led to stronger apoptosis activation, suggesting that endocytosis regulates further caspase activation downstream of the initial DISC. This amplification could be one of the determinants that differentiate type I and II cells, but this requires further investigation.
In summary, we have uncovered a bidirectional relationship between DR-induced apoptosis and clathrin-dependent endocytosis: DR activation leads to caspase-mediated destruction of important components of the clathrin endocytic machinery, thereby halting this process. Disruption of clathrin-dependent endocytosis augments caspase activation downstream of DRs and increases apoptosis, suggesting a positive feedback loop that amplifies caspase activation and apoptosis execution.
Methods
Materials and Imaging Methods.
The materials and imaging methods used are described in Supporting Methods, which is published as supporting information on the PNAS web site.
Identification of the AP2α Caspase Cleavage Site.
BJAB cells (5 × 108) were stimulated with Apo2L/TRAIL for 30 min and then lysed, immunoprecipitated by using C-terminal-specific anti-AP2α (AP6, MA1-064 from Affinity BioReagents), and collected on protein A/G agarose (Pierce). The C-terminal fragment was resolved by SDS/PAGE and either eluted for tryptic cleavage and analysis by liquid chromatography–electrospray ionization–ion trap tandem mass spectrometry or, in a separate immunoprecipitation, transferred to a polyvinylidene membrane for N-terminal sequencing.
Endocytosis, Uptake, and Surface Down-Regulation Assays.
For routine Tf measurements, 106 suspended cells were pretreated for indicated times at 37°C with 1–10 μg/ml Apo2L/TRAIL. In some studies, cells were pretreated with 0.25% DMSO ± 50 mM zVAD-fmk for 30 min at 37°C before adding Apo2L/TRAIL. After Apo2L/TRAIL stimulation, cells were equilibrated for 30 min on ice with 5 μg/ml fluorescent Tf in binding medium (serum-free DME/3% BSA/20 mM Hepes, pH 7.2), shifted to 37°C for 0–5 min as indicated, and rapidly chilled on ice to halt endocytosis. Sedimented cells were resuspended in cold 2% paraformaldehyde/PBS, and the mean fluorescence intensity of 10,000 cells was quantified with a Coulter EPICS Elite ESP flow cytometer. Endocytic rates were determined from linear slopes of steady-state internal/surface plots (24). For temperature-shift experiments, ice binding incubations were avoided. Instead, cells were preconditioned for 20 min and maintained at 30°C or 38°C in binding medium, incubated for an additional 0 (control) or 20 min with 647Tf, chilled on ice for 30 min, and processed for flow cytometry to determine internal/surface 647Tf ratios.
For routine DR5 measurements, cells were saturated on ice with 5 μg/ml 6475C7 in binding medium, washed three times with cold binding medium, and incubated for 30 min on ice with or without 10 μg/ml Apo2L/TRAIL. Cells were then shifted to 37°C for the indicated time, rapidly chilled, and either acid-stripped or not three times with cold 2 M urea/50 mM glycine/150 mM NaCl (pH 2.4). Cells were processed for flow cytometry and internalization was calculated similarly to the method described in ref. 25 with the equation: % uptake = 100 × [(St − S0)/(N − S0)], where St and S0 are the values of acid stripped samples incubated for time = t and time = 0, respectively, and N is nonstripped fluorescence, which remained essentially unchanged during the course of the 37°C incubation. For temperature shift, cells were preconditioned for 20 min and maintained at 30°C or 38°C, preequilibrated with 5 μg/ml 6475C7 for 3 min, treated with 10 μg/ml Apo2L/TRAIL for 0 (control) or 20 min, and chilled on ice 30 min. Cells were acid-stripped or not and quantified by flow cytometry.
For surface DR5 down-modulation assays, suspended cells were preequilibrated at 37°C with unlabeled 5C7 for 30 min to bind surface and recycling DR5 pools, treated for the indicated time with or without 10 μg/ml Apo2L/TRAIL, and chilled on ice. Surface 5C7 was probed with CY5 anti-mouse IgG and quantified by flow cytometry. This technique was used to avoid reduced surface probing efficiency attributable to Apo2L/TRAIL-induced receptor clustering, as seen upon direct probing with 6475C7 (data not shown).
Caspase Activity Assays.
The caspase-3/7 assay was performed by using the Apo-ONE Homogenous Caspase-3/7 assay (Promega). Cells were harvested, counted, aliquoted at equal numbers for each treatment, treated with Apo2L/TRAIL at varying doses for 4 h at 30°C or 38°C, and lysed in Homogeneous Caspase-3/7 reagent (containing the caspase-3/7 substrate Z-DEVD-R110). Lysates were incubated at room temperature for 1 h before reading in a fluorometer at 485/530 nM.
Apoptosis Assays.
Cells were stained with propidium iodide after ethanol fixation and RNase treatment, and the amount of subdiploid DNA was analyzed by flow cytometry as described in ref. 19, using expo32 software with an EPICS XL-MCL cytometer (Beckman Coulter).
DISC Immunoprecipitation.
BJAB cells (1 × 107 per time point) were treated with Flag-Apo2L/TRAIL cross-linked by anti-FLAG M2 Abs, collected, lysed, and immunoprecipitated, and the DISC was analyzed as described in ref. 13.
Supplementary Material
Acknowledgments
We thank S. Schmid (The Scripps Research Institute, La Jolla, CA) for kindly providing mutant dynamin-1 cDNAs, J. Blenis (Harvard University, Boston) for Jurkat cell lines, B. Vogelstein (Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore) for HCT116 cell lines, D. Gray and D. Davis (Genentech, Inc.) for pHUSH-ProEx expression vector, A. Paler-Martinez for help with flow cytometry, S. van Dijk for technical support, and W. Mallet for helpful comments.
Abbreviations
- Apo2L/TRAIL
Apo2 ligand/TNF-related apoptosis-inducing ligand
- CHC
clathrin heavy chain
- DISC
death-inducing signaling complex
- DR
death receptor
- PM
plasma membrane
- Tf
transferrin
- zVAD-fmk
N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Danial N. N., Korsmeyer S. J. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 2.Strasser A., O’Connor L., Dixit V. M. Annu. Rev. Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- 3.Thornberry N. A., Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 4.Nagata S. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 5.LeBlanc H. N., Ashkenazi A. Cell Death Differ. 2003;10:66–75. doi: 10.1038/sj.cdd.4401187. [DOI] [PubMed] [Google Scholar]
- 6.Kischkel F. C., Hellbardt S., Behrmann I., Germer M., Pawlita M., Krammer P. H., Peter M. E. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boatright K. M., Renatus M., Scott F. L., Sperandio S., Shin H., Pedersen I. M., Ricci J. E., Edris W. A., Sutherlin D. P., Green D. R., Salvesen G. S. Mol. Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 8.Scaffidi C., Schmitz I., Krammer P. H., Peter M. E. J. Biol. Chem. 1999;274:1541–1548. doi: 10.1074/jbc.274.3.1541. [DOI] [PubMed] [Google Scholar]
- 9.Conner S. D., Schmid S. L. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 10.Bonifacino J. S., Traub L. M. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 11.Owen D. J., Collins B. M., Evans P. R. Annu. Rev. Cell Dev. Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- 12.McMahon H. T., Mills I. G. Curr. Opin. Cell Biol. 2004;16:379–391. doi: 10.1016/j.ceb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Kischkel F. C., Lawrence D. A., Chuntharapai A., Schow P., Kim K. J., Ashkenazi A. Immunity. 2000;12:611–620. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 14.Stennicke H. R., Renatus M., Meldal M., Salvesen G. S. Biochem. J. 2000;350:563–568. [PMC free article] [PubMed] [Google Scholar]
- 15.Blanchard H., Donepudi M., Tschopp M., Kodandapani L., Wu J. C., Grutter M. G. J. Mol. Biol. 2000;302:9–16. doi: 10.1006/jmbi.2000.4041. [DOI] [PubMed] [Google Scholar]
- 16.Damke H., Baba T., van der Bliek A. M., Schmid S. L. J. Cell Biol. 1995;131:69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damke H., Baba T., Warnock D. E., Schmid S. L. J. Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley R. F., Totpal K., Lindstrom S. H., Mathieu M., Billeci K., Deforge L., Pai R., Hymowitz S. G., Ashkenazi A. J. Biol. Chem. 2005;280:2205–2212. doi: 10.1074/jbc.M410660200. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson D. W. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 20.Owen D. J., Vallis Y., Noble M. E., Hunter J. B., Dafforn T. R., Evans P. R., McMahon H. T. Cell. 1999;97:805–815. doi: 10.1016/s0092-8674(00)80791-6. [DOI] [PubMed] [Google Scholar]
- 21.Berdnik D., Torok T., Gonzalez-Gaitan M., Knoblich J. A. Dev. Cell. 2002;3:221–231. doi: 10.1016/s1534-5807(02)00215-0. [DOI] [PubMed] [Google Scholar]
- 22.Lee K. H., Feig C., Tchikov V., Schickel R., Hallas C., Schutze S., Peter M. E., Chan A. C. EMBO J. 2006;25:1009–1023. doi: 10.1038/sj.emboj.7601016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santambrogio L., Potolicchio I., Fessler S. P., Wong S. H., Raposo G., Strominger J. L. Nat. Immunol. 2005;6:1020–1028. doi: 10.1038/ni1250. [DOI] [PubMed] [Google Scholar]
- 24.Wiley H. S., Cunningham D. D. J. Biol. Chem. 1982;257:4222–4229. [PubMed] [Google Scholar]
- 25.Austin C. D., De Maziere A. M., Pisacane P. I., van Dijk S. M., Eigenbrot C., Sliwkowski M. X., Klumperman J., Scheller R. H. Mol. Biol. Cell. 2004;15:5268–5282. doi: 10.1091/mbc.E04-07-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.