Abstract
We cloned the genes encoding the ribosomal proteins Ph (Pyrococcus horikoshii)-P0, Ph-L12 and Ph-L11, which constitute the GTPase-associated centre of the archaebacterium Pyrococcus horikoshii. These proteins are homologues of the eukaryotic P0, P1/P2 and eL12 proteins, and correspond to Escherichia coli L10, L7/L12 and L11 proteins respectively. The proteins and the truncation mutants of Ph-P0 were overexpressed in E. coli cells and used for in vitro assembly on to the conserved domain around position 1070 of 23S rRNA (E. coli numbering). Ph-L12 tightly associated as a homodimer and bound to the C-terminal half of Ph-P0. The Ph-P0·Ph-L12 complex and Ph-L11 bound to the 1070 rRNA fragments from the three biological kingdoms in the same manner as the equivalent proteins of eukaryotic and eubacterial ribosomes. The Ph-P0·Ph-L12 complex and Ph-L11 could replace L10·L7/L12 and L11 respectively, on the E. coli 50S subunit in vitro. The resultant hybrid ribosome was accessible for eukaryotic, as well as archaebacterial elongation factors, but not for prokaryotic elongation factors. The GTPase and polyphenylalanine-synthetic activity that is dependent on eukaryotic elongation factors was comparable with that of the hybrid ribosomes carrying the eukaryotic ribosomal proteins. The results suggest that the archaebacterial proteins, including the Ph-L12 homodimer, are functionally accessible to eukaryotic translation factors.
Keywords: archaebacterium, elongation factor, ribosome, ribosomal protein, ribosomal stalk, translation elongation
Abbreviations: eEF, eukaryotic elongation factor; Ph, Pyrococcus horikoshii
INTRODUCTION
The ‘GTPase-associated centre’ is a functional site in the large ribosomal subunit that is responsible for GTPase-related events, which are dependent upon translation factors in protein biosynthesis. Protein components of this site in most eubacterial ribosomes constitute a characteristic pentameric complex, L10(L7/L12)2(L7/L12)2 [1,2] in which two L7/L12 homodimers bind to the C-terminal region of L10 [3], although recent studies have discovered a heptameric form, L10(L7/L12)2(L7/L12)2(L7/L12)2, in a few eubacterial species [4,5]. The L10·L7/L12 complex, together with another protein, L11, binds to the highly conserved domain around position 1070 (Escherichia coli numbering is used throughout) of 23S rRNA through interaction with the L10 moiety [6–8]. The protein complex constitutes a highly flexible and functionally important protuberance, the so-called ‘stalk’, in the ribosome [9]. Although the ribosomal stalk can be observed by cryo-electron microscopy [10], the detailed structure within the ribosome has not been resolved by X-ray crystallography [11–13].
The eukaryotic ribosomal phosphoproteins P0 and P1/P2 (P proteins) are counterparts of eubacterial L10 and L7/L12 respectively [14]. Unlike eubacterial, L7/L12, the eukaryotic stalk proteins, P1 and P2, are related, but are different proteins [15–17]. Previous biochemical and genetic studies indicate that P1 and P2 form a heterodimer [18–22], and that the interaction between the two proteins is important for their assembly on to P0 in order to constitute the pentameric complex [21,23], designated as ‘P0·P1-P2’. This complex not only binds to eukaryotic 28S rRNA, but also cross-binds to prokaryotic 23S rRNA [24] and determines the specificity of the ribosome for eukaryotic elongation factors (eEFs), eEF-1α and eEF-2 [25].
Archaebacteria constitute a third primary biological kingdom, separate from eubacteria and eukaryotes [26]. The archaebacterial translocase, EF-2, functions only with ribosomes from the same kingdom [27], although the ribosomes from methanogens, but not from Thermoplasma acidophilum or Sulfolobus solfataricus, showed some accessibility to eEF-2 [27,28]. There is, however, limited information about the functional structure of the ribosomal GTPase-associated centre in archaebacteria. Comparison of the components constituting the archaebacterial active site with those in eubacteria and eukaryotes has shown that: (i) the secondary structure of the position 1070 domain of 23S/28S rRNA is highly conserved among the three kingdoms [28,29], although the base at position 1067, one of the EF-G (elongation factor-G)/eEF-2-binding sites [30,31], is adenine in archaebacteria and eubacteria, but is guanine in eukaryotes [32,33] (see Supplementary Figures S1 and S2 at http://www.BiochemJ.org/bj/396/bj3960565add.htm) and the amino acid sequences and molecular mass of L7/L12- and L10-like proteins in archaebacteria show greater similarity with eukaryotic proteins than eubacterial proteins [34] (see Supplementary Figures S2–S4 at http://www.BiochemJ.org/bj396/bj3960565.add.htm).
The most inscrutable and interesting point is that only one species of archaebacterial L7/L12-like protein has been detected, despite its structural similarity to eukaryotic P1 or P2 proteins. A chemical cross-linking study of S. solfataricus ribosomes has suggested that the L7/L12-like protein is present as a homodimer in the 50S subunit, and presumably two homodimers are anchored to the L10-like protein [35]. The truncation study of the S. solfataricus L7/L12-like protein has shown that the N-terminal region is important for its incorporation into the ribosome [36]. In a functional study, it was shown that S. solfataricus ribosomes that were treated with ethanol to remove the L7/L12-like proteins retained significant activity in polyphenylalanine synthesis [36].
To clarify the mode of assembly of the archaebacterial ribosomal GTPase-associated centre and its function, we cloned and expressed the L7/L12-, L10- and L11-like proteins of Pyrococcus horikoshii, designated Ph (Pyrococcus horikoshii)-L12, Ph-P0 and Ph-L11 respectively, and performed in vitro reconstitution of the functional site, together with the position 1070 domain of 23S rRNA. The functionality of these P. horikoshii proteins was tested in a previously developed hybrid ribosome system [25], in which the E. coli L10·L7/L12 complex and L11 on the 50S subunit were replaced with the equivalent proteins from the other species. By using the hybrid ribosome system, the present study clearly shows that the P. horikoshii proteins, including the Ph-L12 homodimer, make the E. coli ribosomes accessible to eEFs, as well as archaebacterial elongation factors, but not eubacterial elongation factors. The eukaryotic P1-P2 heterodimers associated with P0 appear to be compatible with the archaebacterial stalk homodimers bound to the anchor protein, without significant loss of ribosomal function.
EXPERIMENTAL
Plasmid construction, protein expression and purification
Genomic DNA from the archaebacterium P. horikoshii was prepared as described by Blin and Stafford [37]. The coding regions for ribosomal proteins Ph-L12, Ph-L11, Ph-P0 and the C-terminal truncation mutants of Ph-P0, CΔ105 and CΔ131 were amplified by PCR [38] and inserted into an E. coli expression vector, pET-3a or pET-28c (Novagen). Proteins expressed in E. coli cells were purified as follows. The expressed Ph-P0 was present in the insoluble fraction and was solubilized in Buffer A, which consisted of 20 mM sodium acetate (pH5.5), 7 M urea and 5 mM 2-mercaptoethanol, dialysed against Buffer A, and was then applied to a column of CM-cellulose (Whatman) equilibrated with the same buffer. Ph-P0 was eluted in Buffer A containing 0.06 M LiCl. The Ph-P0 fraction was dialysed overnight against Buffer B, which consisted of 20 mM sodium phosphate (pH 6.5), 6 M urea, 100 mM LiCl and 5 mM 2-mercaptoethanol, and was further purified by HPLC on a DEAE-5PW column (Tosoh) in a linear gradient of 100–280 mM LiCl. The truncation mutants of Ph-P0 were purified by using an AKTA FPLC system with Resource S and Resource Q columns (Pharmacia) under similar conditions. Ph-L11 and Ph-L12 in the soluble fraction were enriched by incubation of the S100 fractions at 90 °C for 10 min followed by removal of insoluble material formed upon the heating by centrifugation. The soluble protein samples were dialysed overnight against Buffer C, which consisted of 20 mM sodium acetate (pH 4.5), 7 M urea and 5 mM 2-mercaptoethanol, and were then applied to a column of CM-cellulose equilibrated with the same buffer. Ph-L12 and Ph-L11 were eluted in Buffer C containing 0 and 0.06 M LiCl respectively. The Ph-L12 fraction was dialysed overnight against Buffer B and purified by HPLC on a DEAE-650S column in a linear gradient of 100–325 mM LiCl. The Ph-L11 fraction was dialysed overnight against Buffer D, which consisted of 20 mM sodium phosphate (pH 6.5), 6 M urea, 5 mM LiCl and 5 mM 2-mercaptoethanol, and was purified by HPLC on a CM-5PW column (Tosoh) with a linear gradient of 5–204 mM LiCl. The silkworm (Bombyx mori) counterparts, P0, P1/P2 and eL12 (a eukaryotic homologue of prokaryotic L11) were also expressed in E. coli cells and purified as described recently [39]. Expression and purification of the E. coli equivalent proteins L10, L7/L12 and L11 were performed as described previously [40].
In vitro RNA synthesis
The P. horikoshii rDNA fragment corresponding to residues 1142–1240 of 23S rRNA, which is similar to residues 1030–1128 of E. coli 23 S rRNA (designated here as the 1070 fragment), was amplified by PCR and inserted into the HindIII and BamHI sites of the pT7Blue T-Vector (Novagen). Synthesis and purification of the RNA fragment was performed as described previously [41] except that T7 RNA polymerase was used instead of SP-6 RNA polymerase. The RNA fragments containing residues 1029–1127 of E. coli 23S rRNA and the equivalent region (residues 1841–1939) in rat 28S rRNA were synthesized as described [41,42].
In vitro reconstitution of Ph-P0·Ph-L12·Ph-L11·rRNA
The putative ‘stalk complex’ corresponding to the eukaryotic P0·P1-P2 complex was reconstituted by mixing isolated Ph-P0 and Ph-L12 at a molar ratio of 1:6 in a buffer containing 7M urea, 300 mM KCl, 5 mM 2-mercaptoethanol and 20 mM Tris/HCl (pH 7.6) and by dialysis against the same buffer without urea, as described previously [21]. This procedure using urea is essential for in vitro reconstitution of the functional eukaryotic protein complex, since isolated eukaryotic P0 is insoluble in the absence of urea. Although isolated Ph-P0 and Ph-L12 were soluble and bound to each other without urea, the Ph-P0·Ph-L12 complex formed by using urea showed slightly higher activity in factor accessibility than the complex formed without urea. Therefore, the stalk protein complexes used in the present study (Ph-P0·Ph-L12, as well as silkworm P0·P1-P2 and E. coli L10·L7/L12) were all reconstituted by the same procedure using urea [21]. The molecular assembly of Ph-P0·Ph-L12 and Ph-L11 on the 1070 rRNA fragment was carried out in a solution containing 20 mM MgCl2, 300 mM KCl and 20 mM Tris/HCl (pH 7.6), as described in [21]. The complexes formed were assayed by 6% (w/v) polyacrylamide (acrylamide/bisacrylamide ratio 39:1) native gel electrophoresis at 6.5 V/cm with a buffer system containing 5 mM MgCl2, 50 mM KCl and 50 mM Tris/HCl (pH 8.0). Samples were electrophoresed for 6 h at constant voltage and 4 °C with buffer recirculation. The gel was subjected to autoradiography.
Ribosomal subunits, 50S core particles and hybrid 50S particles
E. coli ribosomal subunits were prepared as described previously [24,25]. The 50S core, deficient in L10·L7/L12 and L11, was prepared by extraction of the 50S subunits from the L11-deficient E. coli mutant, AM68 [43], in a solution containing 50% ethanol and 0.5 M NH4Cl at 0 °C, as described previously [25]. The P. horikoshii–E. coli or silkworm–E. coli hybrid 50S particle was prepared by mixing the E. coli 50S core with the P. horikoshii ribosomal proteins or silkworm proteins [25] as described in the Figure legends, followed by incubation at 37 °C for 5 min.
Acrylamide/agarose composite gel electrophoresis
The samples of hybrid 50S particles were analysed by electrophoresis on acrylamide/agarose composite gels composed of 3% (w/v) acrylamide (acrylamide/bisacrylamide, 19:1) and 0.5% (w/v) agarose [44], as described previously [25]. Gels were stained with 0.2% Azur B.
EFs and functional assays
eEF-1α and eEF-2 were isolated from pig liver as described by Iwasaki and Kaziro [45]. Ph-EF-1α and Ph-EF-2 from P. horikoshii, were expressed in E. coli cells using plasmids that were constructed by insertion of individual coding sequences into the pET-22b vector (Novagen). The S100 fractions of individual cells were heated at 90 °C for 10 min to remove E. coli proteins. The supernatant containing Ph-EF-1α was applied to a column of CM-cellulose equilibrated with a buffer of 20 mM sodium phosphate (pH 6.8) and 5 mM 2-mercaptoethanol, and Ph-EF-1α was eluted in the same buffer containing 0.02 M LiCl. Ph-EF-2 was isolated by elution from a DEAE-cellulose column equilibrated with a buffer of 20 mM Tris/HCl (pH 8.0) and 5 mM 2-mercaptoethanol. E. coli EF-Tu and EF-G were overexpressed in E. coli cells by using the pET-3a vector (Novagen), and purified from the cell extract by DEAE-sephadex A-50 column chromatography, as described in [46]. The assay for ribosomal GTPase activity dependent on eEF-2 and Ph-EF-2 was performed as described previously [21]. The assay for GTPase activity dependent on E. coli EF-G was described previously [40], except that the reaction mixture contained 7.5 pmol of the E. coli 30S subunit and 10 mM MgCl2. Polyphenylalanine synthesis by the hybrid ribosomes was assayed with eEF-1α and eEF-2, as described previously [21].
RESULTS AND DISCUSSION
Expression and isolation of P. horikoshii ribosomal proteins Ph-P0, Ph-L11 and Ph-L12
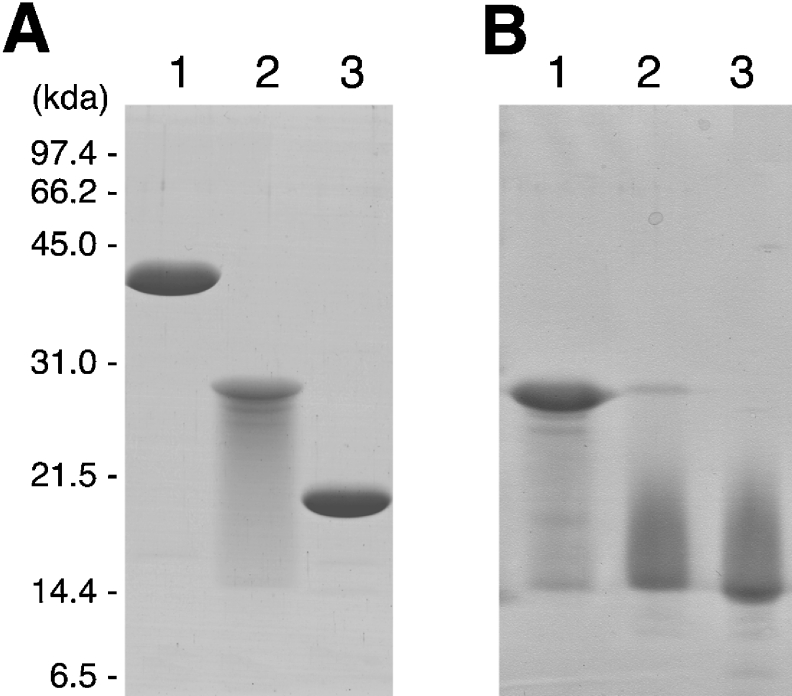
P. horikoshii ribosomal proteins Ph-P0, Ph-L11 and Ph-L12 were expressed in E. coli cells and isolated by HPLC. SDS/PAGE of the isolated proteins (Figure 1A) showed that Ph-P0 (lane 1) and Ph-L11 (lane 3) appeared as single bands of 39.5 kDa and 18.4 kDa, close to the predicted molecular masses of 37.5 kDa and 17.7 kDa respectively. However, Ph-L12 appeared as a major band of 27.9 kDa with smearing (lane 2). This value is close to twice the predicted size of Ph-L12 (11.8 kDa), suggesting that Ph-L12 forms a homodimer, which is extremely stable even after heating at 95 °C in SDS solution. After treatment of Ph-L12 with 1–2% H2O2 and subsequently with the SDS sample solution, a smeared band of approx. 15 kDa appeared, corresponding to the monomer (Figure 1B, lanes 2 and 3). This tight association of the proteins was also observed in the N-terminal halves of Ph-L12 (T. Naganuma and T. Uchiumi, unpublished work), which implies that Ph-L12 monomers bind tightly to each other through interaction with the N-terminal halves. The destabilization of the stalk dimer by treatment with H2O2 has also been reported in E. coli L7/L12 [47,48]. The molecular details of the destabilization of the Ph-L12 homodimer with H2O2 remain to be clarified.
Figure 1. Isolated ribosomal proteins constituting the archaebacterial GTPase-associated centre.
(A) Isolated Ph-P0 (lane 1), Ph-L12 (lane 2) and Ph-L11 (lane 3) were subjected to SDS/gel electrophoresis (5 μg of each). (B) Ph-L12 (5 μg) was incubated with 0% (lane 1), 1% (lane 2) or 2% (lane 3) H2O2 at 30 °C for 1 h, and then subjected to SDS/PAGE. The gel was stained with Coomassie Brilliant Blue R-250.
In vitro reconstitution of Ph-P0·Ph-L12 and Ph-P0·Ph-L12·Ph-L11·rRNA complexes
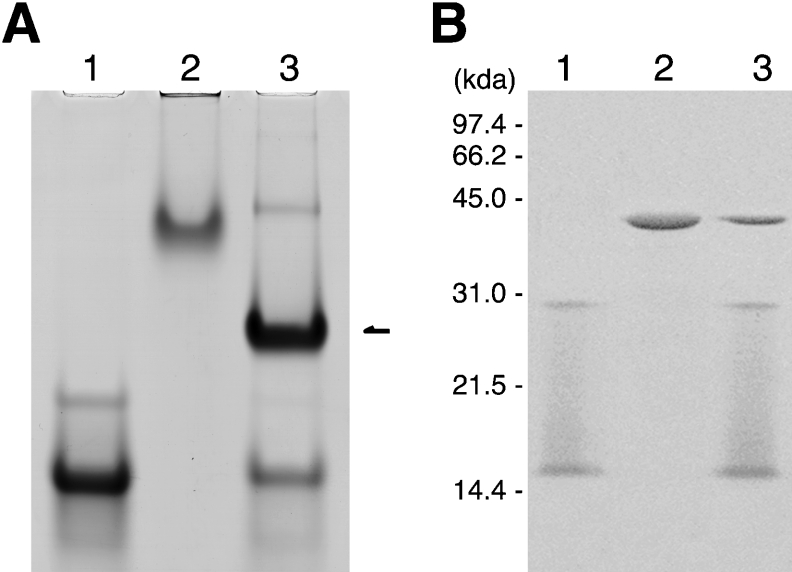
Ph-P0 was mixed with 6 M-fold amount of Ph-L12 and analysed by non-denaturing gel electrophoresis (Figure 2A). An intense band appeared (lane 3) with different mobility from those of isolated Ph-L12 (lane 1) and Ph-P0 (lane 2). To confirm that the new band in lane 3 of Figure 2(A) corresponded to the Ph-P0·Ph-L12 complex, the band was cut out of the gel and subjected to SDS/gel electrophoresis (Figure 2B). The new band contained three protein components (lane 3) corresponding to Ph-P0 (lane 2), the Ph-L12 dimer, and its monomer dissociated presumably by oxidation of the dimer during native gel electrophoresis (lane 1). The results indicate that the expressed Ph-P0 can form a complex with Ph-L12 homodimers in vitro.
Figure 2. Formation of a complex between Ph-P0 and Ph-L12.
(A) Isolated Ph-L12 (600 pmol, lane 1), Ph-P0 (100 pmol, lane 2) and the complex (lane 3) formed with both proteins was subjected to non-denaturing gel electrophoresis. The gel was stained with Coomassie Brilliant Blue R-250. The band that appeared as a result of mixing the two proteins is indicated with an arrow. (B) Major bands that appeared in (A), lanes 1, 2 and 3 (arrow), were cut out of the gel, incubated in SDS sample solution and then used as samples for SDS/gel electrophoresis. The gel was stained with the fluorescent reagent SYPRO Orange (Molecular Probes, Invitrogen) and the pattern was visualized with a STORM 860 PhosphorImager (Amersham Biosciences).
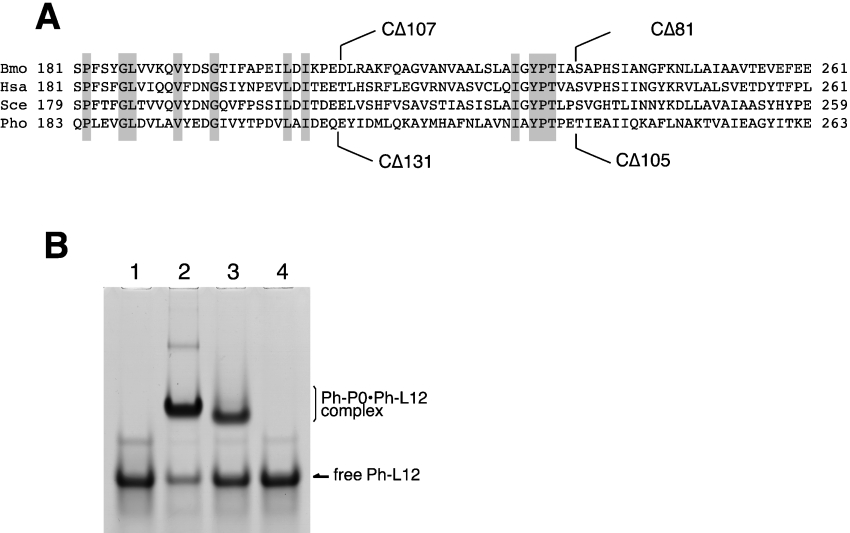
We recently analysed the binding sites for eukaryotic P1-P2 heterodimers within silkworm P0 [39]. The truncation mutant of silkworm P0 (Figure 3A), CΔ107, which lacks the C-terminal amino acid residues 210–316 failed to bind the two P1-P2 dimers, whereas the mutant lacking residues 236–316 (CΔ81) retained the ability to bind a single dimer. By using various truncation mutants including CΔ81 and CΔ107, we concluded that the region containing residues 210–235 in silkworm P0 is involved in binding one of the two P1-P2 dimers and the region spanning residues 236–261 is for binding another dimer. An alignment of sequences around the putative stalk binding sites of P0-like proteins shows some similarity between eukaryotic and archaebacterial proteins, although the sequence identity is low (Figure 3A).
Figure 3. Effect of C-terminal truncation of Ph-P0 on Ph-P0·Ph-L12 complex formation.
(A) Amino acid sequence alignment of the C-terminal parts of P0-like proteins from silkworm (Bombyx mori, Bmo), human (Homo sapiens, Hsa), yeast (Saccharomyces cerevisiae, Sce) and P. horikoshii (Pho). The sites of truncation in our previous study [39] are marked on the sequence of Bmo-P0. The truncation sites of P. horikoshii in the present study are also indicated below the Pho-P0 sequence. (B) The complexes were reconstituted by mixing 600 pmol of Ph-L12 with 100 pmol of wild-type Ph-P0 (lane 2), CΔ105 (lane 3) or CΔ131 (lane 4) as described in the Experimental section. The sample in lane 1 was Ph-L12 alone. Each sample was subjected to native gel electrophoresis. The gel was stained with Coomassie Brilliant Blue. Free proteins, CΔ105 and CΔ131, were not detected on the gel because of the failure of these proteins that were in the free state to run into the gel.
To determine whether archaebacterial Ph-P0 also has binding sites for Ph-L12 homodimers in this region, we constructed Ph-P0 mutants in which the C-terminal amino acids 238–342 (CΔ105) and 212–342 (CΔ131) were truncated (Figure 3A). The CΔ105 and CΔ131 truncation mutants of Ph-P0 correspond to the CΔ81 and CΔ107 truncation mutants of the silkworm P0, which could bind only one and no P1-P2 heterodimer respectively [39]. Binding of Ph-L12 homodimers to the truncated Ph-P0 mutants was tested by non-denaturing gel electrophoresis (Figure 3B). After mixing the CΔ105 Ph-P0 mutant with Ph-L12, a band that migrated slightly faster (lane 3) than that of the wild-type Ph-P0·L12 complex appeared (lane 2). However, the CΔ131 Ph-P0 mutant did not form a complex with Ph-L12 (lane 4), suggesting that binding sites for Ph-L12 homodimers are present within the C-terminal 131-amino-acid sequence of Ph-P0, and that there may be a conserved feature, at least in part, in binding to P0-like proteins between the archaebacterial Ph-L12 homodimer and the eukaryotic P1-P2 heterodimer. Further biochemical, biophysical and crystal structure analyses will elucidate the details of the interaction between Ph-P0 and Ph-L12 dimers.
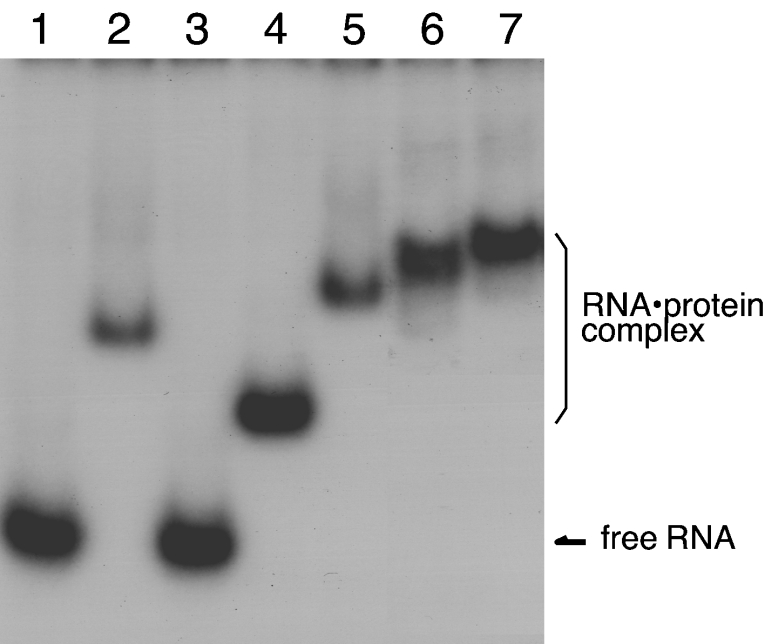
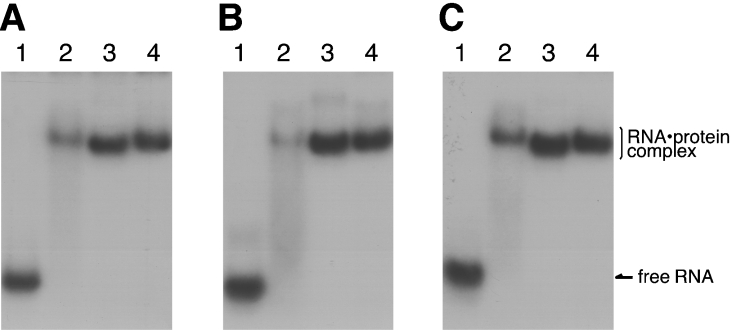
To test the ability of the expressed archaebacterial ribosomal proteins to bind the 1070 domain of 23S rRNA, gel mobility-shift assays were performed using the [32P]rRNA fragment (Figure 4). Ph-P0 (lane 2) and Ph-L11 (lane 4), but not Ph-L12 (lane 3), showed the ability to bind directly to the 1070 rRNA fragment. Supershifts of the complex bands were observed by mixing Ph-P0 with Ph-L11 (lane 5), Ph-P0 with Ph-L12 (lane 6), and Ph-P0, Ph-L11 and Ph-L12 together (lane 7), implying that all of the proteins assemble on to the 1070 rRNA region in vitro. Binding of the archaebacterial proteins, Ph-P0·Ph-L12 and Ph-L11, to the 1070 rRNA fragment of P. horikoshii 23S rRNA (Figure 5A, lane 3) was compared with that of E. coli L10·L7/L12 and L11 (lane 2) and with that of silkworm P0·P1-P2 and eL12 (lane 4). These cross-bindings were also tested with the equivalent RNA fragments of rat 28S rRNA (Figure 5B) and E. coli 23S rRNA (Figure 5C). These protein homologues bound equally well to the 1070 rRNA fragments from members of the three kingdoms, suggesting that there are highly conserved protein–RNA interactions at the GTPase-associated centre among the three kingdoms.
Figure 4. Binding ability of the P. horikoshii ribosomal proteins to the 1070 rRNA fragment.
An aliquot (20 pmol) of each of isolated Ph-P0 (lane 2), Ph-L12 (lane 3), Ph-L11 (lane 4), Ph-P0+Ph-L11 (lane 5), Ph-P0·Ph-L12 complex (lane 6), and Ph-P0·Ph-L12+Ph-L11 (lane 7) was mixed with 5 pmol of the [32P]RNA fragment covering the 1070 domain of P. horikoshii 23S rRNA (lane 1, RNA alone), separated by native gel electrophoresis and then subjected to autoradiography.
Figure 5. Conserved rRNA–protein binding property in the GTPase-associated centre.
RNA fragments of the 1070 rRNA domain from P. horikoshii 23S (A), rat 28S (B) and E. coli 23S (C). rRNAs (lanes 1) were mixed with the E. coli L10·L7/L12 complex and L11 (lanes 2), the Ph-P0·Ph-L12 complex and Ph-L11 (lanes 3), and the silkworm P0·P1-P2 complex and eL12 (lanes 4), and were analysed as described in Figure 4.
Functional properties of the P. horikoshii ribosomal proteins assembled on to the E. coli 50S particles
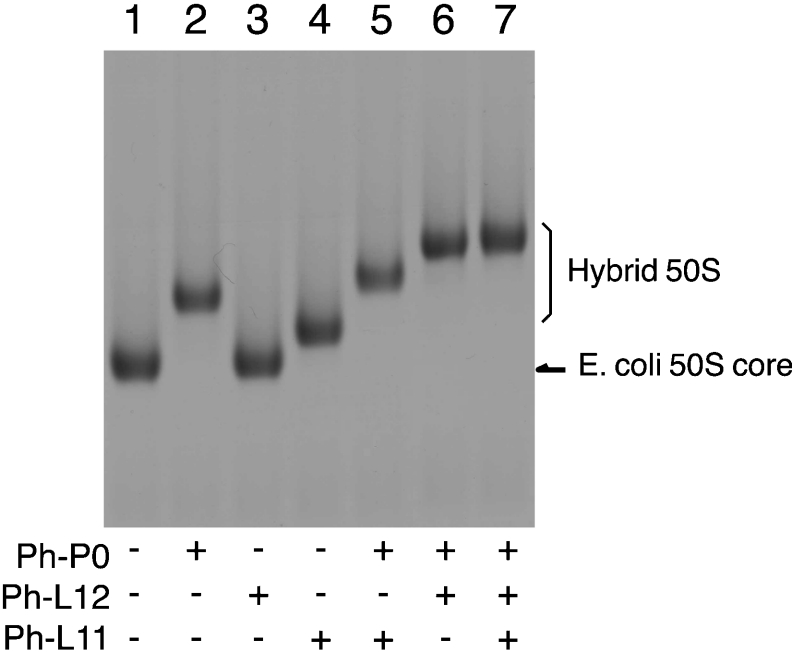
We have established in vitro conditions under which to replace the E. coli ribosomal protein L11 and the L10·L7/L12 complex on the 50S subunit with eukaryotic counterparts eL12 and P0·P1-P2 respectively [25]. The resultant hybrid ribosome showed polyphenylalanine synthesis, as well as GTPase activity, that was dependent on eEF-1α and eEF-2, but not on prokaryotic EF-Tu and EF-G [25]. This hybrid system is therefore useful for studying the functional role of the stalk protein complex at the GTPase-associated centre and also the structural determinants responsible for the kingdom-dependent specificity between ribosomes and translation factors. We attempted to examine the functional characteristics of the archaebacterial stalk-constituting proteins, Ph-P0, Ph-L11 and Ph-L12, expressed in E. coli cells. The formation of the hybrid 50S particles that resulted from mixing E. coli 50S cores with archaebacterial proteins was confirmed by acrylamide/agarose composite gel electrophoresis (Figure 6). The gel mobility of E. coli 50S cores (lane 1) was shifted upwards by the addition of recombinant Ph-P0 (lane 2) and Ph-L11 (lane 4), but not Ph-L12 (lane 3). Supershifts were observed after mixing Ph-P0 with Ph-L11 (lane 5), Ph-P0 with Ph-L12 (lane 6), or all three proteins (lane 7), implying that the three proteins are bound to E. coli 50S cores.
Figure 6. Assembly of P. horikoshii ribosomal proteins on to the GTPase-associated centre of E. coli 50S subunits.
An aliquot (10 pmol) of the 50S core particles deficient in L10·L7/L12 and L11 (lane 1) was incubated with 20 pmol of Ph-P0 (lane 2), Ph-L12 (lane 3), Ph-L11 (lane 4), Ph-P0+Ph-L11 (lane 5), the Ph-P0·Ph-L12 complex (lane 6), and Ph-P0·Ph-L12+Ph-L11 (lane 7), and then analysed by acrylamide/agarose composite gel electrophoresis, as described in the Experimental section.
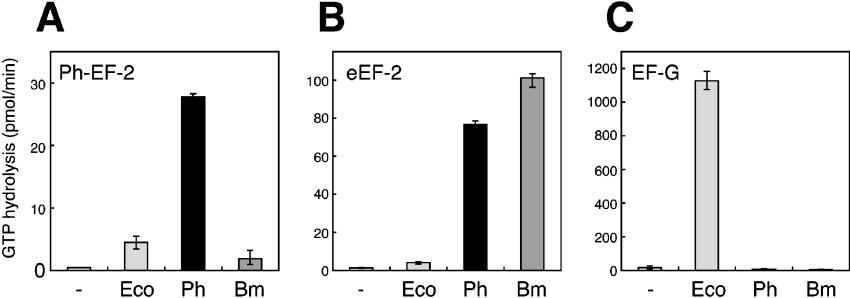
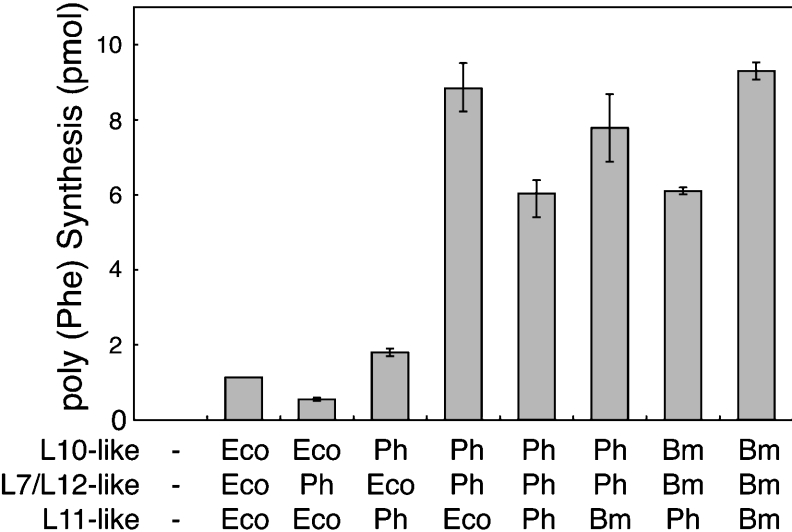
The hybrid 50S particles, together with E. coli 30S subunits, were tested for their function in enhancing accessibility to translocases from P. horikoshii (Figure 7A), pig liver (Figure 7B) and E. coli (Figure 7C). The GTPase activity of the hybrid ribosome carrying Ph-P0·Ph-L12 and Ph-L11 (Ph) was compared with that of the hybrid carrying silkworm P0·P1-P2 and eL12 (Bm), and also the E. coli ribosome reconstituted with L10·L7/L12 and L11 (Ec). The Ph-hybrid ribosome showed GTPase activity not only with P. horikoshii EF-2 (Figure 7A), but also with animal eEF-2 (Figure 7B). No activity of the Ph-hybrid was detected with E. coli EF-G (Figure 7C). Low P. horikoshii EF-2-dependent activity with the Ph-hybrid ribosome assayed at 37 °C could be predicted, considering that the optimum temperature for growth of P. horikoshii is extremely high (98 °C). Rather, it was unexpected that the Ph-hybrid showed high GTPase activity with animal eEF-2 at 37 °C, which was comparable with that of the silkworm hybrid ribosome carrying silkworm P0, P1/P2 and eL12 (Figure 7B). The functional accessibility of the hybrid ribosome carrying P. horikoshii proteins to eEFs was also tested in the polyphenylalanine synthesis assay, which is dependent on both eEF-1α and eEF-2 (Figure 8). The hybrids carrying the L10-like and L7/L12-like proteins from P. horikoshii and silkworm had comparable activity, although the addition of Ph-L11 instead of E. coli L11 or the silkworm homologue eL12 slightly decreased the activity. These results imply that the archaebacterial ribosomal proteins at the GTPase-associated centre have the potential to make the ribosome accessible to eEFs. The results also suggest that the L10-like and L7/L12-like proteins, but not the L11-like protein, are responsible for the specificity between ribosomes and translation factors. This conclusion is consistent with previous data regarding direct contact between the stalk protein and EF-G [49], but is inconsistent with another report on direct L11–EF-G contact [10]. In the present study, it is particularly noteworthy that Ph-L12 homodimers are compatible with eukaryotic P1-P2 heterodimers. We infer that functionally important structural elements of the stalk are preserved from archaebacterial homodimers to eukaryotic heterodimers.
Figure 7. The GTPase activity of the hybrid ribosomes dependent on P. horikoshii EF-2 (A), pig eEF-2 (B) and E. coli EF-G (C).
E. coli 50S core particles (2.5 pmol) deficient in L10, L7/L12 and L11 were pre-incubated with 10 pmol of each of L10·L7/L12 and L11 (Eco). The 50S cores were also incubated with 10 pmol of each of Ph-P0·Ph-L12, and Ph-L11 (Ph) or silkworm P0·P1-P2 and eL12 (Bm). The resulting 50S particle samples were assayed for GTPase activity in the presence of E. coli 30S subunits (7.5 pmol).
Figure 8. Polyphenylalanine synthesis of hybrid ribosomes dependent on eukaryotic elongation factors.
The 50S core particles (10 pmol) were pre-incubated with L10- (20 pmol), L7/L12- (120 pmol) and L11-like proteins (20 pmol) from E. coli (Eco), P. horikoshii (Ph) or Bombyx mori (Bm, silkworm) in various combinations as indicated, and then assayed for polyphenylananine synthesis dependent on pig eEF-1α and eEF-2, in the presence of E. coli 30S subunits (50 pmol).
Conclusions
The present study demonstrated the in vitro reconstitution of the P. horikoshii ribosomal GTPase-associated centre from purified components, which were cloned and expressed individually. The stalk protein, Ph-L12, was strongly bound as a dimer presumably through interactions between the N-terminal halves. The Ph-L12 homodimers bound to the C-terminal half of Ph-P0 in a similar manner to eukaryotic P1-P2 heterodimers [39], although the number of dimers bound to Ph-P0 has not been determined in this study. The Ph-P0·Ph-L12 complex and Ph-L11 bound to the 1070 rRNA domain from the three kingdoms in the same manner as the equivalent proteins from eukaryotes and eubacteria. The Ph-P0·Ph-L12 complex and Ph-L11 could replace the L10·L7/L12 complex and L11 respectively, on the E. coli 50S subunit. The P. horikoshii–E. coli hybrid ribosome showed functional accessibility for eEF-1α and eEF-2. The levels of GTPase and polyphenylalanine synthesis activity were comparable with those of another hybrid ribosome carrying the eukaryotic P0·P1-P2 complex and eL12 instead of Ph-P0·Ph-L12 and Ph-L11. These results suggest that the archaebacterial ribosomal proteins have the potential to function with eEFs. These results are reasonable, when considering that the amino acid sequences of the archaebacterial proteins resemble eukaryotic rather than eubacterial counterparts [see Supplementary Figures S2–S4 at http://www.BiochemJ.org/bj/396/bj3960565add.htm]. On the other hand, the present evidence is not consistent with previous observations that archaebacterial ribosomes (with the exception of those from methanogens) show no accessibility to eEFs [27,28]. This discrepancy may be explained as follows. Archaebacteria grow in extremely hot, acidic or salty conditions. It is probable that the overall structure of the archaebacterial ribosomal machinery does not work with eEFs that function under normal conditions. However, some archaebacterial ribosomal proteins may, in part, have functional compatibility with the equivalent eukaryotic ribosomal proteins. By using the hybrid system with E. coli ribosomes, we have demonstrated that the proteins constituting the archaebacterial GTPase-associated centre are compatible with their eukaryotic counterparts, at least for translation elongation.
Online data
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research (number 14035222 to T.U. and number 16760633 to T.N.), a research grant from the National Project on Protein Structural and Functional Analyses (to T.U.), and Grant-in-Aid for 21st Century COE Program (to T.N.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This study was also supported in part by a Grant for Promotion of Niigata University Research Projects (to T.U.).
References
- 1.Pettersson I., Hardy S. J. S., Liljas A. The ribosomal protein L8 is a complex of L7/L12 and L10. FEBS Lett. 1976;64:135–138. doi: 10.1016/0014-5793(76)80267-0. [DOI] [PubMed] [Google Scholar]
- 2.Gudkov A. T., Tumanova L. G., Venyaminov S. Y., Khechinashvilli N. N. Stoichiometry and properties of the complex between ribosomal proteins L7 and L10 in solution. FEBS Lett. 1978;93:215–218. doi: 10.1016/0014-5793(78)81106-5. [DOI] [PubMed] [Google Scholar]
- 3.Griaznova O., Traut R. R. Deletion of C-terminal residues of Escherichia coli ribosomal protein L10 causes the loss of binding of one L7/L12 dimer: ribosomes with one L7/L12 dimer are active. Biochemistry. 2000;39:4075–4081. doi: 10.1021/bi992621e. [DOI] [PubMed] [Google Scholar]
- 4.Ileg L. L., Vldeler H., McKay A. R., Sobott F., Fucini P., Nierhaus K. H., Robinson C. V. Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8192–8197. doi: 10.1073/pnas.0502193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaconu M., Kothe U., Schlünzen F., Fischer N., Harms J. M., Tonevitsky A. G., Stark H., Rodnina M. V., Wahl M. C. Structural basis for the function of the ribosomal L7/L12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Beauclerk A. A. D., Cundliffe E., Dijk J. The binding site for ribosomal protein complex L8 within 23S ribosomal RNA of Escherichia coli. J. Biol. Chem. 1984;259:6559–6563. [PubMed] [Google Scholar]
- 7.Egebjerg J., Douthwaite S. R., Liljas A., Garrett R. A. Characterization of the binding sites of protein L11 and the L10.(L12)4 pentameric complex in the GTPase domain of 23S ribosomal RNA from Escherichia coli. J. Mol. Biol. 1990;213:275–288. doi: 10.1016/S0022-2836(05)80190-1. [DOI] [PubMed] [Google Scholar]
- 8.Rosendahl G., Douthwaite S. Ribosomal proteins L11 and L10.(L12)4 and the antibiotic thiostrepton interact with overlapping regions of the 23S rRNA backbone in the ribosomal GTPase centre. J. Mol. Biol. 1993;234:1013–1020. doi: 10.1006/jmbi.1993.1655. [DOI] [PubMed] [Google Scholar]
- 9.Möller W., Maassen J. A. Structure, Function, and Genetics of Ribosomes. In: Hardesty B., Kramer G., editors. NY: Springer-Verlag; 1986. pp. 309–325. [Google Scholar]
- 10.Agrawal R. K., Linde J., Sengupta J., Nierhaus K. H., Frank J. Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-independent translocation. J. Mol. Biol. 2001;311:777–787. doi: 10.1006/jmbi.2001.4907. [DOI] [PubMed] [Google Scholar]
- 11.Ban N., Nissen P., Hansen J., Moore P. B., Steiz A. T. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 12.Yusupov M. M., Yusupova G. Z., Baucom A., Lieberman K., Earnest T., Cate J. H., Noller H. F. Crystal structure of the ribosome at 5.5Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 13.Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A. High resolution structure of the large ribosomal subunit from a Mesophilic Eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 14.Newton C. H., Shimmin L. C., Yee J., Dennis P. P. A family of genes encode the multiple forms of the Saccharomyces cerevisiae ribosomal proteins equivalent to the Escherichia coli L12 protein and a single form of the L10-equivalent ribosomal protein. J. Bacteriol. 1990;172:579–588. doi: 10.1128/jb.172.2.579-588.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maassen J., Schop E. N., Brands J. H., van Hemert F. J., Lenstra J. A., Möller W. Molecular cloning and analysis of cDNA sequences for two ribosomal proteins from Artemia. The coordinate expression of genes for ribosomal proteins and elongation factor 1 during embryogenesis of Artemia. Eur. J. Biochem. 1985;149:609–616. doi: 10.1111/j.1432-1033.1985.tb08968.x. [DOI] [PubMed] [Google Scholar]
- 16.Rich B. E., Steitz J. A. Human acidic ribosomal phosphoproteins P0, P1, and P2: analysis of cDNA clones, in vitro synthesis, and assembly. Mol. Cell. Biol. 1987;7:4065–4074. doi: 10.1128/mcb.7.11.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wool I. G., Chan Y.-L., Glück A. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 1995;73:933–947. doi: 10.1139/o95-101. [DOI] [PubMed] [Google Scholar]
- 18.Tchórzewski M., Boldyreff B., Issinger O. G., Grankowski N. Analysis of the protein-protein interactions between the human acidic ribosomal P-proteins: evaluation by the two hybrid system. Int. J. Biochem. Cell Biol. 2000;32:737–746. doi: 10.1016/s1357-2725(00)00017-0. [DOI] [PubMed] [Google Scholar]
- 19.Garinos E., Remacha M., Ballesta J. P. G. Asymmetric interactions between the acidic P1 and P2 proteins in the Saccharomyces cerevisiae ribosomal stalk. J. Biol. Chem. 2001;276:32474–32479. doi: 10.1074/jbc.M103229200. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalo P., Lavergne J. P., Reboud J. P. Pivotal role of the P1 N-terminal domain in the assembly of the mammalian ribosomal stalk and in the proteosynthetic activity. J. Biol. Chem. 2001;276:19762–19769. doi: 10.1074/jbc.M101398200. [DOI] [PubMed] [Google Scholar]
- 21.Shimizu T., Nakagaki M., Nishi Y., Kobayashi Y., Hachimori A., Uchiumi T. Interaction among silkworm ribosomal proteins P1, P2 and P0 required for functional protein binding to the GTPase-associated domain of 28S rRNA. Nucleic Acids Res. 2002;30:2620–2627. doi: 10.1093/nar/gkf379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tchórzewski M., Krokowski D., Boguszewska A., Liljas A., Grankowski N. Structural characterization of yeast acidic ribosomal P proteins forming the P1A-P2B heterocomplex. Biochemistry. 2003;42:3399–3408. doi: 10.1021/bi0206006. [DOI] [PubMed] [Google Scholar]
- 23.Hanson C. L., Videler H., Santos C., Juan P. G., Ballesta J. P. C., Carol V., Robinson C. V. Mass spectrometry of ribosomes from Saccharomyces cerevisiae: implications for assembly of the stalk complex. J. Biol. Chem. 2004;279:42750–42757. doi: 10.1074/jbc.M405718200. [DOI] [PubMed] [Google Scholar]
- 24.Uchiumi T., Hori K., Nomura T., Hachimori A. Replacement of L7/L12.L10 protein complex in Escherichia coli ribosomes with the eukaryotic counterpart changes the specificity of elongation factor binding. J. Biol. Chem. 1999;274:27578–27582. doi: 10.1074/jbc.274.39.27578. [DOI] [PubMed] [Google Scholar]
- 25.Uchiumi T., Honma S., Nomura T., Dabbs E. R., Hachimori A. Translation elongation by a hybrid ribosome in which proteins at the GTPase center of the Escherichia coli ribosome are replaced with rat counterparts. J. Biol. Chem. 2002;277:41401–41409. doi: 10.1074/jbc.M107730200. [DOI] [PubMed] [Google Scholar]
- 26.Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U.S.A. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klink F., Schümann H., Thomsen A. Ribosome specificity of archaebacterial elongation factor 2. FEBS Lett. 1983;155:173–177. doi: 10.1016/0014-5793(83)80233-6. [DOI] [PubMed] [Google Scholar]
- 28.Beauclerk A. A. D., Hummel H., Holmes D. J., Böck A., Cundliffe E. Studies of the GTPase domain of archaebacterial ribosomes. Eur. J. Biochem. 1985;151:245–255. doi: 10.1111/j.1432-1033.1985.tb09095.x. [DOI] [PubMed] [Google Scholar]
- 29.Gutell R. R., Fox G. E. A compilation of large subunit RNA sequences presented in a structural format. Nucleic Acids Res. 1988;16:r175–r269. doi: 10.1093/nar/16.suppl.r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sköld S.-E. Chemical crosslinking of elongation factor G to the 23S RNA in 70S ribosomes from Escherichia coli. Nucleic Acids Res. 1983;11:4923–4932. doi: 10.1093/nar/11.14.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moazed D., Robertson J. M., Noller H. F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature (London) 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 32.Thompson J., Cundliffe E., Dahlberg A. E. Site-directed mutagenesis of Escherichia coli 23 S ribosomal RNA at position 1067 within the GTP hydrolysis centre. J. Mol. Biol. 1988;203:457–465. doi: 10.1016/0022-2836(88)90012-5. [DOI] [PubMed] [Google Scholar]
- 33.Uchiumi T., Kominami R. A functional site of the GTPase-associated center within 28S ribosomal RNA probed with an anti-RNA autoantibody. EMBO J. 1994;13:3389–3394. doi: 10.1002/j.1460-2075.1994.tb06641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimmin L. C., Ramirez C., Matheson A. T., Dennis P. P. Sequence alignment and evolutionary comparison of the L10 equivalent and L12 equivalent ribosomal proteins from Archaebacteria, Eubacteria, and Eucaryotes. J. Mol. Evol. 1989;29:448–462. doi: 10.1007/BF02602915. [DOI] [PubMed] [Google Scholar]
- 35.Casiano C., Traut R. R. Protein topography of Sulfolobus solfataricus ribosomes by cross-linking with 2-iminothiolane. J. Biol. Chem. 1991;266:21578–21583. [PubMed] [Google Scholar]
- 36.Köpke A. K., Leggatt P. A., Matheson A. T. Structure function relationships in the ribosomal stalk proteins of archaebacteria. J. Biol. Chem. 1992;267:1382–1390. [PubMed] [Google Scholar]
- 37.Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976;3:2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saiki R. K., Gelfand D. H., Stoffel S., Sharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 39.Hagiya A., Naganuma T., Maki Y., Ohta J., Tohkairin Y., Shimizu T., Nomura T., Hachimori A., Uchiumi T. A mode of assembly of P0, P1 and P2 proteins at the GTPase-associated center in animal ribosome: in vitro analyses with P0 truncation mutants. J. Biol. Chem. 2005;280:39193–39199. doi: 10.1074/jbc.M506050200. [DOI] [PubMed] [Google Scholar]
- 40.Nomura T., Mochizuki R., Dabbs E. R., Shimizu Y., Ueda T., Hachimori A., Uchiumi T. A point mutation in ribosomal protein L7/L12 reduces its ability to form a compact dimer structure and to assemble into the GTPase center. Biochemistry. 2003;42:4691–4698. doi: 10.1021/bi027087g. [DOI] [PubMed] [Google Scholar]
- 41.Uchiumi T., Sato N., Wada A., Hachimori A. Interaction of the sarcin/ricin domain of 23 S ribosomal RNA with proteins L3 and L6. J. Biol. Chem. 1999;274:681–686. doi: 10.1074/jbc.274.2.681. [DOI] [PubMed] [Google Scholar]
- 42.Uchiumi T., Wada A., Kominami Y. A base substitution within the GTPase-associated domain of mammalian 28S ribosomal RNA causes high thiostrepton. J. Biol. Chem. 1995;270:29889–29893. doi: 10.1074/jbc.270.50.29889. [DOI] [PubMed] [Google Scholar]
- 43.Dabbs E. R. Selection for Escherichia coli mutants with proteins missing from the ribosome. J. Bacteriol. 1979;140:734–737. doi: 10.1128/jb.140.2.734-737.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tokimatsu H., Strycharz W. A., Dahlberg A. E. Gel electrophoretic studies on ribosomal proteins L7/L12 and the Escherichia coli 50S subunit. J. Mol. Biol. 1981;152:397–412. doi: 10.1016/0022-2836(81)90250-3. [DOI] [PubMed] [Google Scholar]
- 45.Iwasaki K., Kaziro Y. Polypeptide chain elongation factors from pig liver. Methods Enzymol. 1979;60:657–676. doi: 10.1016/s0076-6879(79)60062-9. [DOI] [PubMed] [Google Scholar]
- 46.Arai K., Kawakita M., Kaziro Y. Studies on polypeptide elongation factors from Escherichia coli. II. J. Biol. Chem. 1972;247:7029–7037. [PubMed] [Google Scholar]
- 47.Caldwell P., Luk D. C., Weissbach H., Brot N. Oxidation of the methionine residues of Escherichia coli ribosomal protein L12 decreases the protein's biological activity. Proc. Natl. Acad. Sci. U.S.A. 1978;75:5349–5352. doi: 10.1073/pnas.75.11.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gudkov A. T., Behlke J. The N-Terminal sequence protein of L7/L12 is responsible for its dimerization. Eur. J. Biochem. 1978;90:309–312. doi: 10.1111/j.1432-1033.1978.tb12605.x. [DOI] [PubMed] [Google Scholar]
- 49.Datta P. P., Sharma M. R., Qi L., Frank J., Agrawal R. K. Interaction of the G' domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol. Cell. 2005;20:723–731. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.