Abstract
The alarming increase of antibiotic-resistant bacterial pathogens points to the need for novel therapeutic approaches to combat infection. To discover novel antimicrobials, we devised a screen to identify compounds that promoted the survival of the model laboratory nematode Caenorhabditis elegans infected with the human opportunistic pathogen Enterococcus faecalis. E. faecalis colonizes the nematode intestinal tract, forming a persistent lethal infection. Infected nematodes were rescued by antibiotic treatment in a dose-dependent manner, and antibiotic treatment markedly reduced the number of bacteria colonizing the nematode intestine. To facilitate high throughput screening of compound libraries, we adapted a previously developed agar-based C. elegans-E. faecalis infection assay so that it could be carried out in liquid medium in standard 96-well microtiter plates. We used this simple infection system to screen 6,000 synthetic compounds and 1,136 natural product extracts. We identified 16 compounds and 9 extracts that promoted nematode survival. Some of the compounds and extracts inhibited E. faecalis growth in vitro, but, in contrast to traditional antibiotics, the in vivo effective dose of many of these compounds was significantly lower than the minimum inhibitory concentration needed to prevent the growth of E. faecalis in vitro. Moreover, many of the compounds and extracts had little or no affect on in vitro bacterial growth. Our findings indicate that the whole-animal C. elegans screen identifies not only traditional antibiotics, but also compounds that target bacterial virulence or stimulate host defense.
Keywords: antibiotic resistance, chemical genetics, Enterococcus faecalis, antiinfectives
The growing problem of antibiotic-resistant bacteria (1–4) and the imminent threat of biowarfare agents (5) point to a need for new antiinfective therapies. However, the rate of new antimicrobial discovery is unlikely to meet the expected need for the foreseeable future (6–10). Specific problems include the overmining of cultivable microorganisms (11), a high background of toxic compounds or compounds with poor pharmacokinetic properties in synthetic compound libraries (7, 12), the inability of most synthetic leads to penetrate across the multidrug resistance (MDR) barrier of Gram-negative bacteria (13, 14), and the use of in vitro assays for small-molecule discovery that bear little resemblance to the biological systems in which the drugs need to function (12, 15, 16). Reasoning that some of these obstacles might be overcome by screening directly in a live-animal infection model, we developed an assay for identifying compounds that promote the survival of the nematode Caenorhabditis elegans persistently infected with the human opportunistic bacterial pathogen Enterococcus faecalis, an infection that leads to nematode death.
C. elegans is a useful and simple model host that can be infected and killed by a remarkably large number of human pathogens, including the Gram-negative bacteria Pseudomonas aeruginosa and Salmonella enterica, the Gram-positive bacteria E. faecalis and Staphylococcus aureus, and the fungal pathogen Cryptococcus neoformans (17, 18). Each of these organisms has been studied in the C. elegans model by simply replacing the normal food source (the lawn of Escherichia coli) with the pathogen in question and monitoring the survival of the nematodes. This simple feeding-based pathogenicity assay facilitates high-throughput screening and genetic analysis. Previous work has shown that there is broad overlap between the bacterial virulence factors required for pathogenesis in mammals and for C. elegans killing (18, 19). Additionally, the C. elegans pathogenesis models demonstrate that key features of the innate immune response have been conserved between C. elegans and mammals (20, 21).
Here, we show that nematodes infected with the persistent colonizer E. faecalis die when transferred to liquid medium in 96-well plates but can be rescued by treatment with antibiotics. We used this infection system to identify synthetic compounds and natural product extracts that promoted host survival. Advantages of this model are the ability to discover compounds with no antimicrobial activity in vitro, including prodrugs or compounds that target functions important only for in vivo survival or virulence, or activators of innate immunity. Because the readout of this in situ assay is nematode survival, an additional advantage of the screen is that it decreases the large background of compounds that are toxic or ineffective in vivo due to poor pharmacokinetics. Thus, this assay not only identifies compounds with novel antimicrobial activities but also has the potential to solve the bottleneck of toxicity/efficacy testing in drug development.
Results
Antibiotic Treatment Rescues Infected Nematodes.
When C. elegans are transferred from a lawn of E. coli, their normal laboratory food, to a lawn of E. faecalis growing on agar medium, the E. faecalis forms a persistent and lethal infection in the C. elegans intestine (22), surviving digestion and accumulating to levels up to 105 colony-forming units (cfu) per worm. The infection persists in the worms even after they are transferred to a benign bacterial food source such as Enterococcus faecium that does not kill worms (22). We established an agar-based persistent infection model to determine whether antibiotic treatment could cure worms of an E. faecalis infection. In these experiments, Enterococcus faecium, which accumulates in the C. elegans intestine, but which does not kill the worms or persistently colonize (22), was used as a negative control. Worms were fed on lawns of E. faecalis or E. faecium and transferred to fresh lawns of E. faecalis or E. faecium. The rationale for transferring the worms from E. faecalis to E. faecium was that worms could feed on the nonpathogenic E. faecium strain but would remain colonized with E. faecalis.
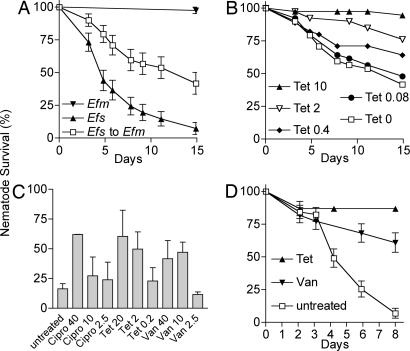
Worms transferred from E. faecalis to E. faecalis died with similar kinetics as worms left on an E. faecalis lawn for the entire duration of the experiment; half of the worms died within 5 days (Fig. 1A and data not shown). Worms transferred from E. faecalis to E. faecium died because of the persistent infection, although the killing rate was slower compared with worms exposed only to E. faecalis. In contrast, control worms transferred from E. faecium to E. faecium survived for >15 days (Fig. 1A).
Fig. 1.
Curing of a nematode E. faecalis infection on solid medium by antibiotic treatment. (A) Kaplan–Meier survival curves of WT N2 C. elegans feeding continuously on lawns of E. faecalis (Efs) V583 (▴) or E. faecium (Efm) DO (▾). Nematodes feeding for 24 h on E. faecalis and subsequently transferred to lawns of E. faecium (□) die with an LT50 of 14.9 days. Error bars equal SEM. (B) Kaplan–Meier survival curves of E. faecalis V583-infected N2 C. elegans transferred to lawns of E. faecium on media containing tetracycline at 10 μg/ml (▴, P < 0.0001), 2 μg/ml (▿, P < 0.0014), 0.4 μg/ml (♦, P < 0.06), 0.08 μg/ml (●, P = 0.56), or no tetracycline (□). (C) Antibiotic concentrations required to promote rescue of E. faecalis VS583 infected N2 nematodes. Survival was measured 13 days postinfection after treatment with tetracycline (Tet), vancomycin (Van), or ciprofloxacin (Cipro). The MICs for the E. faecalis strain VS583 are 0.39 μg/ml Cipro, 0.27 μg/ml Tet, and 3.1 μg/ml Van. Note that VS583 is a vancomycin-sensitive derivative of V583, the well-studied vancomycin-resistant E. faecalis strain. Error bars are standard deviations. (D) Kaplan–Meier survival curve of glp-4;sek-1 nematodes infected for 12 h on E. faecalis OG1RF and transferred to BHI media containing 20 μg/ml tetracycline (▴), 40 μg/ml vancomycin (▾), or no additional antibiotic (□). Error bars equal SEM.
We used the infection protocol described in the previous paragraph to determine whether worms persistently infected with E. faecalis could be cured by antibiotic treatment. We found that E. faecalis-infected worms were rescued by antibiotic treatment in a dose-dependent manner. Varying concentrations of tetracycline were added to the media that were used to grow the lawns of E. faecium to which the E. faecalis-infected worms were transferred. The E. faecium strain DO is tetracycline-resistant. The E. faecalis strain V583 is tetracycline-sensitive, with a minimum inhibitory concentration (MIC) of 0.24 μg/ml in brain–heart infusion (BHI) broth. As shown in Fig. 1B, 10 μg/ml tetracycline completely prevented the death of E. faecalis-infected nematodes after transfer to an E. faecium plate. Tetracycline at 2 μg/ml also rescued to a significant extent. Tetracycline at 0.08 or 0.4 μg/ml did not significantly rescue E. faecalis-infected nematodes. Similar results were obtained with other E. faecalis strains (data not shown).
Several different antibiotics, including ampicillin, ciprofloxacin, and vancomycin, to which a particular strain of E. faecalis was susceptible, also rescued worms infected with E. faecalis VS583 [a vancomycin-sensitive derivative of V583 (23)]. However, gentamycin, kanamycin, and nalidixic acid, which are not active against the E. faecalis strain, did not promote survival of VS583-infected worms (Fig. 1C and data not shown). In all of the cases tested in which a particular antibiotic cured the worms, the antibiotic had to be at a concentration severalfold above its MIC to promote the survival of the worms. In the case of E. faecalis strain VS583, the antibiotic concentration that promoted survival relative to the MIC was ≈4-fold above that for ampicillin (1.5 μg/ml), 100-fold above the MIC for ciprofloxacin (40 μg/ml), 7-fold above that for tetracycline (2 μg/ml), and 3-fold above that for vancomycin (10 μg/ml) (Fig. 1C and data not shown). A useful comparator for the efficacy of tested antibiotics in curing the infected worms is the therapeutic human blood plasma concentration of these compounds, which is 2–20 μg/ml ampicillin, 2.4–4 μg/ml ciprofloxacin, 2.4–4 μg/ml tetracycline, and 5–10 μg/ml vancomycin (24). Thus, for three of the four antibiotics (ciprofloxacin being the exception), the concentrations that promoted nematode survival are equivalent to the desired plasma levels.
Increasing the Rate of Killing Using an Immunocompromised C. elegans Mutant.
We wished to shorten the duration of the curing assay, primarily because we were concerned about the stability of small molecules. Indeed, we found that the glycopeptide antibiotic vancomycin becomes ineffective in the curing assay after ≈5 days. We were also concerned that some nematode killing was a consequence of eggs being retained in the uterus and hatching internally. To accelerate worm killing and to prevent internal hatching of progeny, we substituted glp-4;sek-1 mutant worms for WT. sek-1 mutants contain a mutation in a mitogen-activated protein kinase (MAPK) kinase (MAPKK) of the C. elegans innate immune p38 MAPK signaling cascade and are more susceptible to a variety of pathogens (25). glp-4 temperature-sensitive sterile mutants do not make a germ line at the restrictive temperature and survive without a bacterial food source, whereas WT C. elegans die by internal hatching of progeny when transferred to bacteria-free media. As shown in Fig. 1D, when glp-4;sek-1 worms were infected with E. faecalis OG1RF and transferred to BHI agar without an E. faecium lawn, they died with an LT50 (time for half to die) of 4.2 days, which is almost as fast as worms left on an E. faecalis lawn (data not shown) and is one-third the time required to kill WT N2 worms, as shown in Fig. 1A. Incorporating tetracycline at 20 μg/ml or vancomycin at 40 μg/ml into the BHI medium (to which the E. faecalis-infected worms were transferred) without any lawn effectively rescued the infected worms, with 80–90% of the worms surviving for >6 days (Fig. 1D).
Development of an E. faecalis Curing Assay in Liquid Media.
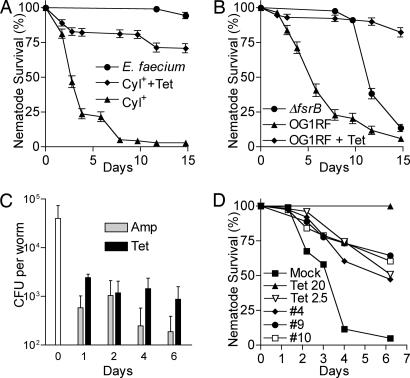
C. elegans are typically grown (and infected with pathogens) on agar medium. However, it is difficult to automate an agar-based assay. The multiple steps involved in an agar-based assay, including pouring agar, growing a bacterial lawn, adding compounds, and seeding worms, prompted us to develop a liquid pathogenicity assay amenable to automation that could be used to screen for antiinfective compounds. After infection with E. faecalis on agar plates, the infected worms are resuspended in a liquid medium consisting of 10–20% BHI in M9 buffer. After transfer to microtiter plates, the worms are incubated without agitation, which allows both the worms and any free-living bacteria that are transferred with the worms to sink to the bottom of the wells. Nevertheless, worms that are not infected with a pathogen (e.g., worms feeding on E. faecium strain 11M12; Fig. 2A) or worms infected with E. faecalis that are transferred into BHI containing an appropriate antibiotic (see below) live at least 14 days. In contrast, worms infected with E. faecalis strain OG1RF that are transferred to BHI without antibiotics die with an LT50 of 5.8 days (Fig. 2B).
Fig. 2.
The liquid infection assay gauges differences in nematode killing due to E. faecalis strains with varying degrees of pathogenicity or due to antibiotic treatment. (A) Kaplan–Meier survival curves of glp-4;sek-1 nematodes that were infected with the cytolysin (Cyl) producing E. faecalis strain MMH594 (▴, ♦) or the E. faecium strain 11M12 (●). Cyl-infected nematodes were treated with 0 (▴) or 20 μg/ml (♦) tetracycline (Tet). Error bars equal SEM. (B) Kaplan–Meier survival curves of glp-4;sek-1 nematodes that were infected with E. faecalis OG1RF (▴, ♦) or the two-component quorum-sensing regulator mutant OG1RF ΔfsrB (●). OG1RF-infected nematodes were treated with 0 (▴) or 20 μg/ml (♦) tetracycline. (C) The bacterial load in the nematode intestinal tract after antibiotic treatment. E. faecalis-infected nematodes were treated in liquid media containing 20 μg/ml ampicillin (Amp) or tetracycline, and the number of cfu per worm was determined. Error bars equal standard deviations. (D) Curing kinetics of selected hit compounds. Shown are Kaplan–Meier survival curves of infected nematodes treated with 25 μg/ml compound 4 (♦), 50 μg/ml compound 9 (●), 25 μg/ml compound 10 (□), 2.5 μg/ml tetracycline (▿), 20 μg/ml tetracycline (▴), or mock treatment (■). In pairwise comparisons to mock treatment using log-rank tests, the difference for all of the treatments was significant, with P < 0.0001.
To determine whether the liquid killing assay behaved similarly to the standard agar assay, we compared killing mediated by OG1RF to killing mediated by an OG1RF ΔfsrB mutant, which is defective in a two-component, quorum-sensing pathway (26). In the agar assay, the ΔfsrB mutant is severely attenuated (22). Similarly, the ΔfsrB mutant is highly attenuated in the liquid assay, with an LT50 of 11.7 days (Fig. 2B). As on solid media, E. faecium did not kill nematodes in the liquid assay, and E. faecalis strains that produce cytolysin killed more quickly in the liquid assay. The strain that showed the fastest killing was the cytolysin-positive strain MMH594 (27). The LT50 of glp-4;sek-1 worms infected with MMH594 was 2.8 days (Fig. 2A), and this strain was used in the compound screening assays described below. Importantly, worms infected with the three different E. faecalis strains could be rescued with tetracycline, as illustrated in Fig. 2 A and B.
We also examined the effect of antibiotic treatment on the colonization of E. faecalis in the nematode intestinal tract. After feeding on E. faecalis OG1RF for 16 h, an average of 4.0 × 104 cfu was recovered from each worm (Fig. 2C). The worms were then transferred to liquid media containing 20 μg/ml ampicillin or tetracycline or no antibiotic. Although the number of viable bacteria that colonized the untreated worms remained relatively constant for 2 days of incubation in liquid media (data not shown), the number of cfu per worm in ampicillin or tetracycline-treated worms declined dramatically. After 24 h of treatment, the cfu per worm in ampicillin- and tetracycline-treated worms were only 1.5% and 6.1% of the cfu in untreated worms, respectively. After 6 days of treatment, the percentage dropped to 0.6% and 3.6% for the ampicillin- and tetracycline-treated worms, respectively (Fig. 2C). Interestingly, the small numbers of worms that appeared sickly or that died with antibiotic treatment were colonized with the same numbers of E. faecalis as untreated worms.
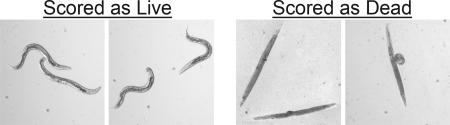
In the liquid assay, live worms maintain a sinusoidal posture (Fig. 3) and can be seen moving their body and pumping their pharyngeal muscles. In contrast, dead worms become straight and rigid as they become bloated with E. faecalis cells. Frequently, the dead and dying worms become so bloated that the body of the worms is stretched and lengthened or the vulva of the worm bursts out (Fig. 3). These obvious differences in appearance between live and dead worms greatly facilitated the scoring of the assay.
Fig. 3.
Scoring live/dead worms in the liquid killing assay. Living nematodes in the liquid infection assay maintain a sinusoidal shape, whereas dead nematodes in the liquid infection assay appear as straight, rigid rods as the corpse becomes filled with bacteria.
Screening for Antiinfectives That Promote Survival of E. faecalis-Infected Worms.
We performed a pilot screen in a 96-well plate format using the E. faecalis–C. elegans liquid infection assay for antiinfective substances that permitted the survival of the infected worms. We screened 6,000 individually synthesized compounds obtained from ChemBridge (San Diego), representing a rich variety of structures, and 1,136 natural product extracts from a National Cancer Institute (NCI) library that primarily contains extracts from tropical plants as well as extracts from marine invertebrates and algae. We manually examined the assay plates using a dissecting microscope after 6–8 days of incubation when 80–85% of the untreated worms have died. The false-positive rate of the screen was 2.7%. The false-negative rate using antibiotic controls or worms infected with the attenuated ΔfsrB strain was 1.7%.
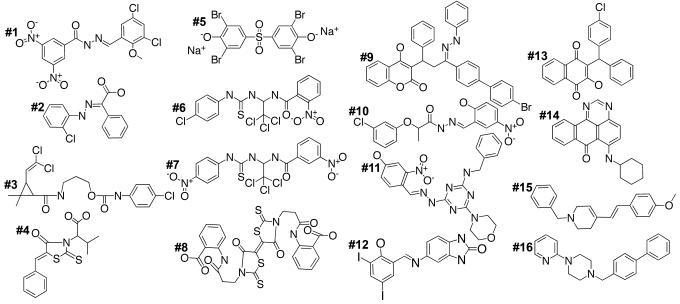
The screen of the 6,000 ChemBridge compounds and 1,136 NCI extracts was carried out in duplicate, and a compound was scored as a “hit” if >50% of the worms survived in both duplicates or if one of the duplicate wells showed a high level of rescue (approximately >80% rescue). The 50% cutoff for this primary screen was not stringent, to ensure that substances with at least some activity would not be excluded. The primary screen identified 90 small molecule compounds (1.5% hit rate) and 58 extracts (5.8% hit rate). In a secondary screen, 18 of the small molecule compounds (0.3%) and 9 of the extracts (0.9%) were reconfirmed to promote survival of infected worms. For further testing, additional samples were purchased from ChemBridge. The structures, activity in the worm infection assay, and MIC values of 16 of these hits are shown in Table 1 and Fig. 4.
Table 1.
Compounds that promote the survival of nematodes infected with E. faecalis
| Compound no. | Compound ID | Fold survival vs. untreated* | Therapeutic concentration, μg/ml | Percentage colonization vs. untreated† | MIC, μg/ml |
|---|---|---|---|---|---|
| 1 | 5113013 | 3.0 | 25 | 22 | >125 |
| 2 | 5117999 | 2.6 | 12.5 | 13 | >125 |
| 3 | 5139008 | 1.6 | 6.3 | 2 | >30 |
| 4 | 5140317 | 2.7 | 50 | 7 | >125 |
| 5 | 5142855 | 2.3 | 12.5 | 1 | >125 |
| 6 | 5142992 | 2.2 | 100 | 8 | >125 |
| 7 | 5143237 | 3.0 | 25 | 1 | 31.3 |
| 8 | 5146442 | 3.1 | 25 | 3 | >125 |
| 9 | 5151562 | 3.3 | 25 | 6 | 31.3 |
| 10 | 5189005 | 2.6 | 25 | 5 | 3.9 |
| 11 | 5200063 | 2.6 | 25 | 8 | 15.6 |
| 12 | 5202043 | 3.0 | 50 | 1 | 15.6 |
| 13 | 5248942 | 2.7 | 6.3 | 9 | 7.8 |
| 14 | 5253943 | 2.1 | 6.3 | 11 | 2.0 |
| 15 | 5255640 | 1.9 | 100 | 61 | >125 |
| 16 | 5260504 | 1.8 | 25 | 101 | >125 |
| Tet control | 4.0 | 1.6 | 5 | 0.24 |
Tet, tetracycline.
*Note that 24% of the untreated worms survived 4 days postinfection.
†Untreated worms were colonized with 3.3 × 104 cfu per worm.
Fig. 4.
Structures of the compounds that promote the survival of nematodes infected with E. faecalis. The compound number in bold corresponds to the numbering in Table 1.
Among the 16 ChemBridge compounds, 15 did not exhibit any obvious toxicity toward C. elegans, at least with respect to overall appearance and health of the worms. The only compound displaying toxicity (compound 6) resulted in some growth retardation. However, none of the 16 exhibited cytotoxicity against sheep erythrocytes when tested in a hemolysis assay (data not shown). Importantly, 13 of the 16 compounds exhibited significant activity, increasing survival by at least 2-fold when retested in the curing assay (Table 1 and Fig. 4). Five of the compounds (compounds 1, 7, 8, 9, and 12) increased survival 3-fold or more, a curing rate that is significantly above the threshold sensitivity of the assay. Fig. 2D shows example curing rates with three selected ChemBridge compounds. Although none of the ChemBridge compounds cured as effectively as 20 μg/ml tetracycline, these compounds were as effective as 2.5 μg/ml tetracycline. Note that tetracycline is one of the best penetrating antibiotics and is often used against intracellular pathogens. In this regard, vancomycin, an efficacious anti-E. faecalis antibiotic, is probably a more realistic benchmark comparator for our hit compounds. Indeed, the hit compounds have comparable worm-curing activity to vancomycin (Fig. 1).
Of the 13 compounds that increased worm survival by at least 2-fold, 7 of them inhibited the growth of E. faecalis with MICs lower than 31 μg/ml (Table 1). The most potent antimicrobial compounds were compounds 10 and 14, with MICs of 3.9 and 2.0 μg/ml, respectively (Table 1). In contrast to the compounds with in vitro antimicrobial activity, 6 of the compounds had an MIC that was greater than the highest concentration tested (125 μg/ml) or had an MIC greater than the aqueous solubility limit of the compound (30 μg/ml for compound 3). Nevertheless, at least 5 of these compounds with high MICs (compounds 3, 4, 5, 6, and 8) were comparable with tetracycline in dramatically reducing the level of intestinal colonization. In contrast, two compounds, 15 and 16, had moderate activity in the curing assay and had only a modest or no effect on colonization. Similarly to the ChemBridge compounds, at least two of the NCI extracts did not significantly inhibit E. faecalis growth in vitro (at 200 μg/ml), even though they were effective at curing the worms in vivo.
Discussion
Our screen of a synthetic compound library and of natural product extracts for substances that cure C. elegans of a persistent E. faecalis infection suggests that, in contrast to a traditional antibiotic screen, the E. faecalis assay not only identifies compounds that block pathogen replication in vitro but also identifies compounds that may be prodrugs, that affect the virulence of the pathogen, that suppress pathogen survival, or that enhance the immune response of the host. Because some of the identified compounds and extracts have significant activity only in vivo in the whole-animal assay, these data provide proof-of-principle for using a whole-animal screen in a drug discovery program to identify novel antimicrobial compounds.
Compounds with a Lower Effective Dose in Vivo than in Vitro.
One of the most interesting features of many of the identified compounds is their unusual ability to promote nematode survival at concentrations that were much lower than their MIC value in vitro. In contrast, the effective dose in the C. elegans curing assay of all of the known antibiotics that we tested was severalfold higher than the in vitro MIC. The effective dosage in the nematode model is similar to the therapeutic concentration of most antibiotics in human blood serum, which is typically 5- to 20-fold higher than the MIC (24). Possible explanations for the discovery of highly effective in vivo compounds with the nematode model include the possibilities that (i) the screen selects for compounds that are concentrated in the worm gut, (ii) the C. elegans immune system makes the bacteria more susceptible to inhibition, or (iii) the compounds weaken the integrity of the bacteria so that they are more readily digested. However, it seems likely that each of these explanations would also apply to traditional antibiotics. An alternative explanation is that the C. elegans curing assay specifically identifies compounds that target functions mainly important for in vivo survival or virulence, or activators of innate immunity, and that compounds with these activities may be more common than traditional antibiotics. Natural or synthetic compounds that block the virulence of pathogenic microbes (“virulence antiinfectives”) are a largely unexplored class of antimicrobial agents. The successful targeting of a virulence product is demonstrated in a recent paper by Hung et al. (28), in which a high-throughput screen was used to identify compounds that inhibit the activity of the Vibrio cholerae transcriptional regulator ToxT, which is required for expression of cholera toxin.
Compounds that inhibit E. faecalis virulence could potentially target expression or function of cytolysin, serine protease, gelatinase, the quorum-sensing pathway, or colonization of the bacteria in the worm gut. We did not detect inhibition of proteolytic or cytolytic activity of the bacteria grown on milk agar or blood agar plates by any of the 10 compounds that we identified that had a high MIC value (data not shown). A possible mode of action for these compounds is the inhibition of bacterial colonization, but this activity is difficult to distinguish from antibiotic activity, which also reduces bacterial colonization.
Compounds that function as immune enhancers may activate the C. elegans immune pathway downstream of a conserved p38 MAPK cascade. A cascade consisting of the C. elegans PMK-1, SEK-1, and NSY-1 proteins, corresponding to the p38 MAPK, and upstream MAPK kinase (MAPKK) and MAPK kinase kinase (MAPKKK), is required for the response to a variety of bacterial and fungal pathogens, and loss of any of these signaling components results in nematodes that have enhanced susceptibility to the pathogens (25). The C. elegans-E. faecalis curing assay used a sek-1 mutant worm strain that dies more quickly when exposed to E. faecalis, and a potential activity of a hit would be an activator of the downstream p38 MAPK.
Toxicity Testing.
Fifteen of 16 compounds identified in this study did not show any signs of toxicity against C. elegans or mammalian erythrocytes, indicating that the screen was able to select against toxic compounds. In a separate study using the nematode infection assay, we found that the indole derivative INF55, which is an effective inhibitor of multidrug resistance (MDR) pumps, is toxic to C. elegans, in agreement with the finding that this compound is also toxic to HeLa cells at similar concentrations (P. Markham, personal communication). These results indicate that the worm infection model will be able to select against at least some compounds that exhibit toxicity.
In other studies, C. elegans has been used as an indicator of toxicity from heavy metals, environmental pollutants, organic solvents, and neurotoxins (29). Toxicity against nematodes has been quantified based on nematode survival, growth, reproduction, expression of stress response proteins, feeding behavior, and movement. The utility of C. elegans in toxicology testing greatly depends on how it correlates with toxicity in mammalian models. Williams and Dusenbery (30) determined that toxicity of heavy metals against C. elegans as measured by the LC50 values correlates well with toxicity against mice or rats in rank order tests. Additionally, Cole et al. (31) reported a significant correlation from rank order toxicity tests from organophosphates between C. elegans and rodents.
Conclusions.
The advent of genomics and combinatorial chemistry brought with them the promise of using defined targets to identify new antibiotics (32). Follow-up chemistry was then supposed to produce modifications enabling an inhibitor to effectively penetrate pathogen cells. This approach, however, has proven to be a formidable problem. Our results suggest that C. elegans whole-animal antimicrobial screening may be an effective new drug-discovery platform.
There are a number of advantages of the C. elegans whole-animal screening method:
-
1
Nematodes can be infected by a variety of different pathogens, making the model amenable to identification of both narrow- and broad-spectrum antibiotics.
-
2
Compounds are automatically tested for host toxicity because the assay requires that the compound does not adversely affect the normal physiology of the nematodes.
-
3
Compounds are automatically tested for in vivo efficacy.
-
4
The screen automatically identifies prodrugs that have to be modified by the host to form an active antimicrobial.
-
5
The screen identifies compounds that target previously unidentified virulence factors (or other functions important for in vivo survival) but may not interfere with the growth of the pathogen in vitro.
-
6
The screen identifies compounds that enhance host immunity.
The data shown in Table 1 provide proof-of-principle that the C. elegans antimicrobial screen will produce hits that are overlooked in a conventional in vitro screen and may yield better-quality hits that have direct in vivo activity.
Materials and Methods
Bacterial and Nematode Strains.
WT Bristol N2 (33) and glp-4(bn2ts);sek-1(km4) (34, 35) C. elegans strains were maintained by using standard practices (36). E. faecalis strains MMH594 (27), OG1RF (37), OG1RF ΔfsrB (26), V583 (38), VS583 (23), and E. faecium strains DO (39) and 11M12 (23) were grown on BHI media (Difco/Becton Dickinson) at 37°C.
Nematode Killing and Rescue.
N2 or glp-4(bn2ts);sek-1(km4) worms were synchronized by isolating eggs from gravid adults, hatching the eggs overnight in M9 buffer, and plating L1-stage worms onto lawns of E. coli on nematode growth medium (NGM) agar media. Worms were grown to sterile, young adults by incubation at 25°C for 48–52 h, washed off the plates with M9 buffer, resuspended and washed in M9 buffer, deposited onto lawns of E. faecalis grown on BHI agar plates containing kanamycin at 80 μg/ml to inhibit E. coli growth, incubated for 8–12 h at 25°C, and resuspended in M9 buffer. For assays using agar media, ≈35 infected worms were washed and then deposited onto 35-mm plates containing the appropriate antibiotics. Plates were incubated at 25°C and scored for worm survival at regular intervals. Worms were considered dead if they were unresponsive to touch with a platinum wire pick. For the initial assays using liquid media, ≈80 infected worms were transferred to wells of a six-well plate containing 2 ml of media consisting of 10% BHI 80 μg/ml kanamycin, 90% M9 buffer, and the appropriate antibiotics. The plates were incubated without agitation at 25°C and 80–85% relative humidity. To score for worm survival, the six-well plates were shaken by hand, and the worms were considered to be dead if they did not move or exhibit muscle tone.
Bacterial Colonization.
Infected worms were washed three times with M9 buffer containing 1 mM sodium azide to inhibit expulsion of bacteria from the worm intestine. Approximately 10 worms were transferred to a 2-ml microcentrifuge tube, and the volume was brought to 250 μl. Fifty microliters of buffer was removed and plated to determine the number of external cfu. Approximately 400 mg of 1.0-mm silicon carbide particles (Catalog no. 11079110sc; Biospec Products, Bartlesville, OK) were added to each tube, the tubes were vortexed at maximum speed for one minute, which disrupts the worms but does not affect bacterial survival, and the resulting suspension was diluted and plated onto selective media to determine cfu.
Screening for Antiinfectives.
Synchronized L4 stage to young adult glp-4(bn2ts);sek-1(km4) worms were infected for 8 h on lawns of MMH594 as described above. The worms were resuspended in media composed of 20% BHI, 80 μg/ml kanamycin, and 80% M9 buffer. Approximately 25 worms in a volume of 50 μl were transferred into 0.3-ml wells of 96-well plates. An equal volume of liquid media containing 125 units/ml nystatin and the compounds or extracts to be tested were mixed into the wells. Each compound or extract was tested in individual wells, and the screen was performed by using duplicate 96-well plates. The final concentration of compounds from ChemBridge Diverset E was 25 μg/ml with DMSO at 1%. The concentration of the extracts from the natural product NCI library was 150 or 200 μg/ml with DMSO at 1.5 or 2%. The plates were sealed with gas-permeable membranes (Breatheasy, Diversified Biotech) and incubated without agitation at 26°C and 80–85% relative humidity.
Each 96-well plate contained 80 test compounds that were tested with worms infected with the cytolysin-positive E. faecalis strain MMH594. The remaining 16 wells contained positive and negative controls to determine whether the assay yielded predictable and reproducible responses to antibiotics or avirulent E. faecalis mutants and a clear threshold between positive and negative responses. Each plate contained eight negative control wells that did not contain any antimicrobial compound and four positive control wells that contained 20 μg/ml ampicillin (two wells) or 20 μg/ml tetracycline (two wells). These control wells were seeded with glp-4;sek-1 infected with E. faecalis MMH594. In addition, each plate contained four positive control wells that were seeded with glp-4;sek-1 worms infected with the E. faecalis ΔfsrB mutant. For the screen of 1136 NCI extracts, each 96-well plate contained 88 samples, four negative control wells, two positive control wells that contained 20 μg/ml ampicillin or 20 μg/ml tetracycline, and two positive control wells seeded with worms infected with the ΔfsrB mutant. In total, there were 1,312 control wells in the ChemBridge and NCI screens. The false-positive rate was 2.7%, and the false-negative rate was 1.9%. False positives were defined as >50% survival of infected worms in wells that did not contain an antibiotic. False negatives were defined as <50% survival of worms treated with an antibiotic or infected with the ΔfsrB mutant.
Worm survival was scored manually after 6 days of incubation. A total of 6,000 compounds from a ChemBridge library Diverset E and 1,136 extracts from the NCI Natural Products Repository (www.dtp.nci.nih.gov/branches/npb/repository.html) were screened. Compounds or extracts that increased worm survival by 2- to 3-fold were retested for activity. MICs were determined against E. faecalis strain MMH594 by using 2-fold dilution in BHI media according to the broth microdilution protocol of the Clinical and Laboratory Standards Institute [CLSI; formerly National Committee for Clinical Laboratory Standards (NCCLS)] (40). The ability of compounds to hemolyze sheep erythrocytes was based on the protocol of Ciornei et al. (41), with the following modifications: sheep erythrocytes (Rockland Immunochemicals) were treated with compounds (100 μg/ml) in PBS with DMSO at 2% for 2 h and the supernatants were monitored at OD540.
Acknowledgments
We thank K. Carniol and M. S. Gilmore (Schepens Eye Research Institute, Boston) for generous gifts of strains and advice. This work was supported by National Institutes of Health Grant R21 AI059483 and by postdoctoral fellowship grant PF-02-130-01-MBC from the American Cancer Society (to T.I.M.).
Abbreviations
- cfu
colony-forming unit
- MIC
minimum inhibitory concentration
- BHI
brain–heart infusion
- MAPK
mitogen-activated protein kinase
- LT50
time for half to die.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Chambers H. F. Emerg. Infect. Dis. 2001;7:178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zinner S. H. Expert Rev. Anti Infect. Ther. 2005;3:907–913. doi: 10.1586/14787210.3.6.907. [DOI] [PubMed] [Google Scholar]
- 3.Molbak K. Clin. Infect. Dis. 2005;41:1613–1620. doi: 10.1086/497599. [DOI] [PubMed] [Google Scholar]
- 4.Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., Edmond M. B. Clin. Infect. Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 5.Lane H. C., Montagne J. L., Fauci A. S. Nat. Med. 2001;7:1271–1273. doi: 10.1038/nm1201-1271. [DOI] [PubMed] [Google Scholar]
- 6.Boggs A. F., Miller G. H. Clin. Microbiol. Infect. 2004;10(Suppl. 4):32–36. doi: 10.1111/j.1465-0691.2004.1008.x. [DOI] [PubMed] [Google Scholar]
- 7.Projan S. J., Shlaes D. M. Clin. Microbiol. Infect. 2004;10(Suppl. 4):18–22. doi: 10.1111/j.1465-0691.2004.1006.x. [DOI] [PubMed] [Google Scholar]
- 8.Silver L. L. IDrugs. 2005;8:651–655. [PubMed] [Google Scholar]
- 9.Walsh C. Nat. Rev. Microbiol. 2003;1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty T. J., Barrett J. F., Pucci M. J. Curr. Pharm. Des. 2002;8:1119–1135. doi: 10.2174/1381612023394782. [DOI] [PubMed] [Google Scholar]
- 11.Osburne M. S., Grossman T. H., August P. R., MacNeil I. A. ASM News. 2000;66:411–417. [Google Scholar]
- 12.Lipinski C., Hopkins A. Nature. 2004;432:855–861. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- 13.Lewis K., Lomovskaya O. In: Bacterial Resistance to Antimicrobials: Mechanisms, Genetics, Medical Practice and Public Health. Lewis K., Salyers A., Taber H., Wax R., editors. New York: Dekker; 2002. pp. 61–90. [Google Scholar]
- 14.Li X. Z., Nikaido H. Drugs. 2004;64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 15.Horrobin D. F. Nat. Rev. Drug Discovery. 2003;2:151–154. doi: 10.1038/nrd1012. [DOI] [PubMed] [Google Scholar]
- 16.Williams M. Curr. Opin. Investig. Drugs. 2004;5:29–33. [PubMed] [Google Scholar]
- 17.Ewbank J. J. Microbes Infect. 2002;4:247–256. doi: 10.1016/s1286-4579(01)01531-3. [DOI] [PubMed] [Google Scholar]
- 18.Sifri C. D., Begun J., Ausubel F. M. Trends Microbiol. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Mylonakis E., Aballay A. Infect. Immun. 2005;73:3833–3841. doi: 10.1128/IAI.73.7.3833-3841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ausubel F. M. Nat. Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 21.Kim D. H., Ausubel F. M. Curr. Opin. Immunol. 2005;17:4–10. doi: 10.1016/j.coi.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Garsin D. A., Sifri C. D., Mylonakis E., Qin X., Singh K. V., Murray B. E., Calderwood S. B., Ausubel F. M. Proc. Natl. Acad. Sci. USA. 2001;98:10892–10897. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moy T. I., Mylonakis E., Calderwood S. B., Ausubel F. M. Infect. Immun. 2004;72:4512–4520. doi: 10.1128/IAI.72.8.4512-4520.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulz M., Schmoldt A. Pharmazie. 2003;58:447–474. [PubMed] [Google Scholar]
- 25.Kim D. H., Feinbaum R., Alloing G., Emerson F. E., Garsin D. A., Inoue H., Tanaka-Hino M., Hisamoto N., Matsumoto K., Tan M. W., Ausubel F. M. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 26.Qin X., Singh K. V., Weinstock G. M., Murray B. E. Infect. Immun. 2000;68:2579–2586. doi: 10.1128/iai.68.5.2579-2586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huycke M. M., Spiegel C. A., Gilmore M. S. Antimicrob. Agents Chemother. 1991;35:1626–1634. doi: 10.1128/aac.35.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung D. T., Shakhnovich E. A., Pierson E., Mekalanos J. J. Science. 2005;310:670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 29.Sochova I., Hofman J., Holoubek I. Environ. Int. 2006;32:374–383. doi: 10.1016/j.envint.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 30.Williams P. L., Dusenbery D. B. Toxicol. Ind. Health. 1988;4:469–478. doi: 10.1177/074823378800400406. [DOI] [PubMed] [Google Scholar]
- 31.Cole R. D., Anderson G. L., Williams P. L. Toxicol. Appl. Pharmacol. 2004;194:248–256. doi: 10.1016/j.taap.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Walsh C. Antibiotics, Actions, Origins, Resistance. Washington, DC: Am. Soc. Microbiol.; 2003. [Google Scholar]
- 33.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beanan M. J., Strome S. Development (Cambridge, U.K.) 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka-Hino M., Sagasti A., Hisamoto N., Kawasaki M., Nakano S., Ninomiya-Tsuji J., Bargmann C. I., Matsumoto K. EMBO Rep. 2002;3:56–62. doi: 10.1093/embo-reports/kvf001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis J. A., Fleming J. T. In: Caenorhabditis elegans Modern Biological Analysis of an Organism. Epstein H. F., Shakes D. C., editors. Vol. 48. San Diego: Academic; 1995. pp. 3–29. [Google Scholar]
- 37.Murray B. E., Singh K. V., Ross R. P., Heath J. D., Dunny G. M., Weinstock G. M. J. Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahm D. F., Kissinger J., Gilmore M. S., Murray P. R., Mulder R., Solliday J., Clarke B. Antimicrob. Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arduino R. C., Jacques-Palaz K., Murray B. E., Rakita R. M. Infect. Immun. 1994;62:5587–5594. doi: 10.1128/iai.62.12.5587-5594.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.National Committee for Clinical Laboratory Standards. Approved Standard M7–A5. Villanova, PA: NCCLS; 2000. [Google Scholar]
- 41.Ciornei C. D., Sigurdardottir T., Schmidtchen A., Bodelsson M. Antimicrob. Agents Chemother. 2005;49:2845–2850. doi: 10.1128/AAC.49.7.2845-2850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]