Abstract
HACN (homoaconitase) is a member of a family of [4Fe-4S] cluster-dependent enzymes that catalyse hydration/dehydration reactions. The best characterized example of this family is the ubiquitous ACN (aconitase), which catalyses the dehydration of citrate to cis-aconitate, and the subsequent hydration of cis-aconitate to isocitrate. HACN is an enzyme from the α-aminoadipate pathway of lysine biosynthesis, and has been identified in higher fungi and several archaea and one thermophilic species of bacteria, Thermus thermophilus. HACN catalyses the hydration of cis-homoaconitate to (2R,3S)-homoisocitrate, but the HACN-catalysed dehydration of (R)-homocitrate to cis-homoaconitate has not been observed in vitro. We have synthesized the substrates and putative substrates for this enzyme, and in the present study report the first steady-state kinetic data for recombinant HACN from T. thermophilus using a (2R,3S)-homoisocitrate dehydrogenase-coupled assay. We have also examined the products of the reaction using HPLC. We do not observe HACN-catalysed ‘homocitrate dehydratase’ activity; however, we have observed that ACN can catalyse the dehydration of (R)-homocitrate to cis-homoaconitate, but HACN is required for subsequent conversion of cis-homoaconitate into homoisocitrate. This suggests that the in vivo process for conversion of homocitrate into homoisocitrate requires two enzymes, in simile with the propionate utilization pathway from Escherichia coli. Surprisingly, HACN does not show any activity when cis-aconitate is substituted for the substrate, even though other enzymes from the α-aminoadipate pathway can accept analogous tricarboxylic acid-cycle substrates. The enzyme shows no apparent feedback inhibition by L-lysine.
Keywords: α-aminoadipate pathway, homoaconitase (HACN), homocitrate, homoisocitrate, cis-homoaconitate, Thermus thermophilus
Abbreviations: ACN, aconitase; DAP, diaminopimelate; HACN, homoaconitase; HCS, (R)-homocitrate synthase; HICDH, (2R,3S)-homoisocitrate dehydrogenase; ICDH, isocitrate dehydrogenase; IPMI, isopropylmalate isomerase; IPMS, (S)-2-isopropylmalate synthase; TCA, tricarboxylic acid
INTRODUCTION
HACN (homoaconitase), or homoaconitate hydratase [1], is an enzyme found in the α-aminoadipate pathway of lysine biosynthesis. Among the twenty common amino acids, only lysine has evolved two completely unrelated biosynthetic pathways: the α-aminoadipate pathway found mainly in lower eukaryotes such as fungi and euglenoids [2,3], and the more widespread DAP (diaminopimelate) pathway, found in plants and most bacteria [4]. The α-aminoadipate pathway has also been identified in archaea, first in Thermoproteus neutrophilus [5], and subsequently in the genomes of several other species [6–10]. A variant of the α-aminoadipate pathway has been found in just one eubacterium, the hyperthermophilic Thermus thermophilus [11–14]. The genes encoding enzymes involved in lysine biosynthesis revealed that the T. thermophilus lysine biosynthetic pathway from homocitrate to α-aminoadipate is similar to that of higher fungi including yeasts, and also similar to that of leucine biosynthesis and the related portion of the TCA (tricarboxylic acid) cycle. On the other hand, the second half of the pathway, the conversion of α-aminoadipic acid into lysine, is completely different from that in lower eukaryotes, but apparently similar to the process of glutamate conversion into ornithine in arginine biosynthesis [14–17]. Thus enzymes involved in T. thermophilus lysine biosynthesis are evolutionarily related to the enzymes involved in biosynthesis of leucine and arginine along with the corresponding part of the TCA cycle.
Because these pathways are not found in animals, lysine biosynthesis has been proposed as a target for anti-microbial therapy [4,18,19]. Previous efforts have centered almost entirely on inhibition of the DAP pathway in order to develop antibiotics; only one report of inhibitors developed to target the α-aminoadipate pathway has appeared [20].
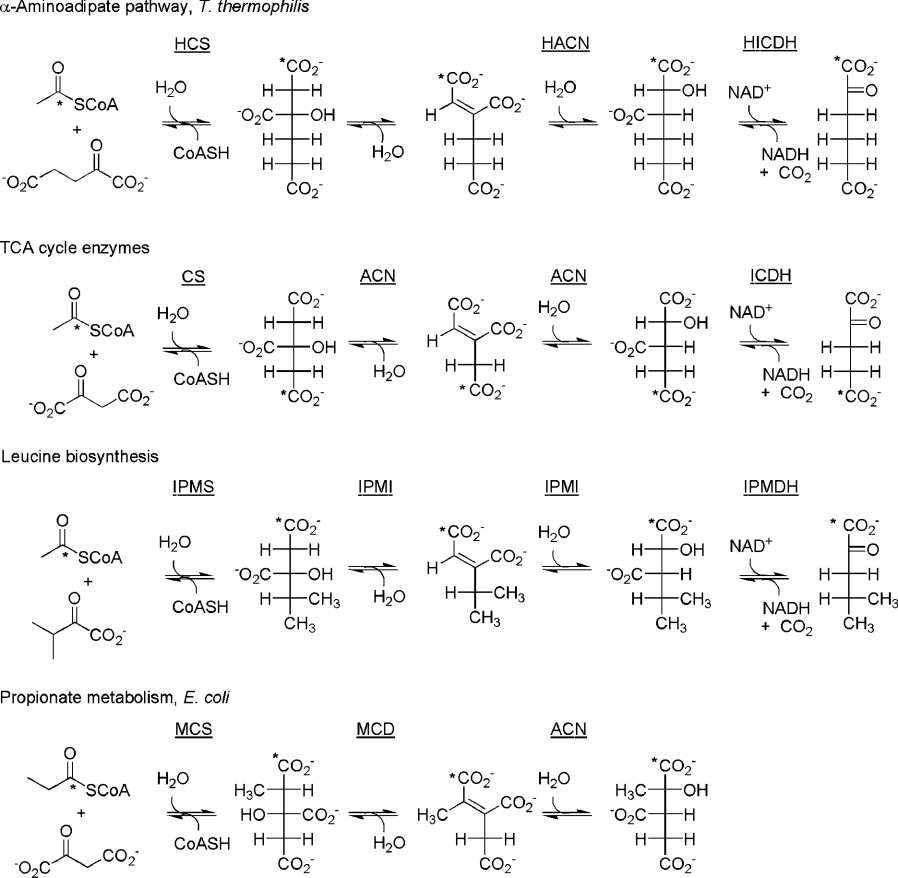
The α-aminoadipate pathway begins with the synthesis of (R)-homocitrate from acetyl-CoA and 2-oxoglutarate by HCS [(R)-homocitrate synthase] [21,22] (Figure 1). (R)-Homocitrate is then dehydrated to cis-homoaconitate, which is subsequently re-hydrated to (2R,3S)-homoisocitrate. HICDH [(2R,3S)-homoisocitrate dehydrogenase] then converts homoisocitrate into 2-oxoadipate and CO2 with the concomitant reduction of NAD+. The reactions are shown in Figure 1 alongside similar metabolic pathways, using Fischer projections and a (hypothetically) labelled carbon atom to make the stereochemistry of the reactions clear. The conversion of cis-homoaconitate into homoisocitrate has been shown to be catalysed by HACN from Saccharomyces cerevisiae [23,24] and Aspergillus nidulans [1] (also called Lys4 and LysF respectively), but the identity of the enzyme responsible for the conversion of homocitrate into cis-homoaconitate is not strictly known. This is somewhat surprising given that the activity was identified almost 40 years ago. The results of accumulation studies using S. cerevisiae lysine auxotrophs in the 1960s indicated that there are two enzymatic steps for the conversion of homocitrate into homoisocitrate: a lys7 mutant was observed to accumulate homocitrate, and a lys4 mutant accumulated homocitrate and cis-homoaconitate. However, Lys7 has subsequently been shown to be a copper chaperone for superoxide dismutase [25]. Predictably, disruption of such a gene induces oxidative stress, disrupting the activity of redox-sensitive enzymes, including Fe-S enzymes like HACN [26]. Thus accumulation of homocitric acid in the lys7 mutant does not rule out Lys4 as a ‘homocitrate dehydratase’. It should be noted that the reported experiments investigating the function of HACN invariably use crude cell extracts or partial purification (e.g. a single ammonium persulphate precipitation), thus experiments using highly purified enzyme are needed.
Figure 1. Relevant reactions from the α-aminoadipate pathway, TCA cycle, leucine biosynthesis pathway, and propionate metabolism pathway.
CS, citrate synthase; IPMS, (S)-2-isopropylmalate synthase; IPMDH, (2R,3S)-3-isopropylmalate dehydrogenase; MCS, (2S,3S)-2-methylcitrate synthase; MCD, 2-methylcitrate dehydratase.
HACN is clearly a homologue of ACN (aconitase) and IPMI (isopropylmalate isomerase) [17,27], and the apparent resemblance of the substrates and the reactions catalysed, including the stereochemistry, is clear from Figure 1. ACN-like enzymes are dependent on a [4Fe-4S]2+ cluster for activity [28]. Multiple sequence alignments of the enzymes demonstrate that all residues known to be in the active site of porcine heart ACN are present in IPMI and HACN [27,29], with the exception of Arg580. This residue forms a salt bridge with the carboxylate of the pro-S acetate moiety of citrate (labelled in Figure 1) in porcine heart ACN, the part of the substrate that varies from pathway to pathway. Modern understanding of enzyme evolution suggests that ACN, IPMI and HACN should share a common fold and active site architecture [30]. The HACN from T. thermophilus consists of two distinct polypeptides, a large and small subunit, designated LysT and LysU [13,14], and this pair of proteins contains all of the predicted catalytic residues. The ACN structure is made up of three small domains and one large C-terminal domain, with the active site located in a cleft at the interface of the large domain with the other three. LysT corresponds to the three small domains of ACN, and LysU to the C-terminal domain. The presence of all the apparent catalytic apparatus required for the isomerization of (R)-homocitrate to (2R,3S)-homoisocitrate has led several investigators to assume that the first step, formation of cis-homoaconitate from homocitrate, is also catalysed by HACN [2,29,31], and has simply gone undetected in vitro. The inherent difficulty in working with [4Fe-4S]2+-dependent proteins, which are rapidly inactivated in the presence of oxygen, is one rationalization for this apparent gap in information. But there is a precedent for two separate enzymes being used for a similar isomerization. For example, bacterial propionate metabolism proceeds through a similar pathway, as depicted in Figure 1, but in E. coli the dehydration of 2-methylcitrate is catalysed by an MCD (2-methylcitrate dehydratase) that is unrelated to the ACN-like enzymes mentioned above, and the hydration of 2-methylaconitate is catalysed by ACN [32]. Rather than a contradiction, this is consistent when one considers that the first transformation, dehydration of 2-methylcitrate, proceeds with opposite stereochemistry. Apparently this difference in stereochemistry was sufficient to require that a different template be recruited as a catalyst. This would only be relevant to HACN if the stereochemistry of the substrate, (R)-homocitrate, were incorrect. The discovery of (S)-homocitrate in the coenzyme B pathway of a hyperthermophilic organism [33] raises the possibility that this enantiomer might be involved in the α-aminoadipate pathway, despite characterization of the product of homocitrate synthase as the (R)-enantiomer.
In the present work, we report the first in vitro study of purified, recombinant HACN using the gene products of lysT and lysU from T. thermophilus. This required the chemical synthesis of all three putative substrates, as well as (S)-homocitrate. We have studied the kinetics of the reaction using a HICDH-coupled assay, and examined the reaction products by HPLC.
EXPERIMENTAL
Materials
Unless otherwise specified, chemical reagents, including porcine heart ACN and ICDH (isocitrate dehydrogenase) were purchased from Sigma–Aldrich Canada Ltd. (Oakville, ON, Canada) or VWR-Canlab (Mississauga, ON, Canada), and were designated molecular biology grade, or the highest grade available. The competent cell set was purchased from Novagen. A QIAprep spin MiniPrep kit was purchased from Qiagen Inc. The Bradford assay kit was obtained from Bio-Rad. Nanosep (10 K) centrifugal ultrafiltration devices were purchased from Pall Life Science. cis-Homoaconitate [34] and (2R,3S)-homoisocitrate [35], and (R)- and (S)-homocitric lactones [36] were synthesized following published procedures, and purified by reverse-phase HPLC before use. HICDH was expressed and purified as reported previously [37].
General methods
Standard molecular biology techniques and media were used unless otherwise stated [38]. Optical rotation values were determined using a Rudolph Instruments Digipol 781 Automatic Polarimeter (1 dm, 1 ml cell) at 589 nm at ambient temperature and all concentrations are given in g/100 ml. NMR spectra were recorded on a Bruker 500 MHz spectrometer. 1H NMR spectra were recorded in CDCl3 or 2H2O and are referenced to the appropriate solvent signal. Chemical shifts are reported in p.p.m. (parts per million) units downfield from tetramethylsilane. Melting points were measured on a Gallencamp melting point apparatus and are uncorrected. HPLC was performed on an Agilent 1100 system with a diode-array detector using a reverse-phase Zorbax SB-C18 column.
Synthesis of (R)- and (S)-homocitrate
(R)-Homocitrate was obtained from the (S)-α-methylbenzylammonium salt of (R)-homocitric lactone [36] (17 mg, 0.055 mmol) by dissolving in 1 ml of H2O. A 3% NaOH solution (0.22 ml, 0.165 mmol) was added and the solution stirred for 10 min. The mixture was extracted with dichloromethane (5 ml) three times. The aqueous solution was evaporated to dryness resulting in the desired product. 1H NMR data (500 MHz) δH (2H2O): 2.61 (1 H, d, J 15.5 Hz), 2.33 (1 H, d, J 15.5 Hz), 2.17 (1 H, ddd, J 4.5, 13, 14 Hz), 1.90 (1 H, m), 1.78 (1 H, ddd, J 4.5, 13, 13.0 Hz), 1.71 (1 H, ddd, J 4.5, 13, 13 Hz). Optical rotation data: [α]29589=−14.5° (c 1.00 in 2H2O). (S)-Homocitrate was isolated by the same procedure as described above [starting with the (S)-α-methylbenzylammonium salt of (R)-homocitric lactone] [36]. The NMR spectrum was indistinguishable from that of the (R)-enantiomer. [α]29589=+14.2 (c 1.05 in 2H2O). The optical activities of the isolated homocitrate trisodium salts matched the value previously reported [35].
Plasmid construction
The plasmid pET-LysTU for the expression of HACN genes was constructed as follows. PCR was performed with KOD polymerase (TOYOBO; Osaka, Japan), using pLYS200 [12] as the template. The PCR conditions used were 98 °C for 15 s, 68 °C for 2 s, and 74 °C for 45 s, with a total of 25 cycles. Two oligonucleotides, 5′-CCGGAATTCCATATGGGACAGACGCTAGCG-3′ and 5′-GCTCTAGAGGATCCCCAGTCGATTAGGCATGGACCTCCTCCTC-3′ (restriction sites underlined), were used to amplify the LysT gene encoding the large subunit of HACN. Another PCR was carried out with 5′-GCTCTAGAGTCGACAAGGAAGTGGGTCCATGCCTAGGGTCTGGAAG-3′ and 5′-CCGCTC-GAGCTCCCCCGGGAAGCG-3′, which were designed to amplify the LysU gene encoding the small subunit of HACN. The former and latter amplified DNA fragments were digested with EcoRI plus XbaI, and XbaI plus XhoI respectively, and the resulting fragments were ligated in tandem with pET26b(+) (Novagen, Madison, WI, U.S.A.) previously digested with EcoRI plus XhoI to yield pET-LysTU. In this construction, 8 amino acid residues, LGHHHHHH, were attached to the C-terminus of LysU.
Preparation of HACN
A 16 ml culture of E. coli BL21-CodonPlus (DE3)-RIL cells (Strategene, La Jolla, CA, U.S.A.) harbouring pET-LysTU in 2×YT [1.6% (w/v) tryptone/1% (w/v) yeast extract/0.5% (w/v) NaCl] medium containing kanamycin (50 μg·ml−1) and chloramphenicol (30 μg·ml−1) was grown for 8 h at 37 °C, then transferred to 1.6 litres of the same medium and the culture was continued for 2 h before induction with isopropyl β-D-thiogalactoside (0.1 mM). The culture was continued for an additional 12 h at 30 °C, harvested by centrifugation, washed with 20 mM Tris/HCl (pH 8.0), and lysed by sonication in 30 ml of 20 mM Tris/HCl (pH 8.0). After centrifugation at 20000 g for 15 min, the supernatant was heated at 70 °C for 30 min and centrifuged. The resulting supernatant was applied onto a column (8 mm×10 cm) packed with 3 ml of Profinity™ IMAC Resin charged with Ni2+ (Bio-Rad Japan, Tokyo) pre-equilibrated with 20 mM Tris/HCl (pH 8.0). The column was washed with 20 mM Tris/HCl (pH 8.0) supplemented with 500 mM NaCl and 20 mM imidazole to remove proteins non-specifically bound to the resin. HACN was eluted with 20 mM Tris/HCl (pH 8.0) supplemented with 500 mM NaCl and 500 mM imidazole. Ammonium sulphate was added to the eluate at 80% saturation, and the purified protein was stored as the ammonium sulphate precipitate.
Reconstitution and activation of HACN
The ammonium sulphate precipitate containing the protein was dialysed in an anaerobic chamber (glovebox). All buffers were autoclaved, then deoxygenated by repeated evacuation followed by saturation with argon. The protein was dissolved in 10 mM Tris/HCl buffer (pH 7.5) containing 0.3 M NaCl (2 ml). The solution was dialysed against 1 litre of the same buffer using the Slyde-A-Lyzer 7000 MWCO cassette system (Pierce) according to the manufacturer's instructions. After 3 h, the buffer was replaced, and the dialysis continued overnight. The dialysed sample was filtered through a 0.2 μm syringe filter, and stored in the glovebox until further use. Reactivation of the protein followed the method reported for thermostable ACN [39].
Kinetics
Initial velocity data were collected on a Beckman DU640 spectrophotometer at 60 °C. All rates were measured relative to a blank which contained everything except HACN. The reaction mixture in the coupled assay contained: 50 mM Hepes/NaOH, (pH 8.0), 200 mM KCl, 5 mM MgCl2, 1 mM NAD+, 29 μg/ml HICDH, 2.5 μg/ml activated HACN and various concentrations of cis-homoaconitate (used in the range of 5–500 μM) in a 1 ml final total volume. All the stock solutions were degassed and saturated with argon. All components were mixed in a 1 ml cuvette under nitrogen gas in the glovebox and then the cuvette was fitted with a rubber septum and transferred outside. The mixture was pre-incubated for 5 min at 60 °C before adding the enzyme, HACN, which was transferred with an air-tight microsyringe. The reaction was monitored by the absorbance increase at 340 nm (NADH molar absorption coefficient ϵ=6220 M−1·cm−1). All data points were the average of at least two experiments and were fitted to the Michaelis–Menten equation by the non-linear least-squares method using Leonora software [40]. Assays of the homoisocitrate conversion into cis-homoaconitate were performed in 50 mM Tris/HCl buffer (pH 8.0, measured at 60 °C), 200 mM KCl, and 20 μg/ml activated HACN, and samples were prepared as described above. The appearance of cis-homoaconitate was measured at 240 nm. The molar absorption coefficient (ϵ=2720 M−1·cm−1) was measured my generating a standard curve from concentrations of 0.05, 0.2, 0.25, 0.5 and 1 M substrate in the same buffer.
The inhibition by lysine was investigated using the same conditions as above, in the presence of 1–20 mM L-lysine. Assays substituting cis-aconitate for cis-homoaconitate used otherwise identical conditions.
HPLC analysis of the reaction products
Separation conditions were based on those reported by Cawthray [41] with slight modifications. Two different satisfactory mobile phase conditions were used over the course of the study. The first was 0.1% trifluoroacetic acid in distilled H2O (pH 2) and 2% MeOH at 25 °C with a flow rate of 1 ml·min−1, resulting in the following retention times: homoisocitric acid, 6.0 min; homocitric acid, 9.1 min; cis-homoaconitic acid, 15.5 min. The second was 98% 25 mM KH2PO4 in distilled H2O (pH 2.5) and 2% MeOH at a flow rate of 0.75 ml·min−1, resulting in the following retention times: homoisocitric acid, 5.5 min; homocitric acid, 8.1 min; cis-homoaconitic acid, 11.6 min. Detector output at 210 nm was used for the quantification of organic acids. Enzymatic reaction mixtures contained 100 μg of HACN or 175 μg of ACN or both, 10 mM substrate (homocitrate, cis-homoaconitate, or homoisocitrate) and 50 mM Hepes, Tris, or phosphate buffer, pH 8.0. Reactions were prepared as described for kinetic studies, and incubated at 37 °C for 7 days in the glovebox, or at 60 °C for 15 min. A blank reaction containing everything except HACN and was incubated in the same conditions as the corresponding enzymatic reaction mixture. Before injection, samples were cooled on ice, acidified to pH 2–3 and the enzyme was removed by centrifugal ultrafiltration. The injection volume was 100 μl. The relative molar absorption coefficients at 210 nm of (R)-homocitrate:cis-homoaconitate:(2R,3S)-homoisocitrate were determined to be 1.0:45.4:1.07, which was determined by preparing and injecting molar ratios of 1:1, 2:1, 5:1, 10:1 and 50:1 of each pair of compounds. This ratio is similar to the reported ratio of cis-aconitate to citrate [41].
RESULTS AND DISCUSSION
Preparation and activity of HACN and HICDH
The genes encoding the large subunit of HACN, LysT, and the small subunit of HACN, LysU, were cloned and expressed in E. coli and the protein purified using a C-terminal histidine tag introduced into LysU. The purification procedure resulted in both subunits being present with no significant contamination by other proteins, as indicated by SDS/PAGE analysis (see Supplementary data, Figure S1 at http://www.BiochemJ.org/bj396/bj3960479add.htm).
Activation of the enzyme in the absence of oxygen is required to ensure that the active site contains the [4Fe-4S]2+ cluster required for activity in ACN and ACN-related enzymes. Two different methods of activation, based on literature methods for ACN activation [39,42] were tested, and both resulted in an active protein (as described below). The enzyme preparation did not require reconstitution of the Fe-S cluster i.e. the isolated protein contained the cluster in its [3Fe-4S]+ form. Protein stripped of its Fe-S cluster could be reconstituted by the method of Kennedy and Beinert [43] and fully activated. Enzyme that was not treated with any activation procedure showed no apparent activity. The activity of the coupling enzyme, HICDH, was routinely assayed using either (2R,3S)-homoisocitrate or isocitrate, since the enzyme accepts either compound as a substrate [37,44].
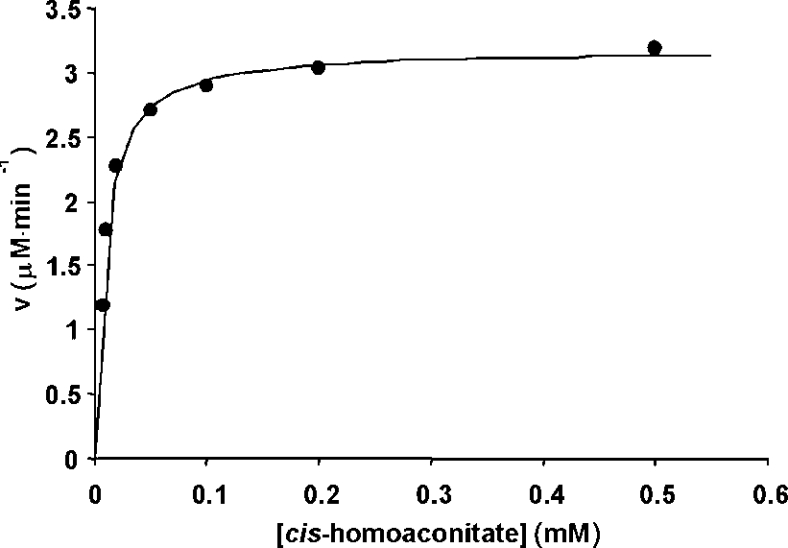
A plot of the dependence of the reaction rate on cis-homoaconitate concentration at pH 8.0 and 60 °C in Hepes buffer is shown in Figure 2. We observed that other pH values resulted in lower activity (results not shown). An alternative activation procedure reported for ACN [42] resulted in lower activity. Analysis of the data results in kinetic constants of kcat=1.3±0.2 s−1 and Km=8.2±0.2 μM. In the reverse direction at this pH, the rate of appearance of cis-homoaconitate could be observed directly at 240 nm. The dehydration reaction also showed normal hyperbolic dependence on the substrate (Supplementary data, Figure S2 at http://ww.BiochemJ.org/bj/396/bj3960479add.htm), and displayed kcat=4.6±0.4 s−1 and Km=36±1 μM. These are the first kinetic constants reported for HACN from any source.
Figure 2. Dependence of the rate of the HACN-catalysed hydration of cis-homoaconitate to homoisocitrate on substrate concentration using the HICDH-coupled assay.
Conditions: 50 mM Hepes/NaOH (pH 8.0), 200 mM KCl, 5 mM MgCl2, 1 mM NAD+, 29 μg/ml HICDH and 2.5 μg/ml activated HACN at 60 °C. The line represents the fit to the Michaelis–Menten equation. V, initial velocity.
The coupled assay indicated no activity in the presence of (R)-homocitrate or (S)-homocitrate as substrates. Both enantiomers were tested as inhibitors of the coupling enzyme HICDH, but this enzyme was fully active on (2R,3S)-homoisocitrate in the presence of either enantiomer of homocitrate.
HPLC analysis of HACN-catalysed reactions
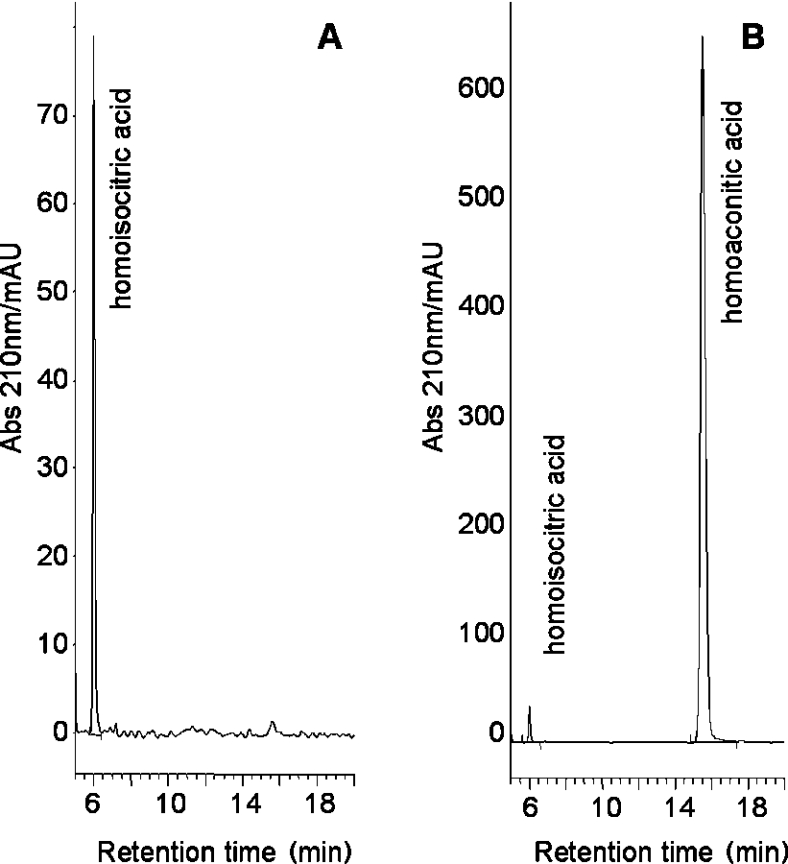
HPLC analysis was essential to determine the equilibrium position of the reaction, and also allowed us to look for any products resulting from the reaction, and to test other substrates as candidates. Thus reaction mixtures containing one of cis-homoaconitate, (2R,3S)-homoisocitrate, (R)-homocitrate and (S)-homocitrate were prepared and examined by HPLC. In each case a control was performed which replicated the experiment in every way, including the activation procedure, but contained no enzyme. The identity of the peaks was confirmed by spiking of the samples with authentic product, resulting in co-elution with the reaction product. In the case of each enantiomer of homocitrate, no change was observed in the chromatogram when the experiment was conducted at room temperature and at 60 °C, which is in agreement with results using the coupled assay. As shown in Figure 3, cis-homoaconitate formation could be observed when (2R,3S)-homoisocitrate was the starting material and (as already indicated by the coupled assay) homoisocitrate was formed from cis-homoaconitate. No homocitrate was formed from cis-homoaconitate. The equilibrium position of the reaction was estimated by the amount of each product present after 7 days of reaction at 37 °C in an anaerobic chamber, starting with either substrate and using various starting concentrations (see Supplementary data at http://www.BiochemJ.org/bj/396/bj3960479add.htm). Based on the relative molar absorption coefficients of the substrates, the integration of the peaks indicate Keq=[(2R,3S)-homoisocitrate]/[cis-homoaconitate]=1.2. This value is in agreement with the ratio of the apparent second order rate constants in the forward and reverse directions, (kcat/Km)fwd/(kcat/Km)rev=1.24, in accordance with the Haldane relationship.
Figure 3. Reverse-phase HPLC chromatograms of the HACN-catalysed dehydration of (2R,3S)-homoisocitrate to cis-homoaconitate, after incubation for 7 days.
Mobile phase 0.1% TFA (trifluoroacetic acid) in distilled H2O (pH 2) and 2% MeOH at 25 °C; flow rate 1 ml·min−1. (A) (2R,3S)-homoisocitrate (10 mM) in a ‘blank’ reaction with no enzyme present (pH 8.0), 50 mM Hepes (pH 8.0) at 37 °C for 7 days in an anaerobic chamber. (B) As above, but in the presence of 100 μg of HACN. The larger scale reflects the larger molar absorption coefficient of cis-homoaconitate. mAU, arbitrary units (×103).
Testing lysine as a feedback inhibitor
Lysine, the end-product of the α-aminoadipate pathway, is known to inhibit the starting point of this pathway, HCS [(R)-homocitrate synthase] [21,22]. The presence of up to 20 mM lysine had no effect on the rate of the reaction as measured by the coupled assay, indicating that lysine inhibits neither HACN nor HICDH.
Testing TCA cycle compounds as substrates for HACN
HICDH is known to recognise isocitrate as a substrate [37,44], and HCS is known to accept oxalacetate as a substrate (although with a Km that is 10-fold higher), thus functioning as a citrate synthase [21,22]. This raises an obvious question: can HACN recognise substrates from the TCA cycle in a similar manner to HCS and HICDH from the α-aminoadipate pathway? Because HICDH recognises isocitrate, the same coupled assay could be used to assay the HACN-catalysed hydration of cis-aconitate. However, no activity was observed in the presence of aconitate, nor did the presence of aconitate inhibit the HACN-catalysed hydration of cis-homoaconitate. No HACN-catalysed activity was observed using citrate as a substrate.
Homoaconitase activity of ACN
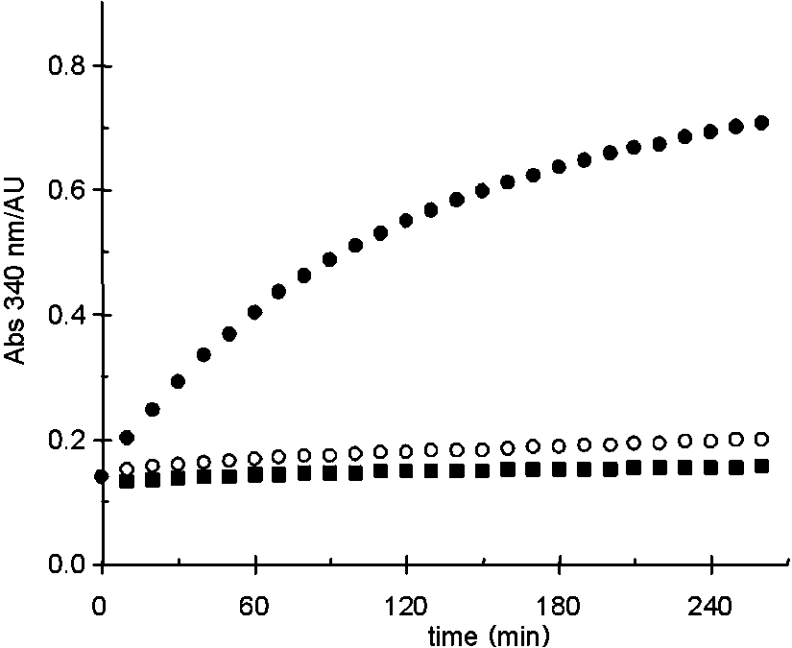
With no apparent ‘homocitrate dehydratase’ activity observed for HACN, the identity of the catalyst for this reaction is unknown. Because the reaction is ACN-like, an ACN from T. thermophilus seems the most likely candidate. Unfortunately, such an enzyme has yet to be identified. To test this hypothesis, commercially available porcine heart ACN was used. The purchased enzyme was activated as described for HACN, and its full activity demonstrated using citrate and aconitate as substrates, and both NADP+-dependent ICDH and HICDH could be used in a coupled assay of isocitrate production. When (R)-homocitrate, (S)-homocitrate, or cis-homoaconitate were used as substrates, the coupled assay indicated that no catalysis was taking place. HPLC analysis of the reaction mixture of (2R,3S)-homoisocitrate and ACN also showed that no reaction was occurring. However, when cis-homoaconitate was used as a substrate, HPLC analysis indicated the ACN-catalysed production of homocitrate. Similarly, cis-homoaconitate was formed from (R)-homocitrate in the presence of ACN (Figure 4). (S)-Homocitrate was not a substrate. Finally, a coupled assay containing (R)-homocitrate, ACN, HACN and HICDH resulted in the reduction of NAD+, but the absence of ACN or HACN resulted in no activity. The assay was performed at 40 °C, a compromise in temperature in order to accommodate the non-thermophilic ACN. Below this temperature the activity of HICDH drops sharply. This result is shown in Figure 5. HPLC analysis of 7 day reactions in the presence of ACN and HACN showed a mixture of all three substrates, with the equilibrium strongly favouring (R)-homocitrate (see Supplementary data at http://www.BiochemJ.org/bj/396/bj3960479.add.htm). This is in agreement with the findings of Bhattacharjee et al. [3,24], using S. cerevisiae lysine auxotrophs.
Figure 4. Reverse phase (C18) HPLC chromatograms of the porcine heart ACN-catalysed dehydration of homocitrate to cis-homoaconitate.
Mobile phase 98% 25 mM KH2PO4 in distilled H2O (pH 2.5) and 2% MeOH, flow rate 0.75 ml·min−1. (A) (R)-homocitrate (10 mM) in a ‘blank’ reaction with no enzyme present (pH 8.0), 50 mM Hepes (pH 8.0) at 37 °C for 7 days in an anaerobic chamber. (B) As above, but in the presence of 175 μg of activated ACN. mAU, arbitrary units (×103).
Figure 5. HICDH-coupled assay of the conversion of (R)-homocitrate into (2R,3S)-homoisocitrate into the presence of 2.5 μg/ml HACN (■), 70 μg/ml ACN (○), and ACN+HACN (●).
Conditions: 50 mM Hepes/NaOH (pH 8.0), 200 mM KCl, 5 mM MgCl2, 1 mM NAD+ and 29 μg/ml HICDH, at 40 °C. AU, arbitrary units.
Conclusions
Our hypothesis upon initiating this work was that HACN could catalyse both steps of the isomerization of (R)-homocitrate to (2R,3S)-homoisocitrate, based on the homology of HACN with ACN and the fact that the reactions in question were similar to those catalysed by ACN and of identical stereochemistry. We cannot prove that HACN does not catalyse both steps of the reaction. We can only demonstrate that our methods can detect such a reaction, but do not. The HPLC and coupled assay results clearly show that we can activate HACN and observe the reversible hydration of cis-homoaconitate to (2R,3S)-homoisocitrate; furthermore, we can activate ACN and observe the ACN-catalysed isomerization of citrate to isocitrate, and the ACN-catalysed reversible dehydration of (R)-homocitrate to cis-homoaconitate. (Consistent with the stereochemical analysis, ACN does not recognise (S)-homocitrate as a substrate.) The results described in the presemt study, especially those shown in Figure 5, demonstrate that in T. thermophilus, HACN alone is inadequate to convert homocitrate into homoisocitrate. Another way in which HACN is inconsistent with the neighbouring α-aminoadipate pathway enzymes is in its inability to recognise a substrate from the TCA cycle, cis-aconitate. One could conjecture that ACN is present in sufficient amounts in the organism to allow HACN to evolve away from ACN-like activity, and away from ‘homocitrate dehydratase’ activity. High-resolution structural analysis of HACN is desirable in order to elucidate the differences in substrate recognition within the ACN family.
Acknowledgments
The authors thank Chantal Levesque for technical contributions. This work was supported by an NSERC (National Science and Engineering Research Council of Canada) Discovery Grant and a New Opportunities Award from the Canadian Foundation for Innovation (to D.R.J.P.), and by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, from Noda Institute for Scientific Research and from the Nagase Science and Technology Foundation (to M.N.).
References
- 1.Weidner G., Steffan B., Brakhage A. A. The Aspergillus nidulans lysF gene encodes homoaconitase, an enzyme involved in the fungus-specific lysine biosynthesis pathway. Mol. Gen. Genet. 1997;255:237–247. doi: 10.1007/s004380050494. [DOI] [PubMed] [Google Scholar]
- 2.Zabriskie T. M., Jackson M. D. Lysine biosynthesis and metabolism in fungi. Natural Products Rep. 2000;17:85–97. doi: 10.1039/a801345d. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharjee J. K. α-Aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes. Crit. Rev. Microbiol. 1985;12:131–151. doi: 10.3109/10408418509104427. [DOI] [PubMed] [Google Scholar]
- 4.Born T. L., Blanchard J. S. Structure/function studies on enzymes in the diaminopimelate pathway of bacterial cell wall biosynthesis. Curr. Opin. Chem. Biol. 1999;3:607–613. doi: 10.1016/s1367-5931(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 5.Schafer S., Paalme T., Vilu R., Fuchs G. 13C-NMR study of acetate assimilation in Thermoproteus neutrophilus. Eur. J. Biochem. 1989;186:695–700. doi: 10.1111/j.1432-1033.1989.tb15262.x. [DOI] [PubMed] [Google Scholar]
- 6.Kawarabayasi Y., Hino Y., Horikawa H., Jin-no K., Takahashi M., Sekine M., Baba S., Ankai A., Kosugi H., Hosoyama A., et al. (2001) Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Research. 8:123–140. doi: 10.1093/dnares/8.4.123. [DOI] [PubMed] [Google Scholar]
- 7.She Q., Singh R. K., Confalonieri F., Zivanovic Y., Allard G., Awayez M. J., Chan-Weiher C. C., Clausen I. G., Curtis B. A., De Moors A., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawarabayasi Y., Hino Y., Horikawa H., Yamazaki S., Haikawa Y., Jin-no K., Takahashi M., Sekine M., Baba S., Ankai A., et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999;6:145–152. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- 9.Cohen G. N., Barbe V., Flament D., Galperin M., Heiling R., Lecompte O., Poch O., Prieur D., Querellou J., Ripp R., et al. An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 2003;47:1496–1512. doi: 10.1046/j.1365-2958.2003.03381.x. [DOI] [PubMed] [Google Scholar]
- 10.Kawarabayasi Y., Sawada M., Horikawa H., Haikawa Y., Hino Y., Yamamoto S., Sekine M., Baba S., Kosugi H., Hosoyama A., et al. Complete sequence and gene organization of the genome of a hyperthermophilic archaeabacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 11.Kosuge T., Hoshino T. Lysine is synthesized through the α-aminoadipate pathway in Thermus thermophilus. FEMS Microbiol Lett. 1998;169:361–367. doi: 10.1111/j.1574-6968.1998.tb13341.x. [DOI] [PubMed] [Google Scholar]
- 12.Kobashi N., Nishiyama M., Tanokura M. Aspartate kinase-independent lysine synthesis in an extremely thermophilic bacterium, Thermus thermophilus: lysine is synthesized via α-aminoadipic acid not via diaminopimelic acid. J. Bacteriol. 1999;181:1713–1718. doi: 10.1128/jb.181.6.1713-1718.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishida H., Nishiyama M., Kobashi N., Kosuge T., Hoshino T., Yamane H. A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: a key to the evolution of amino acid biosynthesis. Genome Res. 1999;9:1175–1183. doi: 10.1101/gr.9.12.1175. [DOI] [PubMed] [Google Scholar]
- 14.Miyazaki J., Kobashi N., Nishiyama M., Yamane H. Functional and evolutionary relationship between arginine biosynthesis and prokaryotic lysine biosynthesis through α-aminoadipate. J. Bacteriol. 2001;183:5067–5073. doi: 10.1128/JB.183.17.5067-5073.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyazaki J., Kobashi N., Fuji T., Nishiyama M., Yamane H. Characterization of a lysK gene as an argE homolog in Thermus thermophilus HB27. FEBS Lett. 2002;512:269–274. doi: 10.1016/s0014-5793(02)02290-1. [DOI] [PubMed] [Google Scholar]
- 16.Sakai H., Vassylyeva M. N., Matsuura T., Sekine S., Gotoh K., Nishiyama M., Terada T., Shirouzu M., Kuramitsu S., Vassylyev D. G., Yokoyama S. Crystal structure of a lysine biosynthesis enzyme, LysX, from Thermus thermophilus HB8. J. Mol. Biol. 2003;332:729–740. doi: 10.1016/s0022-2836(03)00946-x. [DOI] [PubMed] [Google Scholar]
- 17.Velasco A. M., Leguina J. I., Lazcano A. Molecular evolution of the lysine biosynthetic pathways. J. Mol. Evol. 2002;50:445–459. doi: 10.1007/s00239-002-2340-2. [DOI] [PubMed] [Google Scholar]
- 18.Hutton C. A., Southwood T. J., Turner J. J. Inhibitors of lysine biosynthesis as antibacterial agents. Mini Rev. Med. Chem. 2003;3:115–127. doi: 10.2174/1389557033405359. [DOI] [PubMed] [Google Scholar]
- 19.Suvama K., Scab L., Bhattacharjee V., Bhattacharjee J. K. Molecular analysis of the LYS2 gene of Candida albicans: homology to peptide antibiotic synthetases and the regulation of the α-aminoadipate reductase. Curr. Genet. 1998;33:268–275. doi: 10.1007/s002940050336. [DOI] [PubMed] [Google Scholar]
- 20.Palmer D. R. J., Balogh H., Ma G., Zhou X., Kaminskyj S. G. W. Synthesis and antifungal properties of compounds which target the α-aminoadipate pathway. Pharmazie. 2004;59:93–98. [PubMed] [Google Scholar]
- 21.Wulandari A. P., Miyazaki J., Kobashi N., Nishiyama M., Hoshino T., Yamane H. Characterization of bacterial homocitrate synthase involved in lysine biosynthesis. FEBS Lett. 2002;522:35–40. doi: 10.1016/s0014-5793(02)02877-6. [DOI] [PubMed] [Google Scholar]
- 22.Andi B., West A. H., Cook P. F. Kinetic mechanism of histidine-tagged homocitrate synthase from Saccharomyces cerevisiae. Biochemistry. 2004;43:11790–11795. doi: 10.1021/bi048766p. [DOI] [PubMed] [Google Scholar]
- 23.Strassman M., Ceci L. N. Enzymatic formation of cis-homoaconitic acid, an intermediate in lysine biosynthesis in yeast. J. Biol. Chem. 1966;241:5401–5407. [PubMed] [Google Scholar]
- 24.Bhattacharjee J. K., Tucci A. F., Strassman M. Accumulation of α-ketoglutarate in yeast mutants requiring lysine. Arch. Biochim. Biophys. 1968;123:235–239. doi: 10.1016/0003-9861(68)90129-x. [DOI] [PubMed] [Google Scholar]
- 25.Hall L. T., Sanchez R. J., Holloway S. P., Zhu H., Stine J. E., Lyons T. J., Demeler B., Schirf V., Hansen J. C., Nersissian A. M., et al. X-ray crystallographic and analytical ultracentrifugation analyses of truncated and full-length yeast copper chaperones for SOD (LYS7): a dimer-dimer model of LYS7-SOD association and copper delivery. Biochemistry. 2000;39:3611–3623. doi: 10.1021/bi992716g. [DOI] [PubMed] [Google Scholar]
- 26.Wallace M. A., Liou L. L., Martins J., Clement M. H., Bailey S., Longo V. D., Valentine J. S., Gralla E. B. Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis. Cross-compartment protection by CuZn-superoxide dismutase. J. Biol. Chem. 2004;279:32055–32062. doi: 10.1074/jbc.M403590200. [DOI] [PubMed] [Google Scholar]
- 27.Irvin S. D., Bhattacharjee J. K. A unique fungal lysine biosynthesis enzyme shares a common ancestor with tricarboxylic acid cycle and leucine biosynthetic enzymes found in diverse organisms. J. Mol. Evol. 1998;46:401–408. doi: 10.1007/pl00006319. [DOI] [PubMed] [Google Scholar]
- 28.Beinert H., Kennedy M. C., Stout C. D. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 1996;96:2335–2373. doi: 10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- 29.Lombo T., Takaya N., Miyazaki J., Gotoh K., Nishiyama M., Kosuge T., Nakamura A., Hoshino T. Functional analysis of the small subunit of the putative homoaconitase from Pyrococcus horikoshii in the Thermus lysine biosynthetic pathway. FEMS Microbiol. Lett. 2004;233:315–324. doi: 10.1016/j.femsle.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 30.Gerlt J. A., Babbitt P. C. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu. Rev. Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 31.The Kyoto Encyclopedia of Genes and Genomes ( http://www.genome.ad.jp/kegg/) states clearly that homoaconitase catalyses both steps. The ExPASy enzyme nomenclature database ( http://www.expasy.org/enzyme/) suggests that the enzyme catalyses the first step only. BRENDA ( http://www.brenda.uni-koeln.de/) indicates that the enzyme catalyses only the second step.
- 32.Brock M., Maerker C., Schutz A., Volker U., Buckel W. Oxidation of propionate to pyruvate in Escherichia coli. Involvement of methylcitrate dehydratase and aconitase. Eur. J. Biochem. 2002;269:6184–6194. doi: 10.1046/j.1432-1033.2002.03336.x. [DOI] [PubMed] [Google Scholar]
- 33.Howell D. M., Harich K., Xu H., White R. H. A-keto acid chain elongation reactions involved in the biosynthesis of coenzyme B (7-mercaptoheptanoyl threonine phosphate) in methanogenic Archaea. Biochemistry. 1998;37:10108–10117. doi: 10.1021/bi980662p. [DOI] [PubMed] [Google Scholar]
- 34.Massoudi H. H., Cantacuzene D., Wakselman C., Delatour C. B. Synthesis of cis-isomer and trans-isomer of homaconitic and fluorohomoaconitic acid. Synthesis. 1983;1983:1010–1012. [Google Scholar]
- 35.Ma G., Palmer D. R. J. Improved asymmetric syntheses of (R)-(−)-homocitrate and (2R,3S)-(−)-homoisocitrate, intermediates in the α-aminoadipate pathway of fungi. Tetrahedron Lett. 2000;41:9209–9212. [Google Scholar]
- 36.Jia Y., Palmer D. R. J., Quail W. J. The mono-(S)-α-methylbenzylammonium salt of (R)-homocitric lactone. Acta Cryst. E. 2005 in the press. [Google Scholar]
- 37.Miyazaki J., Kobashi N., Nishiyama M., Yamane H. Characterization of homoisocitrate dehydrogenase involved in lysine biosynthesis of an extremely thermophilic bacterium, Thermus thermophilus HB27, and evolutionary implication of β-decarboxylating dehydrogenase. J. Biol. Chem. 2003;278:1864–1871. doi: 10.1074/jbc.M205133200. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J., Fritsch E. F., Maniatis T. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 39.Uhrigshardt H., Walden M., John H., Anemuller S. Purification and characterization of the first archaeal aconitase from the thermoacidophilic Sulfolobus acidocaldarius. Eur. J. Biochem. 2001;268:1760–1771. [PubMed] [Google Scholar]
- 40.Cornish-Bowden A. New York: Oxford University Press; 1995. Analysis of Enzyme Kinetic Data. [Google Scholar]
- 41.Cawthray G. R. An improved reversed-phase liquid chromatographic method for the analysis of low-molecular mass organic acids in plant root exudates. J. Chromatogr. A. 2003;1011:233–240. doi: 10.1016/s0021-9673(03)01129-4. [DOI] [PubMed] [Google Scholar]
- 42.Morrison J. F. The activation of aconitase by ferrous ions and reducing agents. Biochem. J. 1954;56:685–692. doi: 10.1042/bj0580685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kennedy M. C., Beinert H. The state of cluster SH and S2− of aconitase during cluster interconversions and removal. A convenient preparation of apoenzyme. J. Biol. Chem. 1988;263:8194–8198. [PubMed] [Google Scholar]
- 44.Miyazaki K. Bifunctional isocitrate-homoisocitrate dehydrogenase: a missing link in the evolution of β-decarboxylating dehydrogenase. Biochem. Biophys. Res. Commun. 2005;331:341–346. doi: 10.1016/j.bbrc.2005.03.169. [DOI] [PubMed] [Google Scholar]