Abstract
Inflammatory mediators activate the transcriptional complex HIF-1 (hypoxia-inducible factor-1), the key regulator of hypoxia-induced gene expression. Here we report that bacterial LPS (lipopolysaccharide) induces HIF-1α mRNA expression and HIF-1α protein accumulation in human monocytes as well as in non-differentiated and differentiated cells of the human monocytic cell line THP-1 under normoxic conditions. LPS and hypoxia synergistically activated HIF-1. Whereas LPS increased HIF-1α mRNA expression through activation of a NF-κB (nuclear factor κB) site in the promoter of the HIF-1α gene, hypoxia post-translationally stabilized HIF-1α protein. HIF-1α activation was followed by increased expression of the HIF-1 target gene encoding ADM (adrenomedullin). Knocking down HIF-1α by RNA interference significantly decreased ADM expression, which underlines the importance of HIF-1 for the LPS-induced ADM expression in normoxia. Simultaneously with HIF-1 activation, an increase in p44/42 MAPK (mitogen-activated protein kinase) phosphorylation was observed after incubation with LPS. In cells pretreated with the p44/42 MAPK inhibitor PD 98059 or with RNAi (interfering RNA) directed against p44/42 MAPK, LPS-induced HIF-1α accumulation and ADM expression were significantly decreased. From these results we conclude that LPS critically involves the p44/42 MAPK and NF-κB pathway in the activation of HIF-1, which is an important transcription factor for LPS-induced ADM expression.
Keywords: adrenomedullin (ADM), hypoxia-inducible factor-1 (HIF-1), inflammation, macrophages, mitogen-activated protein kinase (MAPK), oxygen sensing
Abbreviations: Act D, actinomycin D; ADM, adrenomedullin; EMSA, electrophoretic-mobility-shift assay; ERK, extracellular-signal-regulated kinase; HIF-1, hypoxia-inducible factor-1; HRE: hypoxia response element; HRP, horseradish peroxidase; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MEK1/2, MAPK/ERK kinase 1/2); NF-κB, nuclear factor κB; pVHL, von-Hippel–Lindau protein; RNAi, interfering RNA; RT-PCR, reverse-transcription PCR; TLR4, toll-like receptor 4; TNFα, tumour necrosis factor-α
INTRODUCTION
The heterodimeric transcription factor HIF-1 (hypoxia-inducible factor-1) regulates the expression of genes involved in angiogenesis, oxygen transport, glucose metabolism and vascular tone [1]. HIF-1 consists of an α- and a β-subunit: whereas HIF-1β is constitutively expressed, HIF-1α is strictly regulated by the cellular oxygen tension. Under normoxic conditions, HIF-1α protein is post-translationally hydroxylated on specific proline residues to enable binding to pVHL (von-Hippel–Lindau protein), which targets HIF-1α for ubiquitinylation and proteasomal degradation [2]. Under hypoxic conditions, proline hydroxylation is blocked, and this leads to an increase in HIF-1α protein and expression of HIF-1 target genes [3,4]. In addition to hypoxia, multiple oncogenic and inflammatory stimuli up-regulate HIF-1α accumulation and activation [5–7]. Recently, an important role for HIF-1 in inflammation and activation of the immune response was proposed [8,9]. Results from these and other groups provide evidence that HIFs play an integrative role in conditions of hypoxia and inflammation.
Nevertheless, the intracellular pathways involved in non-hypoxic HIF-1α activation seem to be cell-specific and stimulus-dependent and are not well characterized. An implication of the p44/42 MAPK (mitogen-activated protein kinase) in hypoxic HIF-1α activation was first discussed by Richard et al. [10] and Minet et al. [11]. Sang and co-workers reported an elevation of HIF-1 activity by p44/42 MAPK activation [12]. The p44/42 MAPKs are activated by a wide variety of proliferative and inflammatory signals [13]. In monocytes and macrophages, bacterial LPS (lipopolysaccharide) strongly induces p44/42 phosphorylation [14,15]. The phosphorylation of p44/42 MAPK is triggered by a cascade of phosphorylation events that can be repressed by specifically designed kinase inhibitors. Activation of NF-κB (nuclear factor κB) is a common end point of signalling cascades in inflammation and the immune response.
Recently, the gene encoding ADM (adrenomedullin) was identified as an HIF-1 target [16]. ADM is a potent hypotensive and immune-modulating peptide initially isolated from extracts of human pheochromocytoma cells [17]. ADM is expressed in endothelial, vascular smooth-muscle[18] and tumour cells [16]. Hypoxia potently stimulates ADM production [16,19]. Analysis of the ADM promoter resulted in the identification of at least 20 putative HREs (hypoxia response elements) located in the 5′-promoter, 5′- and 3′- untranslated regions, intron 1 and exons 3 and 4 of the ADM gene [16]. In addition, elevated levels of ADM were found in septicaemic patients [20], and murine peritoneal macrophages stimulated with LPS express the ADM gene [21]. The mechanisms of LPS-induced ADM expression have not yet been fully elucidated, and it is not known whether LPS-induced ADM expression interferes with hypoxic signaling.
In the present study we show that LPS increases HIF-1α mRNA expression in an NF-κB-dependent way. The p44/42 MAPK was critically involved in this activation under normoxic and hypoxic conditions. We can demonstrate that HIF-1 plays a central role in hypoxia- as well as in LPS-induced ADM expression in human monocytes and macrophages.
MATERIALS AND METHODS
Cell culture
The human monocytic cell lines THP-1 and MonoMac6 were obtained from the German Collection of Micro-organisms and Cell Cultures [DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH]. Cells were cultured in RPMI 1640 medium, supplemented with 10% (v/v) fetal bovine serum, 2 mM L-glutamine, 1 mM sodium pyruvate, penicillin (100 units/ml) and streptomycin (100 μg/ml) in a humidified atmosphere of 5% CO2 in air. For experiments with undifferentiated cells, the cell density was adjusted to 106 cells/ml in 1 ml of serum-free medium in 35-mm-diameter Petri dishes. Differentiation of THP-1 to a macrophage-like phenotype was achieved by treatment of 106 cells/ml in a 35-mm-diameter Petri dish with 10 nM PMA for 5 days. The cells were serum-deprived 24 h before the experiment. Hypoxic conditions were achieved by placing the culture dishes in a Heraeus (Hanau, Germany) incubator under O2/CO2/N2 (3:5:92) for the indicated time periods. For reoxygenation experiments, cells were exposed to hypoxic conditions (3% O2) for 6 h and then transferred to 21% O2 for different times. Bacterial LPS (serotype 0111:B4; Sigma, Deisenhofen, Germany) was added to the cells as indicated for each experiment. The concentrations of LPS, bortezomib (a modified dipeptidyl boronic acid from Ortho Biotech, Neuss, Germany) and PD 98059 (a cell-permeant inhibitor of MAPK kinase from Calbiochem, Bad Soden, Germany) used in the experiments were not toxic for the cells as judged from an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay [22]. At the end of the experiments, cells were centrifuged, lysed in GTC solution (4.2 M guanidine isothiocyanate/0.5% saponin/25 mM sodium citrate), followed by total RNA extraction for the determination of specific mRNAs by RT (reverse transcription)-PCR. Additionally, total cell lysates and nuclear extracts were prepared and submitted to immunoblotting as well as to an NF-κB and HIF-1 DNA-binding EMSA (electrophoretic-mobility-shift assay).
Primary human macrophages
Peripheral-blood monocytes were isolated from 100 ml of buffy coat using NycoPrep™ 1.068 according to the manufacturer's instructions. Isolated monocytes were resuspended in RPMI 1640 medium (BioWhittaker, Verviers, Belgium) supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, penicillin (100 units/ml) and streptomycin (100 μg/ml). Cells were seeded in polystyrene Petri dishes at a density of 2×106 cells/ml and were allowed to attach themselves to the culture dish. After 30 min, the medium, with unattached cells, was removed and monocytes were differentiated in RPMI 1640 with 20% (v/v) human AB serum (Sigma), 2 mM L-glutamine, 1 mM sodium pyruvate, 100 units/ml penicillin and 100 μg/ml streptomycin (all from Invitrogen, Karlsruhe, Germany) for 8 days. Medium was renewed at culture days 3 and 6. Differentiation was confirmed morphologically and by immunohistochemical staining with the macrophage-specific marker CD68 (a 110 kDa transmembrane glycoprotein; 98% of differentiated macrophages were CD68-positive). Cells were serum-deprived 18 h before the start of the experiments performed under serum-free conditions. To achieve hypoxic conditions, culture dishes were placed in a Heraeus incubator under O2/CO2/N2 (3:5:92) for the indicated time periods.
RNA preparation and RT-PCR
Total RNA was isolated by the acid guanidinium thiocyanate/phenol/chloroform extraction method [23]. A 1 μg portion of total RNA was reverse-transcribed into cDNA with oligo(dT)15 as primer for avian-myeloblastosis-virus reverse transcriptase (Promega, Heidelberg, Germany). Qualitative PCRs were performed for the ribosomal protein as housekeeping gene to test the quality of cDNA preparation and for ADM, and HIF-1α to estimate the amount of specific cDNA before real-time PCR. In addition, expression of the cell-surface antigens CD14, and TLR4 involved in LPS binding and signalling were verified. The resulting PCR fragments were visualized on ethidium bromide-stained 1.5% (w/v) agarose gels. Primers for real-time PCR (Invitrogen) were designed with Primer Express software (Applied Biosystems, Weiterstadt, Germany). The amplicon size was 100 bp, the annealing temperature was 60 °C and the CG content was about 60%. The primer sequences used for qualitative and quantitative PCR are listed in Table 1.
Table 1. Primer sequences used for qualitative and real-time PCR.
| Primer | Sequence | GenBank® accession no. |
|---|---|---|
| 5′ribProt | acc agg tgt gca agg agg gc | AF173378 |
| 3′ribProt | gca agt cgt ctc cca tct gc | |
| 5′ADM | gga tgc cgc ccg cat ccg ag | NM_001124 |
| 3′ADM | gac acc aga gtc cga ccc gg | |
| 5′HIF-1α | gct ggc ccc agc cgc tgg ag | XM 007373 |
| 3′HIF-1α | gag tgc agg gtc agc act ac | |
| 5′CD14 | ggt gcc gct gtg tag gaa aga | BC 010507 |
| 3′CD14 | ggt cct cga gcg tca gtt cct | |
| 5′TLR4 | tgg ata cgt ttc ctt ata ag | AF172169 |
| 3′TLR4 | gaa atg gag gca ccc ctt c |
Real-time PCR was performed with SYBR® green as fluorescent dye (Eurogentec, Verviers, Belgium) on the Gene Amp 5700 Sequence Detection System (Applied Biosystems, Weiterstadt, Germany). The cDNA standards for real-time PCR were prepared from the specific PCR products using a DNA purification kit (Roche, Mannheim, Germany). The amount of standard cDNA was determined photometrically. The standard concentrations ranged from 100–0.01 fg/μl. Quantification was done in a two-step real-time PCR with a denaturation step at 95 °C for 10 min and then 40 cycles at 95 °C for 15 s and at 60 °C for 1 min. Amounts of HIF-1α cDNA and ADM cDNA were finally normalized to 1 μg of total RNA.
Whole-cell and nuclear-extract preparation
For preparing whole-cell extracts, cells were centrifuged (1500 g) for 8 s, supernatants were discarded and cells were lysed with 30 μl of lysis buffer [150 mM NaCl, 10 mM Tris, pH 7.9, 1 mM EDTA, 0.1% Igepal (Nonidet P40), 1×protease inhibitor cocktail (Roche)] for 20 min on ice. The lysates were centrifuged at 1500 g at 4 °C for 5 min in a microcentrifuge, and supernatants containing cellular proteins were collected and stored at −80 °C. Protein concentrations of the supernatants were quantified using the Bio-Rad (Munich, Germany) protein assay reagent.
Nuclear proteins were prepared using the method of Schreiber et al. [24]. All procedures were carried out at 4 °C. After incubation, cells were spun down in a microcentrifuge for 8 s and lysed with 150 μl of Buffer A (10 mM Hepes, pH 7.9, 1.5 MgCl2, 10 mM KCl, 0.5 mM PMSF, 0.5 mM dithiothreitol, 0.4% Igepal, 1×protease inhibitor cocktail) and incubated for 20 min on ice. Cell lysates were centrifuged at 1500 g at 4 °C for 5 min. Nuclear pellets were resolved in 80 μl of Buffer B (20 mM Hepes, pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM dithiothreitol and 1×protease inhibitor cocktail) and homogenized on a magnetic stirrer for 30 min. After centrifugation at 1500 g at 4 °C for 15 min, supernatants containing the nuclear proteins were frozen at −80 °C. Protein concentrations were determined using the Bio-Rad kit.
Transfer of RNAi (interfering RNA) to THP-1 cells
RNAi against HIF-1α was selected as previously described [25]. In addition, another HIF-1α RNAi corresponding to nt 1004–1029 of HIF-1α sequence (GeneBank® accession number AF208487) was selected and synthesized by Invitrogen as Stealth RNAi®. RNAi against p44/42 (Validated Stealth RNAi) was obtained from Invitrogen. THP-1 cells were adjusted to a cell density of 1.5×106 cells/ml in PB-sucrose (7 mM sodium pyruvate, 272 mM sucrose and 1 mM MgCl2). RNAi (final concn. 50 μM) was transfected by electroporation using a Bio-Rad Gene Pulser II electroporator with a radio-frequency module. Electroporation provided a transfection efficiency of 70%, as judged from transfection with fluorescein-labelled RNAi, with 60% of the cells alive after electroporation. After electroporation, cells were resuspended in complete medium and cultured for additional 24 h before the experiments were performed.
Transfection of MonoMac6 cells
Cells (2×106/ml) were transfected with a reporter vector in which firefly luciferase is driven by a 800-nt fragment of the HIF-1α promoter (800ntHIFprom in pGL3 (Promega) kindly provided by Dr Carine Michiels, Laboratory of Biochemistry and Cellular Biology, University of Namur, Namur, Belgium). At 6 h after transfection cells were treated for 16 h with LPS to allow sufficient expression of the reporter gene. Luciferase activity was measured in 50 μl of cell lysate and normalized to the expression of the pGL3 control vector.
Western-blot analysis
After addition of 0.25 vol. of sample buffer [50 mM Tris, pH 6.8, 2% (w/v) SDS, 5% (v/v) 2-mercaptoethanol, 0.0125% Bromophenol Blue and 1% glycine), 70 μg of total cell lysate per lane was subjected to SDS/7.5%-(w/v)-PAGE and transferred on to a nitrocellulose membrane (0.2 μm pore size; Schleicher and Schuell, Dassel, Germany). The efficiency of protein transfer and equal loading was confirmed by staining the nitrocellulose membrane with Ponceau S (Sigma). Filters were blocked overnight at 4 °C with 5% (w/v) non-fat dried milk powder in TBS-T (10 mM Tris, pH 7.5, 100 mM NaCl and 0.05% Tween-20). Membranes were washed in TBS-T and incubated with a mouse monoclonal antibody against HIF-1α (Transduction Laboratories, San Diego, CA, U.S.A.), NF-κB (Cell Signaling Technology, Inc., Danvers, MA, U.S.A.) or phospho-specific antibodies against p44/42 MAPK (Cell Signaling Technology) at room temperature for 2 h with gentle shaking. α-Tubulin and RNA polymerase II (both from Sigma) served as markers for equal total protein content. After washing with TBS-T, HRP (horseradish peroxidase)-conjugated anti-mouse IgG antibody (Sigma) or with HRP-linked anti-rabbit IgG antibody (New England Biolabs) were added. Immunoreactive proteins were visualized by chemiluminescence. Membranes were incubated in a detection solution (100 mM Tris/HCl, pH 8.5, 2.65 mM H2O2, 0.45 mM luminol and 0.625 mM p-coumaric acid) for 1 min, followed by exposure to X-ray films (Agfa, Mortsel, Belgium).
HIF-1 DNA binding assay
HIF-1 DNA-binding activity was measured with the DuoSet® IC (IntraCellular) Assay Development System for HIF-1 (R&D Systems, Minneapolis, MN, U.S.A.) according to the manufacturer's instructions. In brief, a 96-well microtitre plate was coated with capture antibody (4 μg/ml in PBS). After repeated washing, the plate was blocked with 5% (w/v) BSA in reagent diluent for 2 h. Nuclear extracts (each sample 80 μg) were incubated with a biotin-labelled double-stranded oligonucleotide containing a specific HIF-1 binding sequence for 30 min at room temperature. Samples (100 μl/well each) were placed on the microtitre plate and incubated for 2 h. Detection of DNA-bound HIF-1 complexes was done using streptavidin-HRP conjugate. Binding activity of HIF-1 from LPS- and hypoxia-treated cells were calculated as a percentage of the value obtained for untreated normoxic controls. Experiments were performed in triplicate and each sample was measured in duplicate.
NF-κB EMSA
[γ-32P]ATP was obtained from ICN (Eschwege, Germany). Oligonucleotides containing the sequence (in boldface) of a NF-κB binding site from the HIF-1α promoter (sense 5′-tcacgaggggtttcccgcctcgca-3′ and antisense: 5′-tgcgaggcgggaaacccctcgtga-3′; corresponding to nt −130 relative to the transcription start site) were synthesized by Invitrogen. After 5′-end-labelling of the sense strand, unincorporated ATP was removed with Bio-Gel G30 columns (Bio-Rad). The annealing reaction was performed in the presence of a 2-fold molar excess of unlabelled antisense oligonucleotide. Binding reactions were set up in a volume of 20 μl. Portions (10 μg) of nuclear extracts were preincubated on ice for 20 min in a binding buffer (12 mM Hepes, pH 7.9, 4 mM Tris/HCl, 60 mM KCl, 1 mM EDTA and 1 mM dithiothreitol) with 100 ng of thymus DNA. For supershift assays the samples were additionally incubated with an anti-NF-κB p65 antibody for 16 h at 4 °C. Samples were subsequently resolved by electrophoresis on native 5%-(w/v)-polyacrylamide gels at room temperature. Gels were dried and analysed directly by autoradiography.
Statistics
Experiments were performed in triplicate. Values of cDNA quantification are given as means±S.D. Dunnett's test and the Tukey–Kramer test were used to calculate whether differences of means between treated and control groups were significantly different.
RESULTS
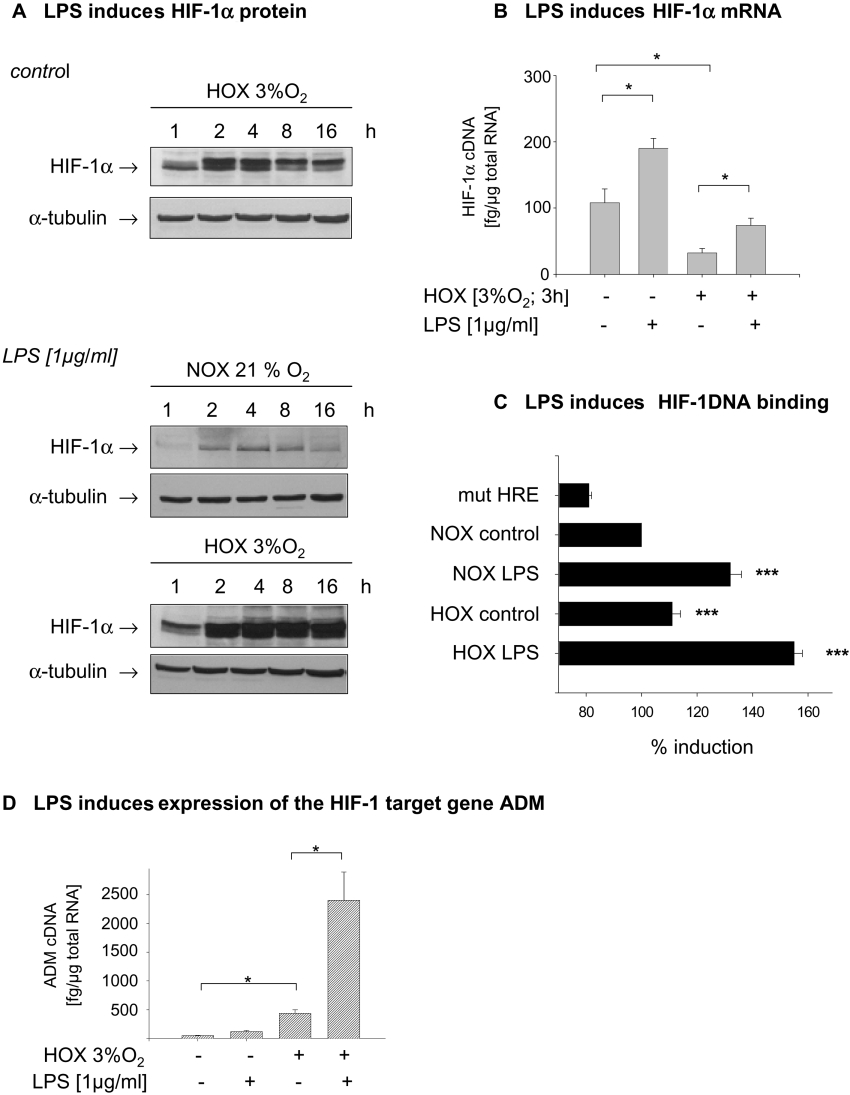
THP-1 cells were incubated under hypoxic conditions (3% oxygen) and the time course of HIF-1α protein accumulation was determined in total cell lysates. Exposure times for all blots were identical to allow better judgment on signal strength, as may be appreciated from equal signal strength for α-tubulin. HIF-1α protein was detected 1 h after the beginning of hypoxic incubation, and maximal induction was achieved after a 4 h incubation period. After more than 4 h the HIF-1α levels declined again. Likewise, the addition of LPS (1μg/ml) to THP-1 cells under normoxic conditions significantly increased HIF-1α accumulation starting after 2 hours of LPS treatment and peaking at 4 h. Treatment of THP-1 cells with LPS under hypoxic conditions enhanced hypoxia-induced HIF-1α accumulation and prolonged the elevation of HIF-1α protein content up to 16 h (Figure 1A). To elucidate the mechanisms underlying the enhanced accumulation of HIF-1α protein by LPS treatment, we analysed HIF-1α mRNA expression. While hypoxia significantly decreased HIF-1α mRNA levels in THP-1 cells, LPS stimulated the expression of HIF-1α under normoxic and hypoxic conditions. (Figure 1B). To analyse the DNA-binding capacity of HIF-1 in THP-1 cells, a ELISA-based HIF-1 binding assay was performed. In nuclear extracts from cells treated with LPS under normoxic conditions a highly significant induction of the HIF-1 DNA binding was observed. Hypoxia alone induced a moderate increase in binding of HIF-1 to DNA, which was significantly enhanced when cells were treated with LPS under hypoxic conditions (Figure 1C). To test whether LPS-induced HIF-1 DNA binding resulted in the expression of an HIF target gene, ADM expression was quantified. Treatment with LPS for 6 h induced ADM expression in THP-1 cells under normoxic and hypoxic conditions, as quantified by real-time PCR. Hypoxia alone induced a 10-fold increase in ADM expression, and LPS significantly enhanced the expression of ADM under both conditions (Figure 1D).
Figure 1. Undifferentiated THP-1 cells accumulate HIF-1α in response to hypoxia and LPS.
(A) THP-1 cells were incubated under normoxic (NOX) and hypoxic (HOX) conditions for up to 16 h in the presence or absence of LPS (1μg/ml). A 70 μg portion of total cell lysate was resolved by SDS/PAGE. HIF-1α protein was detected with an anti-HIF-1α antibody, and anti-α-tubulin antibody was used to demonstrate equal gel loading. (B) THP-1 cells were incubated for 3 h with LPS. Total RNA was prepared, reverse-transcribed into cDNA, and HIF-1α cDNA was quantified by real-time PCR. Amounts of HIF-1α cDNA were normalized to 1 μg of total RNA. Shown are the means±S.D. from four separate experiments, *P<0.05. (C) LPS and hypoxia increase HIF-1 DNA-binding activity. An 80 μg portion of nuclear extract from treated THP-1 cells was incubated with a biotin-labelled double stranded oligonucleotide containing a specific HIF-1 binding sequence or a mutated (mut) HRE for 30 min at room temperature. Detection of DNA-bound HIF-1 complexes was performed using streptavidin–HRP conjugate. Binding activity of HIF-1 from LPS- and hypoxia-treated cells were calculated as a percentage of that exhibited by untreated normoxic controls. n=3, ***P<0.001. (D) LPS induces the expression of the HIF-1 target gene. THP-1 cells were incubated under normoxic and hypoxic condtions in the presence and absence of LPS (1 μg/ml) for 6 h. Total RNA was isolated and reverse-transcribed into cDNA. ADM cDNA was quantifed by real-time PCR. Shown are the means±S.D. from four separate experiments; *P<0.05.
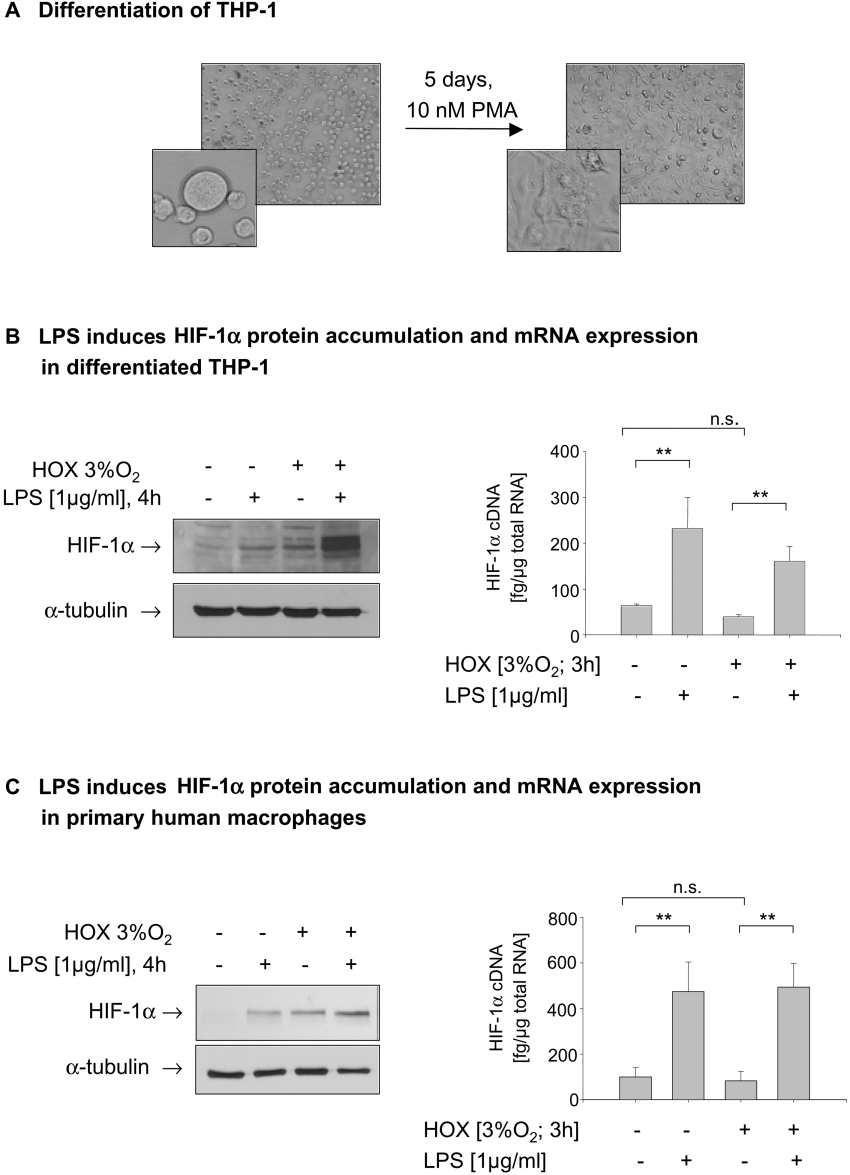
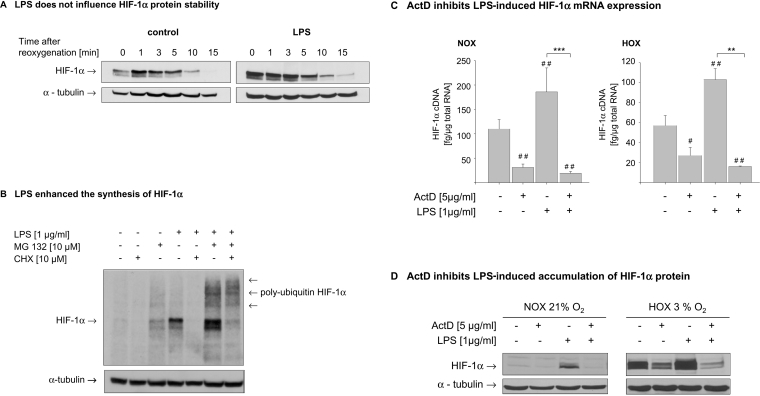
Since a role of HIF-1α during the maturation and differentiation of myeloid cells was suggested [8], we were interested whether the sensitivity to hypoxia and LPS changes during this process. THP-1 cells were differentiated with 10 nM PMA for 5 days to achieve a macrophage-like phenotype (Figure 2A). Because maximal accumulation of HIF-1α in undifferentiated THP-1 cells was observed after an incubation period of 4 h, this time point was chosen for the following experiments. Treatment of differentiated THP-1 with LPS under normoxic conditions induced HIF-1α accumulation comparable to a 4 h hypoxic incubation. Under hypoxic conditions, LPS strongly enhanced HIF-1α protein content (Figure 2B, left panel). In contrast with non-differentiated cells, hypoxia did not reduce the expression of HIF-1α mRNA. As observed with non-differentiated cells, LPS induced an up-to-3-fold higher expression of HIF-1α mRNA under normoxic and hypoxic conditions (Figure 2B, right panel). To confirm these results obtained in the cell line THP-1, we performed experiments with primary human macrophages differentiated from peripheral-blood monocytes by treatment with 20% (v/v) human AB serum for 8 days. In primary macrophages, LPS induced HIF-1α accumulation under normoxic conditions. Hypoxic incubation for 4 h resulted in an accumulation of HIF-1α that was further enhanced by LPS (Figure 2C, left panel). With respect to HIF-1α mRNA expression, hypoxia alone failed to influence HIF-1α mRNA. LPS treatment enhanced HIF-1α mRNA expression under normoxic and hypoxic conditions. No decrease in HIF-1α mRNA was observed in primary human monocytes after hypoxic incubation (Figure 2C, right panel). Similar results for HIF-1α mRNA expression and protein accumulation were obtained in MonoMac cells (results not shown). To further elucidate the mechanisms leading to the LPS-induced HIF-1α accumulation, we first raised the question of whether LPS affects the half-life of HIF-1α protein. THP-1 cells were pre-incubated for 6 h under hypoxic conditions in the presence or absence of LPS, followed by reoxygenation for up to 1 h. As shown in Figure 3(A), HIF-1α protein declined with a half-life of about 5 min. Interestingly, the HIF-1α protein levels measured 1 min after reoxygenation seemed to be higher than the zero-time value. Although the initial protein levels in LPS-treated cells were higher than in the hypoxic control cells, LPS did not alter the half-life of the HIF-1α protein significantly. Adding LPS or cycloheximide immediately before the reoxygenation did also not affect the half-life of HIF-1α protein (results not shown). To test whether LPS interferes with HIF-1α degradation, THP-1 cells were incubated with the proteasome inhibitor MG132 and the protein-synthesis inhibitor cycloheximide for the last 30 min of a 4 h normoxic incubation with LPS. Cycloheximide treatment completely inhibited LPS-induced HIF-1α accumulation. As expected, MG132 alone slightly increased HIF-1α protein as a result of reduced proteasomal degradation. The addition of MG132 to LPS-treated cells further enhanced HIF-1α accumulation. Detection of polyubiquitinated forms of HIF-1α demonstrated that LPS did not interfere with HIF-1α hydroxylation and recognition by pVHL (Figure 3B). Since LPS induced the expression of HIF-1α mRNA under hypoxic and normoxic conditions, the transcriptional inhibitor actinomycin D (ActD) was added to the cells immediately before a 4 h LPS treatment. Quantification of HIF-1α cDNA revealed that the LPS-induced increase in HIF-1α expression was abolished by ActD under normoxic and hypoxic conditions (Figure 3C). Respective changes in HIF-1α protein levels were observed with ActD treatment. (Figure 3D).
Figure 2. LPS induces HIF-1α in differentiated THP-1 cells and in primary human macrophages.
(A) THP-1 cells were treated for 5 days with 10 nM PMA to achieve differentiation from a monocytic to a macrophage-like phenotype. (B) Differentiated cells were incubated with LPS (1μg/ml) under normoxic (NOX) or hypoxic HOX) conditions. Whole-cell lysates were prepared and subjected to SDS/PAGE. HIF-1α and α-tubulin proteins were detected by immunoblotting. In addition, total RNA was extracted and HIF-1α cDNA was quantifed by real-time PCR. Shown are the means±S.D. from four independent experiments, *P<0.05. (C) Primary human macrophages were incubated with LPS (1 μg/ml) under normoxic or hypoxic conditions. Whole-cell lysates were prepared and subjected to SDS/PAGE. A representative immunoblot of HIF-1α and α-tubulin proteins is shown (n=3). After a 3 h treatment with LPS in normoxia and hypoxia, total RNA was extracted and HIF-1α cDNA was quantified by real-time PCR. Shown are the means±S.D. from four independent experiments. *P<0.05.
Figure 3. Mechanisms involved in LPS-induced HIF-1α activation.
(A) THP-1 cells were incubated under hypoxic (HOX) conditions in the presence or absence of LPS (1 μg/ml) for 6 h, followed by reoxygenation for the indicated time periods. Whole-cell lysates were prepared and subjected to SDS/PAGE. HIF-1α and α-tubulin proteins were detected by immunoblotting. A representative immunoblot (n=4) is shown. (B) THP-1 cells were treated with MG132 (‘MG 132’;10 μM) and cycloheximide (CHX; 10 μM) 30 min before the end of 4 h incubation with LPS under normoxic (NOX) conditions. HIF-1α and polyubiquitinated forms of HIF-1α were detected with an anti-HIF-1α antibody; α-tubulin served as control for equal protein loading. (C) Cells were treated with actinomycin D (ActD; 5 μg/ml) immediately before a 4 h incubation with LPS under normoxic or hypoxic conditions. Total RNA was prepared and HIF-1α cDNA quantified by real-time PCR. Shown are the means±S.D. from four independent experiments, **P<0.01; ***P<0.001; # indicates statistically significant difference with respect to the untreated controls. (D) THP-1 cells were pretreated with ActD (5 μg/ml), followed by a 4 h incubation with LPS under normoxic or hypoxic conditions. Total cell lysates were prepared and 70 μg of the lysate were resolved by SDS/PAGE. HIF-1α protein was detected with an anti-HIF-1α antibody; anti-α-tubulin antibody was used to demonstrate equal gel loading.
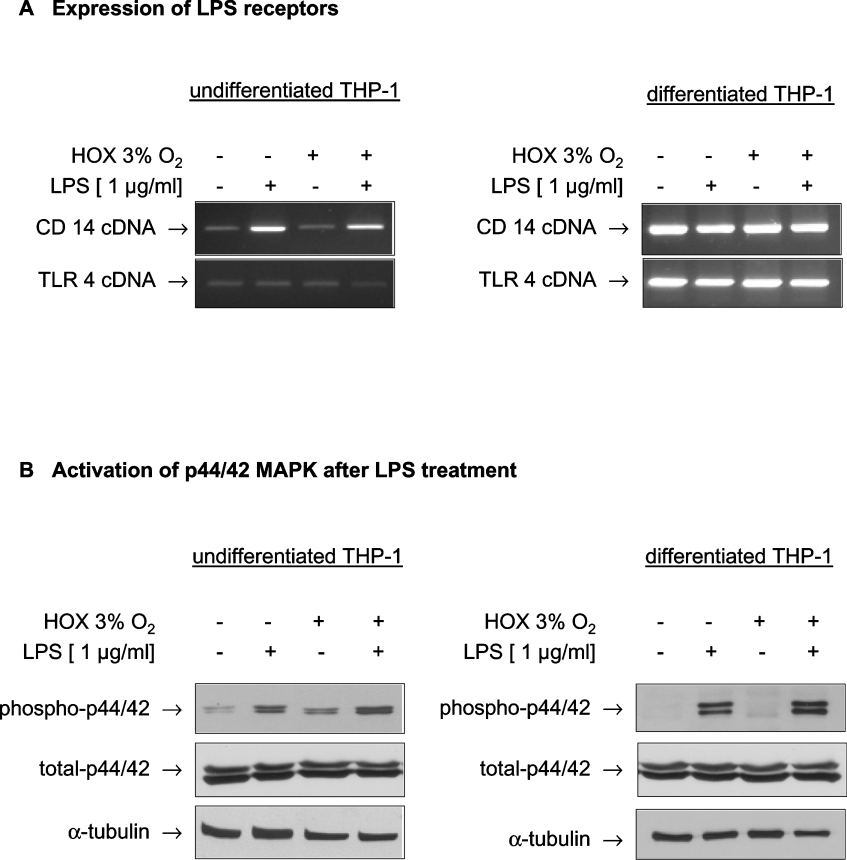
To follow the signalling pathways stimulated by LPS with respect to HIF-1α activation, we screened THP-1 cells for the expression of cell-surface antigens classically involved in the LPS-induced activation of monocytes. RT-PCR for CD14 and TLR4 revealed that differentiation increased the expression level of TLR4 cells, but LPS treatment exerted no effect on TLR4 expression. CD14 expression was strongly increased after incubation with LPS only in undifferentiated THP-1 cells and constitutively expressed at a very high level in differentiated cells (Figure 4A). Downstream signalling of TLR4 includes – depending on the cell type – activation of the ERK (extracellular-signal-regulated kinase) p44/42 MAPK (ERK1/2). By immunoblotting with phosphospecific antibodies phosphorylation of p44/42 was detected in non-differentiated THP-1 cells after stimulation with LPS. In addition, a moderate increase in p44/42 phosphorylation was observed after incubation of THP-1 cells under hypoxic conditions (Figure 4B, left panel). In differentiated THP-1 cells, the phosphorylation status of p44/42 MAPK under control conditions was lower, but a strong increase in phosphorylation was detected after treatment with LPS. Total p44/42 MAPK content was neither influenced by hypoxia nor by LPS (Figure 4B, right panel).
Figure 4. THP-1 cells express CD14 and TLR4, members of the LPS signalling cascade, leading to phosphorylation of p44/42 MAPK after LPS stimulation.
(A) Undifferentiated and differentiated THP-1 cells were treated with LPS under normoxic and hypoxic (HOX) conditions for 6 h. Total RNA was prepared, transcribed into cDNA and expression of the cDNAs for CD14 (‘CD 14’) and TLR4 (‘TLR 4’) were quantified by PCR. (B) THP-1 cells were incubated with LPS (1 μg/ml) under normoxic and hypoxic conditions for 3 h. A 50 μg portion of whole-cell lysate were submitted to SDS/PAGE. Phosphorylated as well as total p44/42 MAPK were detected by immunoblot.
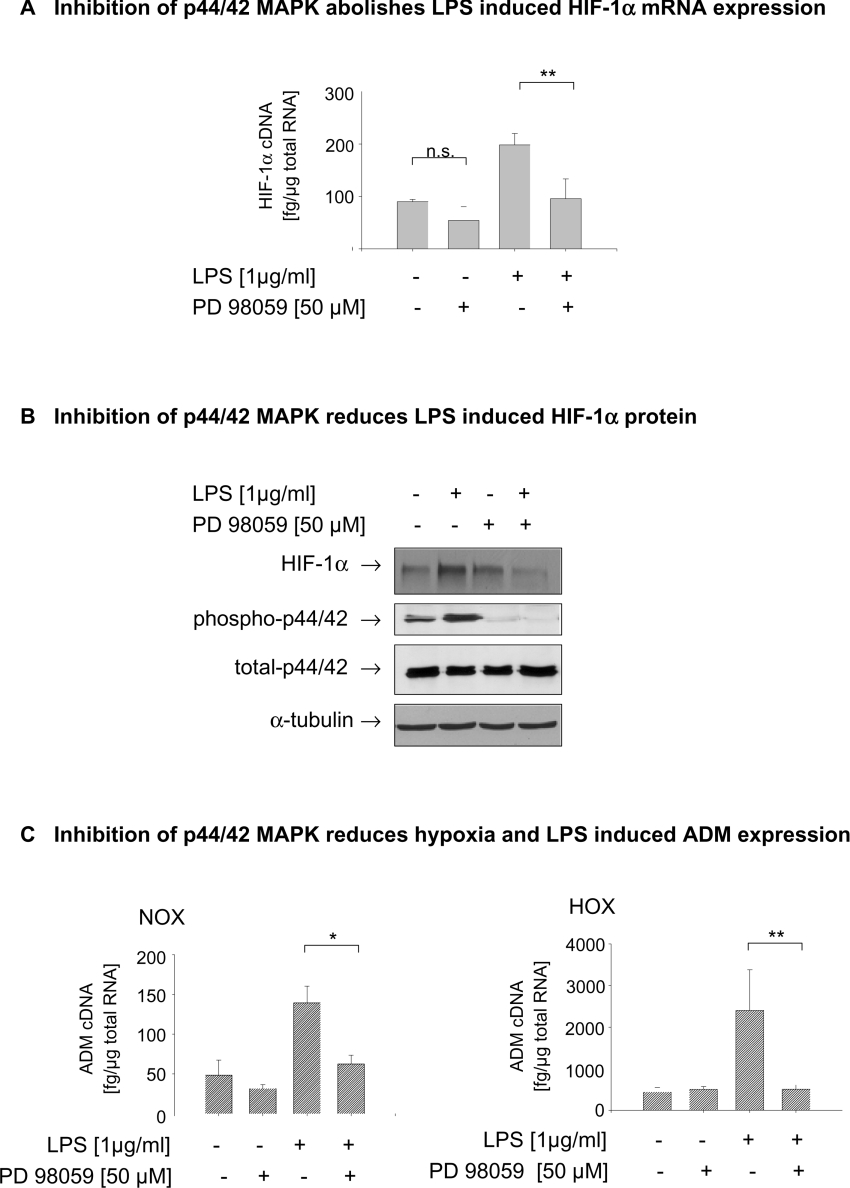
To find out whether activation of p44/42 is critically involved in the LPS-dependent HIF-1α induction, cells were pretreated, 30 min before a 4 h incubation period, with PD 98059, a specific inhibitor of MEK1/2 (MAPK/ERK kinase 1/2), the upstream kinase of p44/42 MAPK. Inhibition of p44/42 MAPK phosphorylation significantly reduced the LPS-induced expression of HIF-1α mRNA (Figure 5A) and of HIF-1α protein (Figure 5B). In a further approach, we transfected THP-1 cells with RNAi directed against p44/42 MAPK 24 h prior to the experiments, which reduced p44/42 proteins to very low levels. As with the pharmacological inhibition of p44/42 MAPK activation, the reduction in p44/42 reduced the LPS-dependent HIF-1α accumulation under normoxic, and to a lesser extent, under hypoxic conditions (results not shown). Quantification of ADM cDNA demonstrated that lowering of p44/42 MAPK resulted in a significant reduction of LPS-induced ADM expression under normoxic and under hypoxic conditions.
Figure 5. Inhibition of p44/42 MAPK phosphorylation reduces the LPS induced HIF-1α mRNA expression and protein accumulation.
(A) THP-1 cells were preincubated with the MEK1/2 inhibitor PD 98059 (50 μM) 30 min before LPS treatment. After 3 h of incubation under normoxic (NOX) or hypoxic (HOX) conditions, total RNA and whole-cell lysates were prepared. Amounts of HIF-1α cDNA were quantified by real-time PCR and normalized to 1 μg of total RNA. Means±S.D. from four independent experiments are shown. **P<0.01. (B) In addition, HIF-1α protein and p44/42 MAPK were analysed by immunoblot in 70 μg of total cell lysate; α-tubulin was used as loading control. (C) Inhibition of p4/42 MAPK activation significantly reduced the LPS-induced expression of the HIF-1 target ADM gene. Amounts of HIF-1α cDNA were quantified by real-time PCR and normalized to 1 μg of total RNA. The means±S.D. for four independent experiments are shown. **P<0.01.
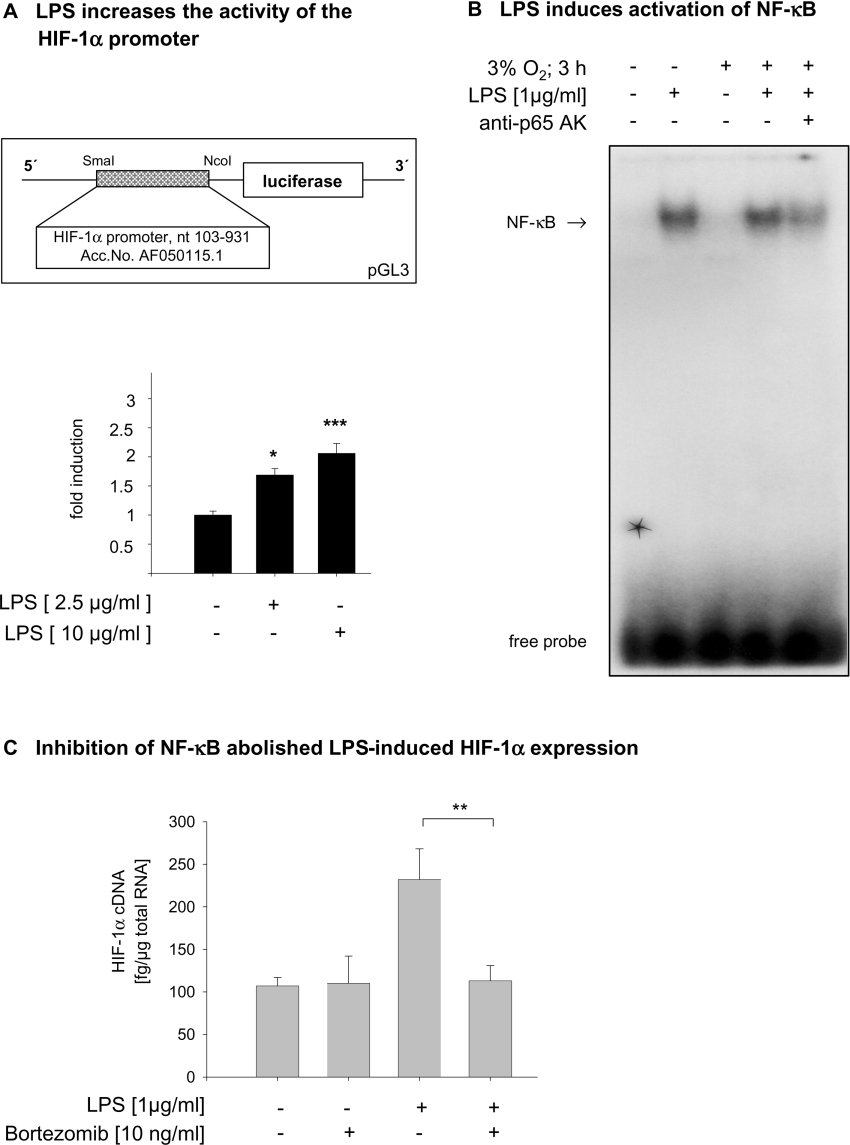
To identify transcription factors involved in LPS induced expression of HIF-1α mRNA, we performed reporter gene assays with a luciferase reporter under the control of a 800 nt fragment of the HIF-1α promoter. The activity of the HIF-1α promoter was dose-dependently stimulated by LPS treatment (Figure 6A). Several potential NF-κB binding sites within this promoter were identified using the program TFSearch version 3.1. Using EMSA, an oligonucleotide including the NF-κB motif located −130 nt of the transcription start site of the HIF-1α promoter was found to specifically bind NF-κB after LPS treatment (Figure 6B). Treatment of THP-1 cells with the NF-κB inhibitor bortezomib completely abolished the increase in HIF-1α mRNA expression by LPS (Figure 6C).
Figure 6. LPS-induced HIF-1α expression depends on the activation of NF-κB.
(A) MonoMac6 cells were transfected with a luciferase reporter vector under the control of the HIF-1α promoter. The HIF-1 promoter activity was dose-dependently increased by LPS. Luciferase activity was normalized to the expression of the corresponding control vector. n=6; P<0.05. The insert shows a schematic drawing of the luciferase reporter construct. (B) Nuclear extracts from LPS-treated THP-1 cells were incubated with an oligonucleotide corresponding to the NF-κB binding motif −130 nt in front of the transcription start site of the HIF-1α promoter. Abbreviation: anti-p65 AK, anti-NF-κB p65 antibody. (C) The LPS-induced expression of HIF-1α mRNA was abolished by the NF-κB inhibitor bortezomib. Total RNA was prepared, and amounts of HIF-1α cDNA were quantified by real-time PCR and normalized to 1 μg of total RNA. Means±S.D. from four independent experiments are shown; **P<0.01.
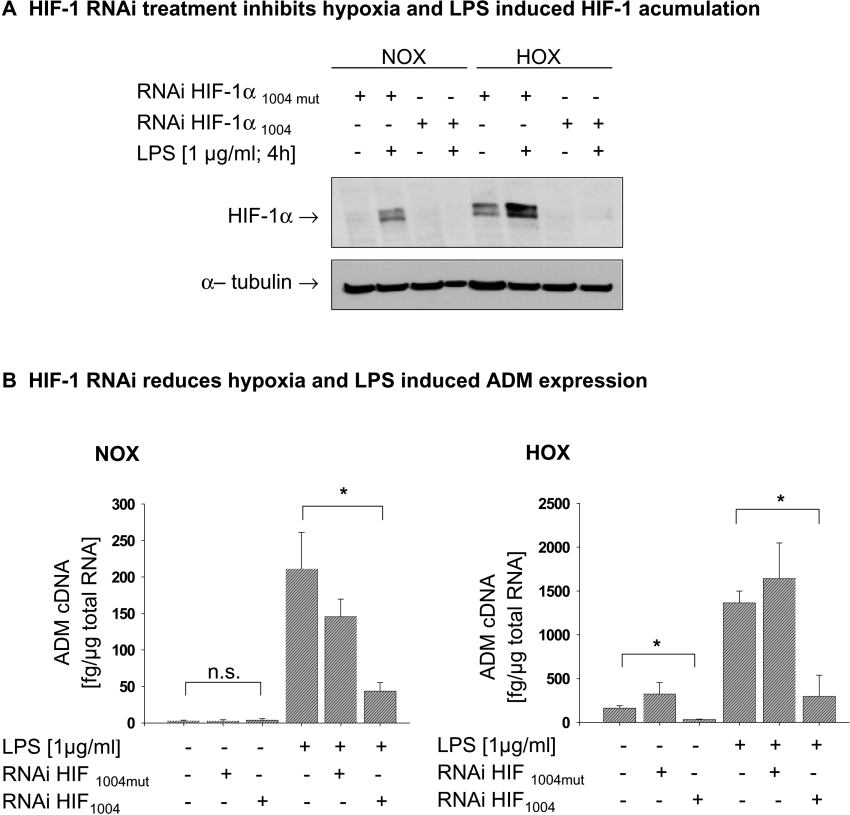
Finally we aimed at elucidating the importance of HIF-1α in the LPS-induced expression of the HIF-1 target ADM. In a first step we established a specific RNAi directed against HIF-1α. THP-1 cells were transfected with HIF-1α RNAi 18 h before the start of the experiments. Specificity of the selected sequence was confirmed by transfecting a mutant HIF-1α RNAi (RNAi HIF-1α1004mut) which had neither an effect on hypoxia- nor on LPS-induced HIF-1α accumulation (Figure 7A). After knocking down HIF-1α using RNAi, quantification of ADM expression revealed a complete loss of hypoxia-induced ADM and a significant decrease in ADM expression after LPS treatment (Figure 7B).
Figure 7. HIF-1α is critically involved in LPS- and hypoxia-induced expression of ADM.
(A) Knocking down HIF-1α by RNAi abolished the LPS and hypoxia-induced HIF-1α protein accumulation. HIF-1α protein was detected by immunoblot in 70 μg of total cell lysate. A mutated HIF-1α RNAi (RNAi HIF 1α 1004 mut) served as control. (B) Treatment of THP-1 with RNAi directed against HIF-1α reduced the LPS induced ADM expression under normoxic (NOX; left panel) and hypoxic (HOX; right panel) conditions. The mutated HIF-1 RNAi has no significant effects on LPS-induced ADM expression. ADM cDNA was quantified by real-time PCR. Results are means±S.D. for four independent experiments; *P<0.05; **P<0.01.
DISCUSSION
Several studies in the recent past have shown that cellular responses in inflammation and hypoxia are closely linked [8,26,27]. During inflammatory processes, monocytes extravasate from the blood into the tissue, where they differentiate into macrophages. HIF-1α has recently been identified as one of the indispensable transcription factors that co-ordinate gene expression in myeloid cells during the process of extravasation [8]. The first aim of the present study was to investigate whether sensitivity to hypoxia is different in monocytes and differentiated macrophages. We mimicked the process of differentiation by treatment of monocytes with phorbol ester. HIF-1α accumulation after hypoxic treatment occurred in both cell types, but was less prominent in differentiated cells. With respect to the HIF-1α mRNA levels a significant decrease of HIF-1α expression after hypoxia was observed only in the undifferentiated cells, whereas in differentiated macrophages no significant differences in HIF-1α mRNA levels in normoxia and hypoxia was detected. Moreover, HIF-1α mRNA levels in differentiated macrophages are about 2-fold higher than in undifferentiated cells, underlining the notion that up-regulation of HIF-1α mRNA expression is an important step in adaptation to an hypoxic environment. Although Blouin et al. [28] reported an LPS-induced increase in HIF-1α mRNA in alveolar-derived rat macrophages, hypoxia alone failed to influence HIF-1α mRNA levels. Our results are a first hint that the expression of HIF-1α mRNA in cells of the innate immune system is influenced by the differentiation status of the cells. The micro-environmental conditions found in areas of inflammation are characterized by low levels of O2 [29]. Likewise invasion of malignant tumour cells causes hypoxia and up-regulation of HIF-1 was observed in tumour-associated macrophages [27]. Activated macrophages in inflamed joints of patients suffering from rheumatoid arthritis express HIF-1α [30], and induction of HIF-1α in primary inflammatory cells of healing wounds was at least partly caused by inflammatory cytokines [31]. Whereas in all these situations hypoxia may also exist to activate HIF-1, hypoxia-independent induction of HIF-1α by inflammatory mediators possibly enhances the activity of the transcription factor. Similar observations have already been made in tumour-cell lines [5,7,25], epithelial cells [32] and vascular smooth-muscle cells [33]. Therefore inflammatory mediators are able to induce HIF-1 under normoxia. In the case of monocytes this possibly prepares the cells for a better survival and activity in a hypoxic environment after extravasation from the blood vessels into the tissue. Interestingly, differentiated THP-1 cells and, in particular, primary human macrophages, do not have to compensate for a hypoxic reduction in HIF-1α mRNA levels in order to reach higher HIF-1α protein levels (Figures 2B and 2C). On the other hand, our non-differentiated THP-1 cells potentially have higher basal HIF-1α mRNA levels on account of their leukaemic origin. However, differentiated and non-differentiated cells can both strongly increase HIF-1α expression when challenged by LPS. Here LPS did not interfere with the hydroxylation of HIF-1α protein, since ubiquitination was not reduced by LPS treatment. From our experiments we cannot exclude the possibility of an additional increase in the translation of HIF-1α mRNA by LPS treatment. Other inflammatory mediators, such as TNFα (tumour necrosis factor α) or interleukin-1β have been found to increase translation of HIF-1α [25,34]. In our THP-1 cells, TNFα is strongly induced after a 4 h LPS treatment (results not shown). TNFα protein released into the culture medium may thus stimulate translation in an autocrine manner [34].
Bacterial LPSs as compounds of the cell wall of Gramnegative bacteria are well known initiators of the inflammatory response. Essential components of the cellular response to LPS are soluble or membrane-bound CD14 and TLR4 (toll-like receptor 4) [35], which were strongly up-regulated in differentiated cells. Thus sensitivity to LPS may increase in differentiated cells. This notion is supported by higher HIF-1α mRNA levels in differentiated THP-1 cells and human monocytes compared with non-differentiated THP-1 cells after LPS treatment under hypoxic conditions.
Downstream signalling of the TLR4 complex leads to the activation of a wide variety of phosphorylation cascades, including activation of the p44/42 MAPK and NF-κB pathways. We focused first on activation of p44/42 as a potential part of the intracellular signaling cascade activated by LPS, on the basis of results obtained in previous work [36]. In our cells we found a significant increase in phosphorylation of p44/42 after LPS treatment or hypoxia, which was followed by increased HIF-1α gene expression and protein accumulation. Activation of p44/42 appears to be critical for LPS-induced HIF-1α activation, since inhibition of the upstream kinase MEK1/2 by PD 98059 and RNAi directed against p44/42 significantly reduced HIF-1α mRNA and protein accumulation.
Analysis of the HIF-1α promoter demonstrated a significant increase in promoter activity after LPS treatment. LPS induced binding of NF-κB to a respective consensus element 130 bp upstream of the transcriptional start site, and inhibition of NF-κB abolished LPS-induced HIF-1 target gene expression. Our data suggest that p44/42 might phosphorylate IκBα (inhibitory κB α-subunit) to up-regulate NF-κB, as it has been shown for prostate cancer cells [37].
Finally, a wide variety of genes in monocytes and macrophages is activated by LPS, of which some are known to be regulated by hypoxia as well. The gene encoding ADM is such a gene, and regulation of its expression is of particular interest in inflammatory diseases. High levels of ADM were found in septic patients, where ADM possibly contributes to the induction and persistence of hypotension in septic shock [20,38]. Macrophages were identified as one source of ADM secretion after stimulation with LPS [39]. On the other hand induction of ADM expression by hypoxia is known from a variety of tumour-cell lines, and the gene encoding ADM has been identified as an HIF-1-dependent [16]. Here LPS-induced ADM expression in normoxia and the enhancement of this activation by hypoxia strongly depend on HIF-1-activating pathways, as shown by lowered ADM expression after inhibition of p44/42 MAPK, which reduced LPS-dependent HIF-1α accumulation. More specifically, RNAi knockdown directed against HIF-1α greatly reduced most of the LPS effect, under both normoxia and hypoxia.
In conclusion, LPS induces HIF-1α in human monocytes and macrophages under normoxic conditions through enhanced transcription of the HIF-1α gene, which appears to require p44/42 MAPK signalling and is at least dependent on NF-κB activation. Under hypoxia, additional stabilization of HIF-1α protein synergistically increases the expression of HIF-1-dependent genes like that encoding ADM. Our data underline the importance of HIF-1 in integrating the inflammatory and hypoxic tissue response.
Acknowledgments
We thank Dr Carine Michiels for the HIF-1α promoter construct. This work was financially supported by the Deutsche Forschungsgemeinschaft, Bonn, Germany (grants FA 225/18-2 and FA 225/20-2).
References
- 1.Bracken C. P., Whitelaw M. L., Peet D. J. The hypoxia-inducible factors: key transcriptional regulators of hypoxic responses. Cell. Mol. Life Sci. 2003;60:1376–1393. doi: 10.1007/s00018-003-2370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masson N., Ratcliffe P. J. HIF prolyl and asparaginyl hydroxylases in the biological response to intracellular O2 levels. J. Cell. Sci. 2003;116:3041–3049. doi: 10.1242/jcs.00655. [DOI] [PubMed] [Google Scholar]
- 3.Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., et al. Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 4.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 5.Metzen E., Zhou J., Jelkmann W., Fandrey J., Brune B. Nitric oxide impairs normoxic degradation of HIF-1α by inhibition of prolyl hydroxylases. Mol. Biol. Cell. 2003;14:3470–3481. doi: 10.1091/mbc.E02-12-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Awad B., Kreft B., Wolber E. M., Hellwig-Burgel T., Metzen E., Fandrey J., Jelkmann W. Hypoxia and interleukin-1β stimulate vascular endothelial growth factor production in human proximal tubular cells. Kidney Int. 2000;58:43–50. doi: 10.1046/j.1523-1755.2000.00139.x. [DOI] [PubMed] [Google Scholar]
- 7.Hellwig-Burgel T., Rutkowski K., Metzen E., Fandrey J., Jelkmann W. Interleukin-1β and tumor necrosis factor-α stimulate DNA binding of hypoxia-inducible factor-1. Blood. 1999;94:1561–1567. [PubMed] [Google Scholar]
- 8.Cramer T., Yamanishi Y., Clausen B. E., Forster I., Pawlinski R., Mackman N., Haase V. H., Jaenisch R., Corr M., Nizet V., et al. HIF-1α is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peyssonaux C., Johnson R. S. An unexpected role for hypoxic response: oxygenation and inflammation. Cell Cycle. 2004;3:168–171. [PubMed] [Google Scholar]
- 10.Richard D. E., Berra E., Gothie E., Roux D., Pouyssegur J. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1α (HIF-1α) and enhance the transcriptional activity of HIF-1. J. Biol. Chem. 1999;274:32631–32637. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- 11.Minet E., Arnould T., Michel G., Roland I., Mottet D., Raes M., Remacle J., Michiels C. ERK activation upon hypoxia: involvement in HIF-1 activation. FEBS Lett. 2000;468:53–58. doi: 10.1016/s0014-5793(00)01181-9. [DOI] [PubMed] [Google Scholar]
- 12.Sang N., Stiehl D. P., Bohensky J., Leshchinsky I., Srinivas V., Caro J. MAPK signaling up-regulates the activity of hypoxia-inducible factors by its effects on p300. J. Biol. Chem. 2003;278:14013–14019. doi: 10.1074/jbc.M209702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldsmith E. J., Cobb M. H., Chang C. I. Structure of MAPKs. Methods Mol. Biol. 2004;250:127–144. doi: 10.1385/1-59259-671-1:127. [DOI] [PubMed] [Google Scholar]
- 14.van der Bruggen T., Nijenhuis S., van Raaij E., Verhoef J., Sweder van Asbeck B. Lipopolysaccharide-induced tumor necrosis factor α production by human monocytes involves the Raf-1/MEK1-MEK2/ERK1-ERK2 pathway. Infect. Immun. 1999;67:3824–3829. doi: 10.1128/iai.67.8.3824-3829.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu A. J., Wang Z. G., Walton M. A., Seto A. Involvement of MAPK activation in bacterial endotoxin-inducible tissue factor upregulation in human monocytic THP-1 cells. J. Surg. Res. 2001;101:85–90. doi: 10.1006/jsre.2001.6271. [DOI] [PubMed] [Google Scholar]
- 16.Garayoa M., Martinez A., Lee S., Pio R., An W. G., Neckers L., Trepel J., Montuenga L. M., Ryan H., Johnson R., et al. Hypoxia-inducible factor-1 (HIF-1) up-regulates adrenomedullin expression in human tumor cell lines during oxygen deprivation: a possible promotion mechanism of carcinogenesis. Mol. Endocrinol. 2000;14:848–862. doi: 10.1210/mend.14.6.0473. [DOI] [PubMed] [Google Scholar]
- 17.Kitamura K., Kangawa K., Kawamoto M., Ichiki Y., Nakamura S., Matsuo H., Eto T. Adrenomedullin: a novel hypotensive peptide isolated from human pheochromocytoma. Biochem. Biophys. Res. Commun. 1993;192:553–560. doi: 10.1006/bbrc.1993.1451. [DOI] [PubMed] [Google Scholar]
- 18.Sugo S., Minamino N., Shoji H., Kangawa K., Kitamura K., Eto T., Matsuo H. Interleukin-1, tumor necrosis factor and lipopolysaccharide additively stimulate production of adrenomedullin in vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 1995;207:25–32. doi: 10.1006/bbrc.1995.1148. [DOI] [PubMed] [Google Scholar]
- 19.Ladoux A., Frelin C. Coordinated up-regulation by hypoxia of adrenomedullin and one of its putative receptors (RDC-1) in cells of the rat blood-brain barrier. J. Biol. Chem. 2000;275:39914–39919. doi: 10.1074/jbc.M006512200. [DOI] [PubMed] [Google Scholar]
- 20.Hirata Y., Mitaka C., Sato K., Nagura T., Tsunoda Y., Amaha K., Marumo F. Increased circulating adrenomedullin, a novel vasodilatory peptide, in sepsis. J. Clin. Endocrinol. Metab. 1996;81:1449–1453. doi: 10.1210/jcem.81.4.8636349. [DOI] [PubMed] [Google Scholar]
- 21.Zaks-Zilberman M., Salkowski C. A., Elsasser T., Cuttitta F., Vogel S. N. Induction of adrenomedullin mRNA and protein by lipopolysaccharide and paclitaxel (taxol) in murine macrophages. Infect. Immun. 1998;66:4669–4675. doi: 10.1128/iai.66.10.4669-4675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen M. B., Nielsen S. E., Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Schreiber E., Matthias P., Muller M. M., Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frede S., Freitag P., Otto T., Heilmaier C., Fandrey J. The proinflammatory cytokine interleukin 1β and hypoxia cooperatively induce the expression of adrenomedullin in ovarian carcinoma cells through hypoxia inducible factor 1 activation. Cancer Res. 2005;65:4690–4697. doi: 10.1158/0008-5472.CAN-04-3877. [DOI] [PubMed] [Google Scholar]
- 26.Demasi M., Cleland L. G., Cook-Johnson R. J., Caughey G. E., James M. J. Effects of hypoxia on monocyte inflammatory mediator production: dissociation between changes in cyclooxygenase-2 expression and eicosanoid synthesis. J. Biol. Chem. 2003;278:38607–38616. doi: 10.1074/jbc.M305944200. [DOI] [PubMed] [Google Scholar]
- 27.Burke B., Giannoudis A., Corke K. P., Gill D., Wells M., Ziegler-Heitbrock L., Lewis C. E. Hypoxia-induced gene expression in human macrophages: implications for ischemic tissues and hypoxia-regulated gene therapy. Am. J. Pathol. 2003;163:1233–1243. doi: 10.1016/S0002-9440(10)63483-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blouin C. C., Page E. L., Soucy G. M., Richard D. E. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1α. Blood. 2004;103:1124–1130. doi: 10.1182/blood-2003-07-2427. [DOI] [PubMed] [Google Scholar]
- 29.Cramer T., Johnson R. S. A novel role for the hypoxia inducible transcription factor HIF-1α: critical regulation of inflammatory cell function. Cell Cycle. 2003;2:192–193. [PubMed] [Google Scholar]
- 30.Hollander A. P., Corke K. P., Freemont A. J., Lewis C. E. Expression of hypoxia-inducible factor 1α by macrophages in the rheumatoid synovium: implications for targeting of therapeutic genes to the inflamed joint. Arthritis Rheum. 2001;44:1540–1544. doi: 10.1002/1529-0131(200107)44:7<1540::AID-ART277>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Albina J. E., Mastrofrancesco B., Vessella J. A., Louis C. A., Henry W. L., Jr, Reichner J. S. HIF-1 expression in healing wounds: HIF-1α induction in primary inflammatory cells by TNF-α. Am. J. Physiol. Cell Physiol. 2001;281:C1971–C1977. doi: 10.1152/ajpcell.2001.281.6.C1971. [DOI] [PubMed] [Google Scholar]
- 32.Sandau K. B., Fandrey J., Brune B. Accumulation of HIF-1α under the influence of nitric oxide. Blood. 2001;97:1009–1015. doi: 10.1182/blood.v97.4.1009. [DOI] [PubMed] [Google Scholar]
- 33.Richard D. E., Berra E., Pouyssegur J. Nonhypoxic pathway mediates the induction of hypoxia-inducible factor 1α in vascular smooth muscle cells. J. Biol. Chem. 2000;275:26765–26771. doi: 10.1074/jbc.M003325200. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J., Callapina M., Goodall G. J., Brune B. Functional integrity of nuclear factor κB, phosphatidylinositol 3′-kinase and mitogen-activated protein kinase signaling allows tumor necrosis factor α-evoked Bcl-2 expression to provoke internal ribosome entry site-dependent translation of hypoxia-inducible factor 1α. Cancer Res. 2004;64:9041–9048. doi: 10.1158/0008-5472.CAN-04-1437. [DOI] [PubMed] [Google Scholar]
- 35.Guillot L., Medjane S., Le Barillec K., Balloy V., Danel C., Chignard M., Si-Tahar M. Response of human pulmonary epithelial cells to lipopolysaccharide involves toll-like receptor 4 (TLR4)-dependent signaling pathways: evidence for an intracellular compartmentalization of TLR4. J. Biol. Chem. 2004;279:2712–2718. doi: 10.1074/jbc.M305790200. [DOI] [PubMed] [Google Scholar]
- 36.Shi L., Kishore R., McMullen M. R., Nagy L. E. Lipopolysaccharide stimulation of ERK1/2 increases TNF-α production via Egr-1. Am. J. Physiol. Cell. Physiol. 2002;282:C1205–C1211. doi: 10.1152/ajpcell.00511.2001. [DOI] [PubMed] [Google Scholar]
- 37.Kukreja P., Abdel-Mageed A. B., Mondal D., Liu K., Agrawal K. C. Up-regulation of CXCR4 expression in PC-3 cells by stromal-derived factor-1α (CXCL12) increases endothelial adhesion and transendothelial migration: role of MEK/ERK signaling pathway-dependent NF-κB activation. Cancer Res. 2005;65:9891–9898. doi: 10.1158/0008-5472.CAN-05-1293. [DOI] [PubMed] [Google Scholar]
- 38.Nishio K., Akai Y., Murao Y., Doi N., Ueda S., Tabuse H., Miyamoto S., Dohi K., Minamino N., Shoji H., et al. Increased plasma concentrations of adrenomedullin correlate with relaxation of vascular tone in patients with septic shock. Crit. Care Med. 1997;25:953–957. doi: 10.1097/00003246-199706000-00010. [DOI] [PubMed] [Google Scholar]
- 39.Kubo A., Minamino N., Isumi Y., Katafuchi T., Kangawa K., Dohi K., Matsuo H. Production of adrenomedullin in macrophage cell line and peritoneal macrophage. J. Biol. Chem. 1998;273:16730–16738. doi: 10.1074/jbc.273.27.16730. [DOI] [PubMed] [Google Scholar]