Abstract
Gelsolin and calponin are well-characterized cytoskeletal proteins that are abundant and widely expressed in vertebrate tissues. It is also becoming apparent, however, that they are involved in cell signalling. In the present study, we show that gelsolin and calponin interact directly to form a high-affinity (Kd=16 nM) 1:1 complex, by the use of fluorescent probes attached to both proteins, by affinity chromatography and by immunoprecipitation. These methods show that gelsolin can form high-affinity complexes with two calponin isoforms (basic h1 and acidic h3). They also show that gelsolin binds calponin through regions that have been identified previously as being calponin's actin-binding sites. Moreover, gelsolin does not interact with calponin while calponin is bound to F-actin. Reciprocal experiments to find calponin-binding sites on gelsolin show that these are in both the N- and C-terminal halves of gelsolin. Calponin has minimal effects on actin severing by gelsolin. In contrast, calponin markedly affects the nucleation activity of gelsolin. The maximum inhibition of nucleation by gelsolin was 50%, which was achieved with a ratio of two calponins for every gelsolin. Thus the interaction of calponin with gelsolin may play a regulatory role in the formation of actin filaments through modulation of gelsolin's actin-binding function and through the prevention of calponin's actin-binding activities.
Keywords: actin cytoskeleton, actin-nucleating activity, calponin, gelsolin, gelsolin–calponin complex
Abbreviations: acrylodan, 6-acryloyl-2-dimethylaminonaphthalene; CH, calponin homology; ERK, extracellular-signal-regulated kinase; F-actin, filamentous actin; G1–G6, the six repeated domains of gelsolin; G-actin, globular actin; GCC, gelsolin–calponin complex; PKC, protein kinase C; TRITC, tetramethylrhodamine β-isothiocyanate
INTRODUCTION
The actin cytoskeleton is a major determinant of cell structure, shape and motility. The properties of actin filaments and the structures that they form are modulated by a host of actin-binding proteins [1]. In addition to the physical role that the microfilaments perform, it is becoming increasingly apparent that these proteins also have important parts to play in cell signalling. The present study focuses on two well-characterized actin-binding proteins, gelsolin and calponin, which have distinct effects on microfilaments.
Gelsolin's actin-binding activity is regulated by Ca2+ ions [2], pH [3,4] phosphorylation [5,6] and phospholipids [7,8]. It is widely expressed in vertebrate cells [9] and is secreted from muscle [10] into the bloodstream. Its function in serum is to sever any actin filaments that may enter the circulation from damaged tissues [11]. In cells, gelsolin severs existing actin filaments, but may also nucleate the polymerization of new filaments from the pointed end, while it caps the barbed end of actin filaments. Increased expression of gelsolin correlates with an increase in cellular locomotion [12]. Gelsolin not only modifies the actin cytoskeleton by severing and capping microfilaments, but it plays additional roles in signal transduction and apoptosis [13]. It is remarkable therefore that a mouse that lacks the single gelsolin gene is viable [14], albeit with a host of identifiable failures such as in wound healing and clotting [14], in podosome assembly [15], and in blood vessel integrity [16]. Gelsolin is known to bind to a number of proteins [5,17], and it is possible that these too regulate gelsolin's interaction with actin.
The calponins are a family of actin-binding proteins that are expressed widely in vertebrate cells [18]. Three major vertebrate calponin isoforms are known: the basic isoform, h1, the neutral isoform, h2, and the acidic isoform, h3. Calponins h1 and h2 are expressed in smooth-muscle cells whereas h3 is expressed in smooth muscle and in non-muscle cells, especially in neurons [19]. It is suggested that calponin may function as an adaptor protein connecting the PKC (protein kinase C) cascade to the ERK (extracellular-signal-regulated kinase) cascade [20] as calponin binds ERK through the CH (calponin homology) domain. Calponin translocates from the cytoskeleton to the plasma-membrane [21,22], together with PKCα [23], when the cell is stimulated with agonists. Calponin is known to bind phospholipids in vitro, particularly those that carry a negative charge such as phosphatidylserine and phosphatidylinositol [24], and is associated with the plasma membrane in a variety of cell types [19,25].
Like gelsolin, calponin is also known to bind a number of protein partners, and at least two of these, actin and tropomyosin, bind gelsolin [17] and calponin [26]. In the present paper, we report that gelsolin and calponin interact with each other to form a tight GCC (gelsolin–calponin complex). This interaction affects the actin-binding activities and may alter the signalling and other properties of both partners.
MATERIAL AND METHODS
Proteins and peptides
Human gelsolin and subdomains of gelsolin were produced in Escherichia coli BL21(de3) cells using the pMW172 vector, after induction of expression with IPTG (isopropyl β-D-thiogalactoside) [27]. Gelsolin was labelled with Oregon Green 488 isothiocyanate (Molecular Probes), and excess reagent was removed by gel filtration using a PD10 column (Amersham Biosciences) equilibrated with 0.1 M NaHCO3 buffer at pH 8.6. The stoichiometry of labelling was determined to be 1.0 using a molar absorption coefficient of 70000 M−1·cm−1 at 496 nm. Basic calponin h1 was isolated from fresh chicken gizzards, as described previously [28]. Calponin was specifically labelled at Cys273 with acrylodan (6-acryloyl-2-dimethylaminonaphthalene). The labelling ratio was estimated spectroscopically using a molar absorption coefficient for acrylodan of 16400 M−1·cm−1 at 387 nm. The labelling stoichiometry was determined to be 0.50 for acrylodan/calponin. Calponin was also coupled to Sepharose 4B using the CNBr procedure according to the manufacturer's instructions (Amersham Biosciences). Recombinant rat acidic calponin h3 (expressed as described in [29]) was a gift from Mario Gimona (Austrian Academy of Sciences, Salzburg, Austria). Actin was made from acetone powder [30] and labelled at Cys374 by N-(1-pyrenyl)iodoacetamide [31]. Synthetic peptides were prepared as described previously and have been widely used [32,33]. Peptide synthesis was performed on a solid-phase support using a 9050 Milligen PepSynthesizer (Millipore) according to the Fmoc (fluoren-9-ylmethoxycarbonyl)/t-butyl ester system. The purified peptides were shown to be homogenous by analytical HPLC.
Cell culture, immunostaining, immunoprecipitation and Western blots
COS-7 cells were cultured in Dulbecco's modified Eagle's medium containing 10% (v/v) FCS (foetal calf serum) and antibiotics as described previously [34]. For immunofluorescence studies, cells were plated on a glass coverslip 24 h before fixation in 3% (w/v) formaldehyde solution, after which they were permeabilized with PBS/0.1% (v/v) Triton X-100 for 5 min and incubated in PBS/10% FCS for 20 min. Coverslips were incubated sequentially with TRITC (tetramethylrhodamine β-isothiocyanate)-conjugated anti-gelsolin and anti-mouse (Sigma), anti-(calponin h3), and FITC-conjugated anti-rabbit (Sigma) antibodies and finally with Hoescht in PBS/0.1% (v/v) Tween 20 for 30 min, as described previously [19]. Coverslips were rinsed in PBS/0.1% (v/v) Tween 20 between each antibody incubation, and three additional washes were performed before coverslips were mounted in Mowiol. Fixed cells were observed with a DMR A oil immersion microscope PL APO 63× (Leica Microsystems). Images were captured with a Micromax camera (Roper Scientific) driven by MetaMorph (version 4.7; Universal Imaging Corp.). A monoclonal anti-gelsolin antibody was purchased from Sigma–Aldrich. The affinity-purified polyclonal anti-(calponin h3) antibody directed to the sequence 311–326 derived from the specific C-terminal tail of the acidic isoform was raised as previously described [19].
For immunoprecipitation studies, COS-7 cells grown to confluence were washed three times in PBS and then lysed in RIPA buffer (20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS and 1 mM EGTA) essentially as described previously [34], except that we included 1 mM EGTA to the lysis buffer to favour the dissociation of the proteins from actin. After 20 min of pre-clearing with Protein A or G beads (Amersham Biosciences), extracts were immunoprecipitated with anti-gelsolin or anti-calponin antibodies. Complexes formed were recovered on Protein G beads (gelsolin) or Protein A beads (calponin). Control immunoprecipitations were performed using both beads in the absence of antibody. Immunoprecipitated proteins were then separated by SDS/9% PAGE and transferred on to nylon membranes (Millipore). After saturation in PBS/1% (w/v) BSA/0.1% (v/v) Tween 20, membranes were incubated with anti-gelsolin or anti-(calponin h3) antibodies and then with horseradish-peroxidase-conjugated anti-mouse or anti-(Protein A) antibodies (Amersham Bioscience) in PBS/1% (w/v) BSA/0.1% (v/v) Tween 20. Proteins were then revealed by chemiluminescence using the ECL® (enhanced chemiluminescence) kit (Amersham Bioscience).
Fluorescence studies
Fluorescence experiments were performed at 21 °C using a LS 50 PerkinElmer luminescence spectrometer. Spectra for Oregon Green-labelled gelsolin were obtained with the excitation wavelength set at 470 nm and fluorescence changes deduced from the corresponding area of the emission spectra between 515 and 530 nm. For acrylodan-labelled calponin, the excitation wavelength was fixed at 388 nm. All fluorescence measurements were performed at least in triplicate.
The parameters Kd and Amax (maximum effect) were calculated by non-linear fitting of the experimental data points by using the following equation:
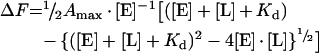 |
where ΔF is the change in fluorescence, [E] is the concentration of the fluorescent protein and [L] is the ligand concentration. Non-linear fitting was performed using the Curvefit software developed by Kevin Raner Software. Additional details on the different experimental conditions are given in the Figure legends.
The number of binding sites (n) and the affinity constant Ka were also determined by another approach [35]. The following relationship was then used:
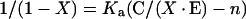 |
where C and E are total concentrations of calponin and Oregon Green-labelled gelsolin respectively and X is the relative fluorescence change A/Amax (corresponding to the fraction of calponin bound to gelsolin).
ELISA
The interaction between coated calponin synthetic peptides and biotinylated gelsolin was monitored using ELISA as described previously [36]. Briefly, peptides (5 μg/ml) in 50 mM NaHCO3/Na2CO3, pH 9.5, were immobilized on plastic microtitre wells which were then saturated with 0.5% gelatin, 3% gelatin hydrolysate in 140 mM NaCl and 10 mM phosphate, at pH 7.5. Binding of biotinylated gelsolin to peptides was monitored at 405 nm using alkaline-phosphatase-labelled streptavidin (dilution 1:1000). Control assays were carried out in wells saturated with gelatin and gelatin hydrolysate alone. Each assay was conducted in triplicate, and the mean value was plotted after subtracting the non-specific absorption. Additional details on the different experimental conditions are given in the Figure legends.
Actin polymerization
Actin polymerization and depolymerization were monitored by fluorescence measurements. Fluorescence increase of pyrenyl–actin [31] was used as an indicator of the state of actin polymerization. Excitation and emission wavelengths for pyrenyl–actin were set at 360 and 386 nm respectively. Actin polymerization was induced by the addition of salts (2 mM MgCl2 and 0.1 M KCl; final concentration). Nucleation enhanced by gelsolin was determined by measuring the rate of actin polymerization. G-actin (globular actin) (4.9 μM) was added to 2 mM MgCl2 and 0.1 M KCl, 20 μM ATP, 0.5 mM CaCl2 and 20 mM Tris/HCl buffer at pH 7.5 in the presence or the absence of gelsolin and/or calponin as indicated in the Figure legends. The severing activity of gelsolin is directly related to the depolymerization rate [37] observed when 34 μM fluorescent F-actin (filamentous actin) (pre-capped by a 1:2000 molar ratio of gelsolin) is diluted to 1.4 μM.
Analytical methods
Protein concentrations were determined by UV absorbency using a Varian MS100 spectrophotometer. Concentrations were determined spectrophotometrically using A280 (1 cm−1) values of 8.93 μM for whole gelsolin, 38.7 μM for calponin h1 and 31.2 μM for calponin h3. These molar absorption coefficients were calculated from tryptophan, tyrosine and cysteine content [38]. SDS/PAGE was carried out on 12.5% (w/v) polyacrylamide slab gels.
RESULTS
A direct interaction between calponin and gelsolin
Gelsolin and calponin form a tight 1:1 complex. This was shown by monitoring fluorescence changes when each partner in turn was labelled covalently with a fluorophore. Gelsolin is also bound by an affinity column of immobilized calponin.
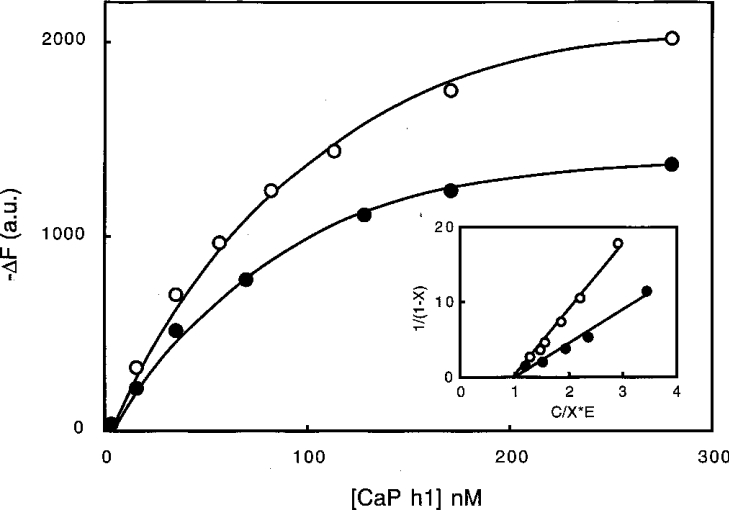
The ability of calponin to interact directly with gelsolin was monitored using two different approaches. In the first set of experiments, the fluorescence of gelsolin labelled with either Oregon Green or acrylodan was used to monitor the interaction with calponin. Binding of calponin h1 to Oregon-Green-labelled gelsolin (in 0.1 M Tris/HCl buffer, pH 7.5, containing 1 mM EGTA) induced a fluorescence decrease (47±5%) concomitant with a weak blue shift in the emission spectrum of approx. 3 nm (Figure 1A and Table 1). The shape of the curve shows that the binding takes place in a saturable manner with an apparent Kd of 16±4 nM. The binding occurs in the presence of EGTA, as well in the presence of Ca2+ (Figure 1A). The interaction induces a similar fluorescence decrease (45±5%) with EGTA, although a slightly weaker affinity (apparent Kd of 30±5 mM) is observed (Table 1). This result suggests that the calponin binding depends, at least in part, upon the conformational state of gelsolin. Since a lowered pH activates gelsolin in a manner that is similar to that of Ca2+ [3,4], we also studied the interaction at pH 6.0. However, although significant binding was observed (Figure 1B), an apparent Kd of 160±25 nM was determined (instead of 16 nM in 1 mM EGTA at pH 7.5). A stoichiometry of 1 mol of calponin per mol of gelsolin was estimated for all of the binding experiments shown in Figure 1(C).
Figure 1. Binding of Oregon-Green-labelled gelsolin to calponin h1.
(A) Interaction of Oregon-Green-labelled gelsolin (85 nM) with calponin (CaP) h1 was monitored by fluorescence. Changes in the intensity of the fluorescence emission spectra (ΔF) of Oregon Green were recorded at various calponin concentrations (0–380 nM) in 0.1 M Tris/HCl buffer, pH 7.5, supplemented with 1 mM EGTA (●) or 1 mM Ca2+ (○). Inset, blue shift in the maximum of the emission spectra was monitored with respect to calponin concentration (0.1 M Tris/HCl buffer, pH 7.5, containing 1 mM EGTA). a.u., arbitrary units. (B) Interaction of Oregon-Green-labelled gelsolin (85 nM) with calponin (CaP) h1 in 0.1 M Mes buffer, pH 6.0. Calponin h1 concentration was varied between 0 and 480 nM. a.u., arbitrary units. (C) Quantitative analysis of the data in (A) and (B) for the interaction between gelsolin and calponin h1 was performed by plotting 1/(1−X) against C/X·E where C is the concentration of calponin expressed in nM and E is the concentration of gelsolin fixed at 85 nM. X, the binding ratio, was determined as described in the Materials and methods section. The experiments were performed in 0.1 M Tris/HCl buffer, pH 7.5, supplemented with EGTA (●) or Ca2+ (○), or in 0.1 M Mes buffer, pH 6.0 (□).
Table 1. Interaction of Oregon-Green-labelled gelsolin with calponin was monitored by fluorescence as described in the legend of Figure 1.
Changes in the intensity of the fluorescence emission spectra of Oregon Green were recorded at various calponin concentrations in 0.1 M Tris/HCl buffer, pH 7.5, supplemented with 1 mM EGTA or 1 mM Ca2+. Percentage fluorescence decreases were extrapolated to infinite calponin concentration.
| Calponin h1 | Calponin h3 | |||
|---|---|---|---|---|
| Buffer conditions | Kd (nM) | Fluorescence decrease (%) | Kd (nM) | Fluorescence decrease (%) |
| pH 7.5 EGTA | 16±4 | 47±5 | 130±20 | 40±4 |
| pH 7.5 Ca2+ | 30±5 | 45±5 | 140±20 | 30±4 |
To confirm the results obtained using labelled gelsolin, similar experiments were then performed in 0.1 M Tris/HCl buffer, pH 7.5, containing 1 mM EGTA, but using acrylodan-labelled calponin h1. The interaction of gelsolin with labelled calponin induced a decrease in the emission spectrum of acrylodan (Figure 2A). An apparent Kd of 14±4 nM was determined (Figure 2B) and a stoichiometry of 1:1 was also observed (Figure 2C).
Figure 2. Interaction of acrylodan-labelled calponin h1 with gelsolin.
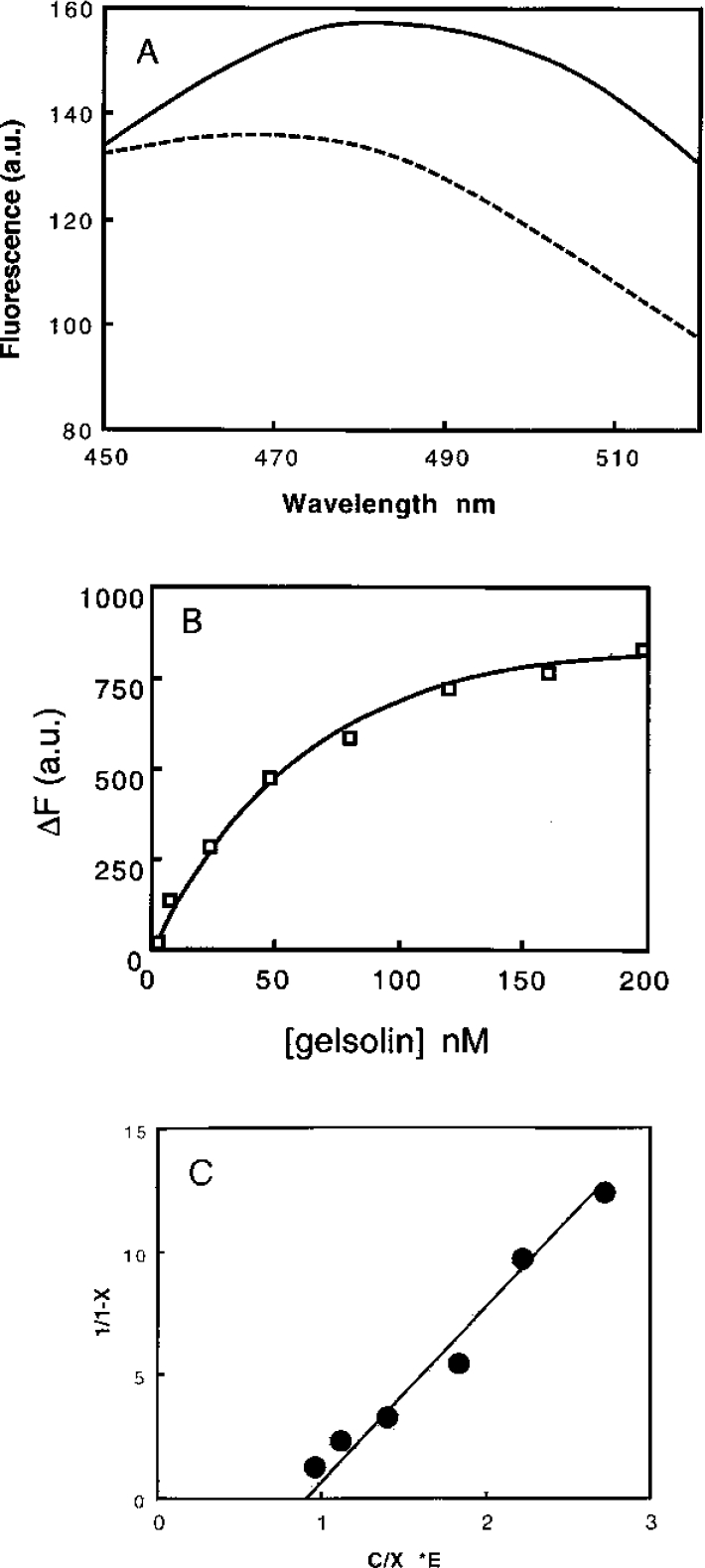
Interaction of gelsolin with calponin was monitored by changes in acrylodan fluorescence intensity. Experiments were carried out in 0.1 M Tris/HCl buffer, pH 7.5, and 1 mM EGTA using a fixed concentration of labelled calponin of 80 nM. a.u., arbitrary units. (A) Emission spectrum of acrylodan-labelled calponin alone (——) or in the presence of 48 nM gelsolin (− − −). The excitation wavelength was 388 nm. (B) Fluorescence decrease is plotted against gelsolin concentrations varied between 0 and 200 nM. (C) Quantitative analysis of the data in (B) for the interaction between acrylodan-labelled calponin h1 and gelsolin was performed by plotting 1/(1−X) against C/X·E, where C is the concentration of gelsolin (in nM) and E is the concentration of calponin h1 fixed at 80 nM.
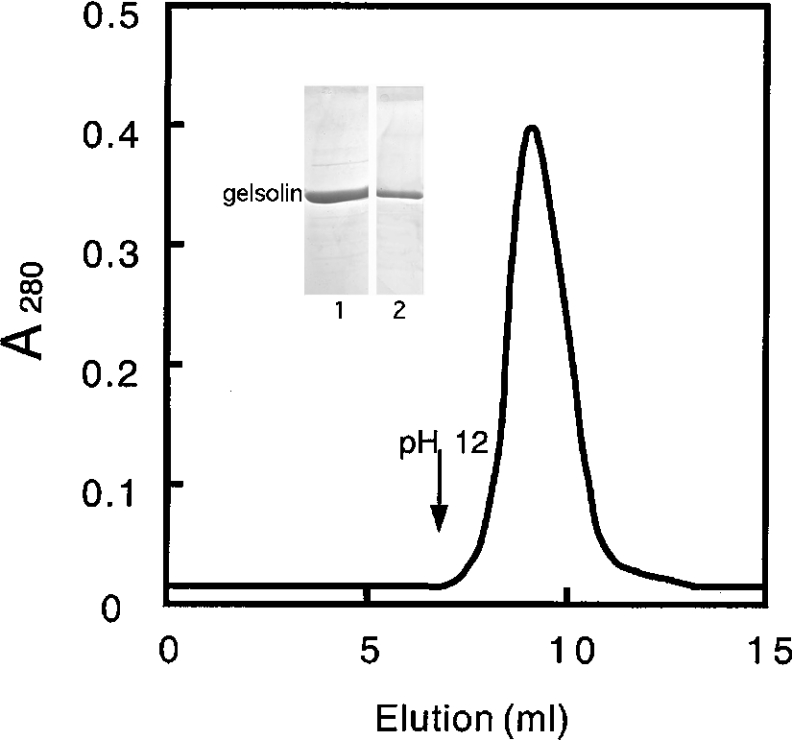
We also detected the GCC by affinity chromatography. Gelsolin was passed over a Sepharose column on to which calponin had been coupled. Figure 3 shows that all of the gelsolin is retained on the column and quantitatively eluted at alkaline pH. This result is in agreement with the high affinity of the interaction determined by fluorescence experiments.
Figure 3. Binding of gelsolin to calponin h1 coupled to Sepharose 4B.
Gelsolin (870 μg) was passed through a column (1.4 cm×3 cm) of Sepharose-4B-linked calponin. The column was washed with 10 mM phosphate buffer, pH 7.4, supplemented with 0.15 M NaCl. The bound material was eluted with 10 mM phosphate buffer, pH 12, supplemented with 10% dioxan. The eluted fraction was quantified from a UV absorption spectrum (in a typical experiment, 780 μg of gelsolin was eluted), pulled-down and analysed by SDS/12.5% PAGE (inset). Lane 1, gelsolin control; lane 2, eluted material.
Similar experiments were conducted to test whether the acidic isoform of calponin (calponin h3) also bound to gelsolin. We observed an equal decrease in the fluorescence of Oregon-Green-labelled gelsolin upon calponin h3 interaction, so that the Ca2+ dependence was much less marked than was the case for calponin h1, but that the apparent Kd was only slightly higher than for calponin h1 (Table 1).
Gelsolin and calponin interact in vivo
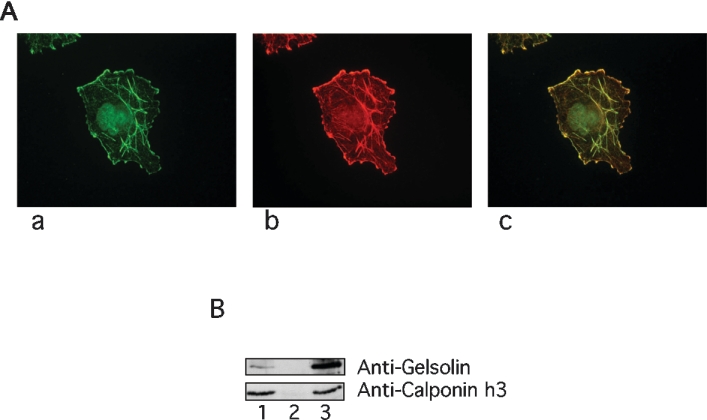
Gelsolin and calponin co-localize in COS-7 cells, as shown by immunofluorescence. Moreover, reciprocal immunoprecipitation of either protein from cell lysates pulls down the other. Gelsolin was found in dynamic actin structures such as lamellipodia present around the cells and along some stress fibres (Figure 4A, panel b), in agreement with previous work [39]. Calponin showed more discrete membrane lamellipodia labelling and more intense stress fibre staining (Figure 4A, panel a), again in agreement with previous work [40]. Superposition of the two images showed marked co-localization of the two proteins especially at the cells' peripheral lamella (Figure 4A, panel c), suggesting that these proteins may interact within these structures. We have shown that gelsolin and calponin bind each other by a number of means. Co-immunoprecipitation studies confirmed the hypothesis that gelsolin binds calponin in vivo (Figure 4B), as the proteins were found to mutually precipitate each other brought about by addition of specific antisera. Calponin was readily recovered when precipitated with anti-gelsolin antibodies (Figure 4B, lane 3), whereas anti-calponin antibodies were less effective in co-precipitating gelsolin (Figure 4B, lane 1). The reason for this inefficiency may be either the restricted repertoire of these anti-peptide antibodies or the structural constraints that impaired recognition of the complex. Control antiserum for an inappropriate target did not precipitate gelsolin nor did gelsolin precipitate when primary antibody was omitted (results not shown).
Figure 4. Gelsolin and calponin interact in vivo.
(A) Co-localization of gelsolin and calponin in COS-7 cells. Endogenous calponin was revealed by an anti-(calponin h3) antibody followed by an FITC-conjugated anti-rabbit antibody (a); endogenous gelsolin was revealed by an anti-gelsolin antibody followed by a TRITC-conjugated anti-mouse antibody (b); co-localization of the two proteins (in yellow) is presented in (c). (B) Co-immunoprecipitation of gelsolin and calponin. RIPA extracts were subjected to immunoprecipitation with an anti-(calponin h3) antibody (lane 1), control (lane 2) and an anti-gelsolin antibody (lane 3) followed by Western blots with the indicated antibodies.
The calponin–gelsolin interface
Calponin binds with high affinity to both the N- and C-terminal halves of gelsolin. The interaction region appears to be mediated through calponin's actin-binding region. The interface between gelsolin and calponin h1 was investigated by three approaches. First, the calponin h1 interaction with the N-terminal [G1–G3 (repeated domains of gelsolin 1–3)] and C-terminal halves (G4–G6) was investigated. We tested whether the individual G1–G3 and G4–G6 domains corresponding to the N- and C-terminal halves of gelsolin induce fluorescence changes in Oregon Green as did the whole gelsolin. As shown in Figure 5 and Table 2, a large decrease in fluorescence was observed in the presence of either half. Analysis of the data shows that these two gelsolin subdomains contain interaction sites for calponin with similar apparent affinities of 20±7 nM and 25±8 nM towards G1–G3 and G4–G6 respectively. A stoichiometry of one is also observed for the two domains (Figure 5, inset). These results were confirmed by an experiment using acrylodan-labelled calponin as a fluorescent reagent (results not shown).
Figure 5. Binding of Oregon-Green-labelled gelsolin domains to calponin h1.
Interaction of Oregon-Green-labelled G1–G3 (140 nM) (○) and G4–G6 (89 nM) (●) with calponin (CaP) h1 was monitored by fluorescence. Changes in the intensity of the fluorescence emission spectra (ΔF) of Oregon Green were recorded at various calponin concentrations (0–280 nM) in 0.1 M Tris/HCl buffer, pH 7.5, supplemented with 1 mM EGTA. Inset, quantitative analysis of the data for the interaction between gelsolin domains and calponin h1 was performed by plotting 1/(1−X) against C/X·E where C is the concentration of calponin (in nM) and E is the concentration of gelsolin domains at the fixed concentration indicated above. Experiments were performed with G1–G3 (○) and G4–G6 (●). a.u., arbitrary units.
Table 2. Interaction of Oregon-Green-labelled gelsolin and gelsolin domains with calponin h1 was monitored by Oregon Green fluorescence.
Changes in the intensity of the fluorescence emission spectra were recorded at various calponin concentrations in 0.1 M Tris/HCl buffer, pH 7.5, supplemented with 1 mM EGTA. Percentage fluorescence decreases were extrapolated to infinite calponin concentration.
| Kd (nM) | Fluorescence decrease (%) | Stoichiometry | |
|---|---|---|---|
| Gelsolin | 16±4 | 45±5 | 1 |
| G1–G3 | 20±7 | 44±5 | 1 |
| G4–G6 | 25±8 | 33±5 | 1 |
Secondly, the interaction of both gelsolin and calponin with F-actin was tested. The experiments were conducted in EGTA to avoid actin severing by gelsolin that would interfere with the binding process. F-actin was used at a saturating concentration [six times the apparent Kd (1.4 μM) reported for the interaction of calponin with F-actin] [41]. Binding was observed by co-sedimentation experiments. As shown in Figure 6, a large amount of calponin is found in the pellets. When the three proteins were mixed together, we observed that calponin is essentially present in the pellet and gelsolin in the supernatant. We conclude from this that gelsolin does not interact with the F-actin–calponin complex.
Figure 6. Binding of calponin to F-actin in the presence of gelsolin.
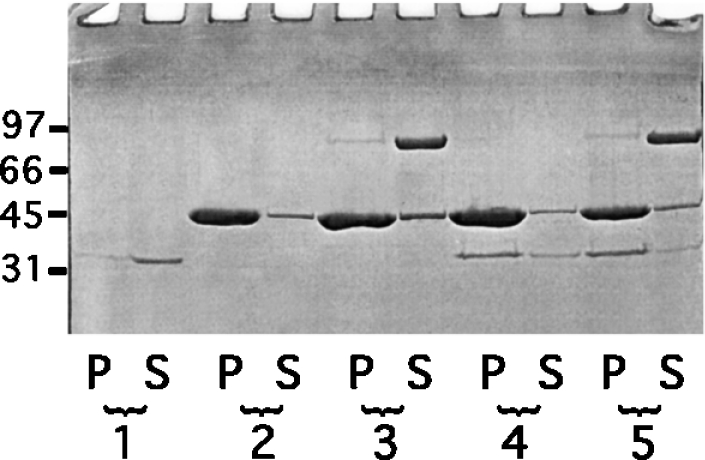
F-actin (8.5 μM) was incubated in the presence of calponin (2.0 μM) and gelsolin (0.22 μM) in 2 mM MgCl2, 0.1 M KCI, 0.1 mM ATP, 1 mM EGTA and 10 mM Tris/HCl, pH 7.5. The samples were ultracentrifuged at 90000 rev./min for 20 min in a Beckman TLA 100.3 rotor, and the supernatant (S) and the pellet (P) were analysed by SDS/12.5% PAGE. Lanes 1, calponin alone; lanes 2, F-actin alone; lanes 3, F-actin+gelsolin; lanes 4, F-actin+calponin; lanes 5, F-actin+calponin+gelsolin. Molecular-mass markers in kDa are rabbit muscle phosphorylase B (97 kDa), BSA (66 kDa), ovalbumin (45 kDa) and bovine carbonate anhydrase (31 kDa).
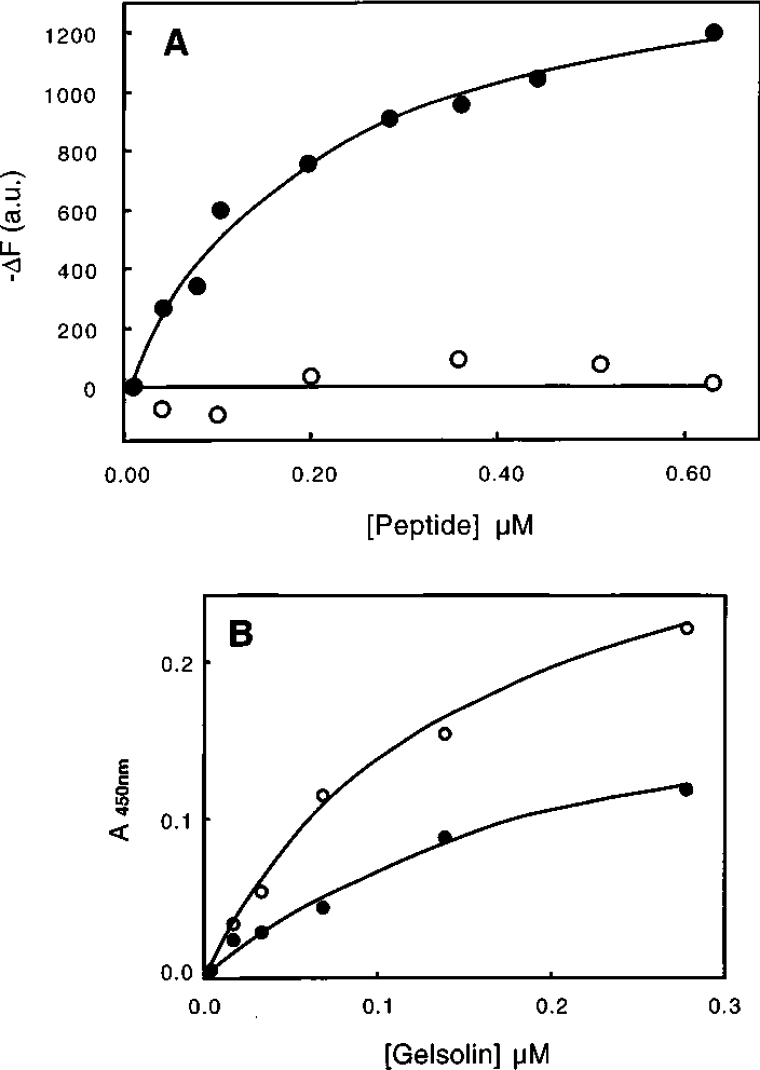
Thirdly, since the above results suggest the possible participation of an actin-binding site in calponin, corresponding synthetic peptides were then tested for their interaction with gelsolin. It has been reported that calponin interaction with filaments is mediated by at least two identified sequences, one adjacent to the CH domain, i.e. sequence 145–163 [33], and the other located in the first repeat region containing the sequence 172–187 [42]. We synthesized these peptides to determine whether either induce conformational changes in gelsolin. Figure 7(A) shows that the 172–187 peptide induces a quenching of the tryptophan residue of gelsolin, whereas the 145–163 peptide did not. However, when tested for gelsolin binding by ELISA (Figure 7B), we found that both peptides (145–163 and 172–187) bound gelsolin. We conclude that gelsolin binds calponin through regions that have been identified previously as being actin-binding regions.
Figure 7. Binding of calponin sequences 145–163 and 172–187 to gelsolin monitored by fluorescence (A) and ELISA (B).
(A) The interaction of the 145–163 (○) and 172–187 (●) peptide fragments in 0.1 M Tris/HCl buffer, pH 7.5, supplemented with 1 mM EGTA was monitored by following the corresponding changes in the tryptophan fluorescence (ΔF) of gelsolin (0.2 μM). a.u., arbitrary units. (B) Coated peptides of sequence 145–163 (●) and 172–187 (○) were reacted in 0.1 M Tris/HCl buffer, pH 7.5, and 1 mM EGTA with biotinylated gelsolin at the concentrations indicated.
Effect of calponin on gelsolin activity
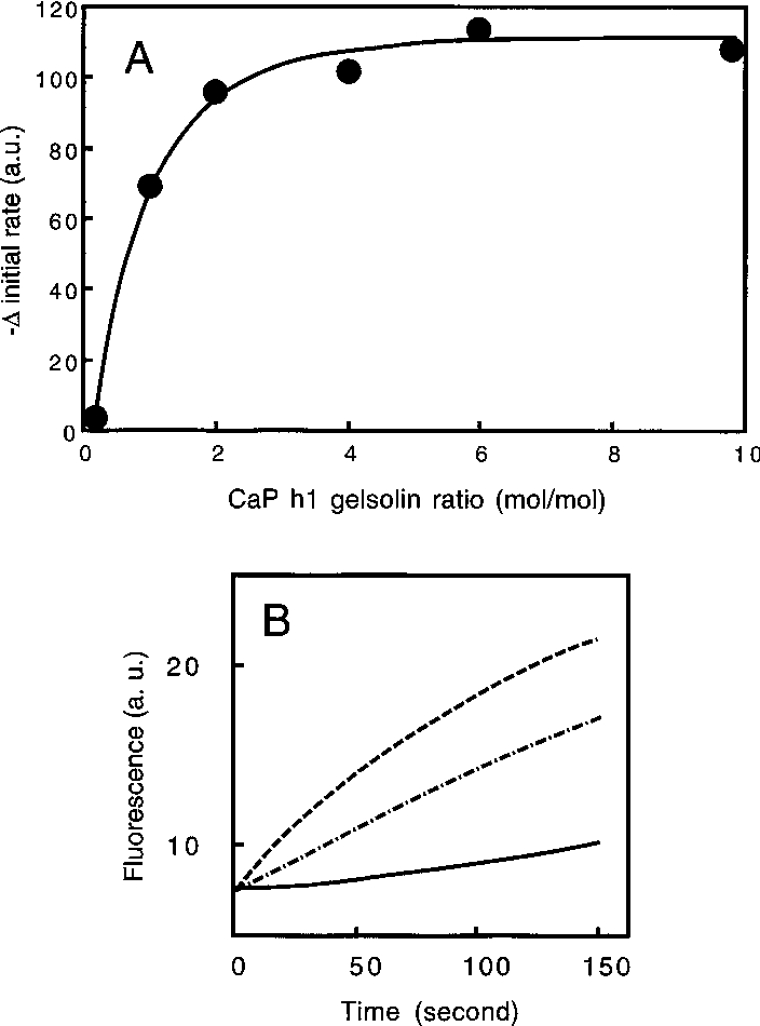
Calponin has little effect on gelsolin's severing activity, but it can reduce its nucleating activity by up to half. Two sets of experiments were performed to test the effect of calponin on the activity of gelsolin. Gelsolin greatly enhanced the initial rate of actin polymerization (results not shown), which is in agreement with the established literature, but no significant effect of calponin on the time course of actin polymerization was seen under the conditions used (2 mM MgCl2 and 0.1 M KCl) (results not shown). More interestingly, the presence of GCCs decreases the initial rate of polymerization induced by gelsolin alone in a concentration-dependent manner (Figure 8A). The maximum inhibition is obtained for ratios of up to two calponins for one gelsolin used at 0.23 μM, a high protein concentration in comparison with the apparent Kd (30 nM) reported for the interaction between these two proteins (Table 1). Therefore further experiments were performed using a calponin/gelsolin ratio of 2:1 (Figure 8B). The means for these experiments correspond to an inhibition of approx. 50±10% (mean for ten experiments). Under the experimental conditions, actin alone was found to have a low initial polymerization rate which was only slightly inhibited by calponin at the concentration found to inhibit gelsolin (results not shown).
Figure 8. Effects of calponin on the activation of actin-nucleating activity by gelsolin.
(A) Labelled G-actin (9.2 μM) was added to 2 mM MgCl2, 0.1 M KCl, 50 μM ATP, 0.5 mM CaCl2 and 5 mM Tris/HCl, pH 7.5, in the presence of gelsolin (0.23 μM) or gelsolin (0.23 μM)+1–10 molar excess of calponin. In each case, the initial polymerization rate was determined and plotted against the calponin (CaP) h1/gelsolin ratio. a.u., arbitrary units. (B) Labelled G-actin (4.9 μM) was added to 2 mM MgCl2, 0.1 M KCl, 50 μM ATP, 0.5 mM CaCl2 and 5 mM Tris/HCl, pH 7.5, in the absence (——) or the presence of 0.12 μM gelsolin (– – –) or 0.12 μM gelsolin+0.26 μM calponin (−·−·−·), as described in the Materials and methods section. The increase in pyrene fluorescence was plotted against time. Results are means for ten experiments. a.u., arbitrary units.
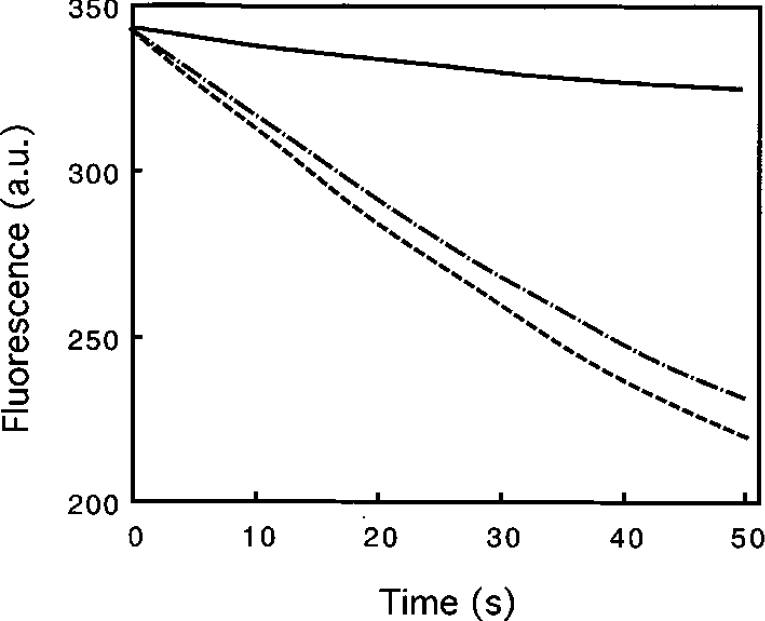
Next, the effect of calponin on the severing activity of gelsolin was examined. In the severing assay, the rate of fluorescence decrease was measured after dilution of gelsolin-capped pyrene-labelled filaments in F-buffer in the presence of gelsolin or GCCs. In these experiments, gelsolin activity was only affected slightly by the presence of calponin. Less than 15% inhibition was determined in several experiments (Figure 9). In addition, calponin alone partially decreases the actin-depolymerization rate induced by dilution (results not shown).
Figure 9. Effects of calponin on the severing activity of gelsolin.
Labelled F-actin (36 μM) (pre-capped by a 1:2000 molar ratio of gelsolin) was diluted to 1.4 μM in 0.5 mM CaCl2, 0.1 mM ATP and 5 mM Tris/HCl, pH 7.5, alone (——) and in the presence of 0.1 μM gelsolin (– – –) or 0.1 μM gelsolin+0.2 μM calponin h1 (−·−·−·). The decrease in fluorescence of pyrenyl actin was plotted against time. a.u., arbitrary units.
DISCUSSION
The present study shows, for the first time, a direct interaction between gelsolin and calponin. The interaction is a relatively high affinity one, with Kd values between 16 and 160 nM, depending on the presence of Ca2+ and on the pH. However, since the effects of Ca2+ and pH are relatively minor, these parameters are unlikely to be important regulators for the formation of the complex in cells. We have found throughout that a simple 1:1 complex is formed and that calponin binds gelsolin at sites within both the N-terminal and the C-terminal halves. Each of these sites binds with approximately the same affinity (Kd=20 nM), whereas the whole molecule binds with a similar Kd (16 nM). It is possible that the two sites on gelsolin partially inhibit each other under the conditions reported in the present paper and so the affinity for calponin is less than that expected to result from two separate interactions at 20 nM. A conformational change may also take place within gelsolin, induced by binding to either of the calponin sites that lowers the affinity for the other. It is not certain whether the binding of calponin functions to regulate gelsolin's actin-binding activity in cells. The main effect of calponin binding is the inhibition of nucleation, whereas the severing action of gelsolin is relatively uninhibited. Although calponin inhibits gelsolin's nucleating activity in vitro, it is not certain how important this activity is for the function of cells. Relatively little is known about how the various actin-binding properties (severing, capping and nucleating) of gelsolin function in cells [13], and further work will demonstrate whether the GCC has other protein partners that might regulate gelsolin's actin-binding activity. Calponin's actin-binding activity was found to be inhibited by gelsolin binding, as gelsolin cannot bind calponin when calponin binds F-actin, and vice versa.
Both calponin and gelsolin have been primarily characterized as being cytoskeletal proteins, and their known properties are very different in this regard. For example, calponin binds and stabilizes actin filaments [18,28,29], whereas gelsolin severs them [2]. Despite this, the effects of deleting calponin and gelsolin expression in mice are remarkably similar. Deletion of calponin h1 [43] and of gelsolin [44] increases the fragility of blood vessels, possibly by weakening endothelial cell adhesions [45] or through disruption of the vascular smooth muscle. Another striking similarity is that deletion of calponin increases bone density [46], as does the deletion of gelsolin [16]. The similarity of all of these effects may be partially explained by our discovery of the GCC that may take part in signalling, so that the complex would be disrupted by the loss of either partner. This interpretation posits, then, that the observed phenotypes arise from the loss of the GCC.
Calponin is a substrate for many kinases and forms direct interactions with others [20,22,23,34], and a more restricted set of kinases are associated with gelsolin [5,6]. Gelsolin [13] and especially calponin [18,26] are known to bind proteins other than kinases, so the GCC may serve as a scaffold or adaptor complex to bring together relevant kinases and substrates. Gelsolin is implicated in the processes of apoptosis and tumorigenesis [13], and an understanding of the possible regulation of gelsolin's actions, including those mediated through calponin complex formation, is an important research goal.
Acknowledgments
This research was supported by grants from AFM (Association Française contre les Myopathies) and the British Council (S.K.M. and C.R.). I.F. is supported by a grant from the Service de Coopération Française within the framework of a Franco-Tunisien co-operation. We are grateful to Mario Gimona (Austrian Academy of Sciences, Salzburg, Austria) for his gift of the recombinant calponin h3, and to Paul McLaughlin (University of Edinburgh, Edinburgh, Scotland, U.K.) for his valuable comments on the manuscript before submission.
References
- 1.dos Remedios C. G., Chhabra D., Kekic M., Dedova I. V., Tsubakihara M., Berry D. A., Noseworthy N. J. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 2.Yin H. L., Stossel T. P. Control of cytoplasmic actin gel–sol transformation by gelsolin, a calcium dependent regulatory protein. Nature (London) 1979;281:583–586. doi: 10.1038/281583a0. [DOI] [PubMed] [Google Scholar]
- 3.Lamb J. A., Allen P. G., Tuan B. Y., Janmey P. A. Modulation of gelsolin function: activation at low pH overrides Ca2+ requirement. J. Biol. Chem. 1993;268:8999–9004. [PubMed] [Google Scholar]
- 4.Lagarrigue E., Ternent D., Maciver S. K., Fattoum A., Benyamin Y., Roustan C. The activation of gelsolin by low pH: the calcium latch is sensitive to calcium but not pH. Eur. J. Biochem. 2003;270:4105–4112. doi: 10.1046/j.1432-1033.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q., Xie Y., Du Q.-S., Wu X.-J., Feng X., Mei L., McDonald J. M., Xiong W.-C. Regulation of the formation of osteoclastic actin rings by proline-rich tyrosine kinase 2 interacting with gelsolin. J. Cell Biol. 2003;160:565–575. doi: 10.1083/jcb.200207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Corte V., Gettemans J., Vandekerckhove J. Phosphatidylinositol 4,5-bisphosphate specifically stimulates PP60c−src catalysed phosphorylation of gelsolin and related actin-binding proteins. FEBS Lett. 1997;401:191–196. doi: 10.1016/s0014-5793(96)01471-8. [DOI] [PubMed] [Google Scholar]
- 7.Janmey P. A., Stossel T. P. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature (London) 1987;325:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- 8.Méré J., Chahinian A., Maciver S. K., Fattoum A., Bettache N., Benyamin Y., Roustan C. Gelsolin binds to polyphosphoinositide-free lipid vesicles and simultaneously to actin microfilaments. Biochem. J. 2005;386:47–56. doi: 10.1042/BJ20041054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin H. L., Albrecht J. H., Fattoum A. Identification of gelsolin, a Ca++-dependent regulatory protein of actin gel–sol transformation and its intracellular distribution in a variety of cells and tissues. J. Cell Biol. 1981;91:901–906. doi: 10.1083/jcb.91.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwiatkowski D. J., Mehl R., Izumo S., Nadal-Ginard N., Yin H. L. Muscle is the major source of plasma gelsolin. J. Biol. Chem. 1988;263:8239–8243. [PubMed] [Google Scholar]
- 11.Lind S. E., Smith D. B., Janmey P. A., Stossel T. P. Role of plasma gelsolin and vitamin D-binding protein in clearing actin from the circulation. J. Clin. Invest. 1986;78:736–742. doi: 10.1172/JCI112634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham C. C., Stossel T. P., Kwiatkowski D. J. Enhanced motility in NIH 3T3 fibroblasts that overexpress gelsolin. Science. 1991;251:1233–1236. doi: 10.1126/science.1848726. [DOI] [PubMed] [Google Scholar]
- 13.Kwiatkowski D. J. Functions of gelsolin: motility, signaling, apoptosis, cancer. Curr. Opin. Cell Biol. 1999;11:103–108. doi: 10.1016/s0955-0674(99)80012-x. [DOI] [PubMed] [Google Scholar]
- 14.Witke W., Sharpe A. H., Hartwig J. H., Azuma T., Stossel T. P., Kwiatkowski D. J. Hemostatic, inflammatory and fibroblast responses are blunted in mice lacking gelsolin. Cell. 1995;81:41–51. doi: 10.1016/0092-8674(95)90369-0. [DOI] [PubMed] [Google Scholar]
- 15.Chellaiah M., Kizer N., Silva M., Alvarez U., Kwiatkowski D., Hruska K. A. Gelsolin deficiency blocks podosome assembly and produces increased bone mass and strength. J. Cell Biol. 2000;148:665–678. doi: 10.1083/jcb.148.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker P. M., Kazi A. A., Wadgaonkar R., Pearse D. B., Kwiatkowski D., Garcia J. G. N. Pulmonary vascular permeability and ischemic injury in gelsolin-deficient mice. Am. J. Respir. Cell Mol. Biol. 2003;28:478–484. doi: 10.1165/rcmb.2002-0024OC. [DOI] [PubMed] [Google Scholar]
- 17.Koepf E. K., Burtnick L. D. Interaction of plasma gelsolin with tropomyosin. FEBS Lett. 1992;309:56–58. doi: 10.1016/0014-5793(92)80738-3. [DOI] [PubMed] [Google Scholar]
- 18.Winder S. J., Walsh M. P. Calponin: Thin filament-linked regulation of smooth muscle contraction. Cell. Signalling. 1993;5:677–686. doi: 10.1016/0898-6568(93)90029-l. [DOI] [PubMed] [Google Scholar]
- 19.Plantier M., Fattoum A., Menn B., Ben-Ari Y., Der Terrosian E., Represa A. Acidic calponin immunoreactivity in postnatal rat brain and cultures: subcellular localization in growth cones, under the plasma membrane and along actin and glial filaments. Eur. J. Neurosci. 1999;11:2810–2812. doi: 10.1046/j.1460-9568.1999.00702.x. [DOI] [PubMed] [Google Scholar]
- 20.Leinweber B. D., Leavis P. C., Grabarek Z., Wang C.-L. A., Morgan K. G. Extracellular regulated kinase (ERK) interaction with actin and the calponin homology (CH) domain of actin-binding proteins. Biochem. J. 1999;344:117–123. [PMC free article] [PubMed] [Google Scholar]
- 21.Parker C. A., Takahashi K., Tao T., Morgan K. G. Agonist-induced redistribution of calponin in contractile vascular smooth muscle cells. Am. J. Physiol. 1994;267:C1262–C1270. doi: 10.1152/ajpcell.1994.267.5.C1262. [DOI] [PubMed] [Google Scholar]
- 22.Menice C. B., Hulvershorn J., Adam L. P., Wang C.-L. A., Morgan K. G. Calponin and mitogen-activated protein kinase signaling in differentiated vascular smooth muscle. J. Biol. Chem. 1997;272:25157–25161. doi: 10.1074/jbc.272.40.25157. [DOI] [PubMed] [Google Scholar]
- 23.Patil S. B., Pawar M. D., Bitar K. N. Direct association and translocation of PKC-α with calponin. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;286:G954–G963. doi: 10.1152/ajpgi.00477.2003. [DOI] [PubMed] [Google Scholar]
- 24.Fujii T., Yamana K., Ogoma Y., Kondo Y. Interaction of calponin with phospholipids. J. Biochem. 1995;117:999–1003. doi: 10.1093/oxfordjournals.jbchem.a124833. [DOI] [PubMed] [Google Scholar]
- 25.Agassandian C., Plantier M., Fattoum A., A R., Der Terrosian E. Subcellular distribution of calponin and caldesmon in rat hippocampus. Brain Res. 2000;887:444–449. doi: 10.1016/s0006-8993(00)03030-4. [DOI] [PubMed] [Google Scholar]
- 26.Childs T. J., Watson M. H., Novy R. E., Lin J. J.-C., Mak A. S. Calponin and tropomyosin interactions. Biochim. Biophys. Acta. 1992;1121:41–46. doi: 10.1016/0167-4838(92)90334-a. [DOI] [PubMed] [Google Scholar]
- 27.Way M., Pope B., Gooch J., Hawkins M., Weeds A. G. Identification of a region of segment 1 of gelsolin critical for actin binding. EMBO J. 1990;9:4103–4109. doi: 10.1002/j.1460-2075.1990.tb07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mezgueldi M., Fattoum A., Derancourt J., Kassab R. Mapping of the functional domains in the amino-terminal region of calponin. J. Biol. Chem. 1992;267:15943–15951. [PubMed] [Google Scholar]
- 29.Burgstaller G., Kranewitter W. J., Gimona M. The molecular basis for the autoregulation of calponin by isoform-specific C-terminal tail sequences. J. Cell Sci. 2002;115:2021–2029. doi: 10.1242/jcs.115.10.2021. [DOI] [PubMed] [Google Scholar]
- 30.Spudich J. A., Watt S. The regulation of rabbit skeletal muscle contraction: biochemical studies of the interaction of the tropomyosin–troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 31.Kouyama T., Mihashi K. Fluorimetry study of N-(1-pyrenyl) iodoacetamide-labelled F-actin: local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur. J. Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- 32.Feinberg J., Kwiatek O., Astier C., Diennet S., Mercy J., Heitz F., Benyamin Y., Roustan C. Capping and dynamic relation between domains 1 and 2 of gelsolin. J. Pept. Sci. 1998;4:116–127. doi: 10.1002/(SICI)1099-1387(199804)4:2%3C116::AID-PSC135%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 33.Mezgueldi M., Mendre C., Calas B., Kassab R., Fattoum A. Characterization of the regulatory domain of gizzard calponin. J. Biol. Chem. 1995;270:8867–8876. doi: 10.1074/jbc.270.15.8867. [DOI] [PubMed] [Google Scholar]
- 34.Abouzaglou J., Benistant C., Gimona M., Roustan C., Kassab R., Fattoum A. Tyrosine phosphorylation of calponins: inhibition of the interaction with F-actin. Eur. J. Biochem. 2004;271:2615–2623. doi: 10.1111/j.1432-1033.2004.04190.x. [DOI] [PubMed] [Google Scholar]
- 35.Valentin-Ranc C., Combeau C., Carlier M. F., Pantaloni D. Myosin subfragment-1 interacts with two G-actin molecules in the absence of ATP. J. Biol. Chem. 1991;266:17872–17879. [PubMed] [Google Scholar]
- 36.Méjean C., Lebart M. C., Poyer M., Roustan C., Benyamin Y. Localization and identification of actin structures involved in the filamin–actin interaction. Eur. J. Biochem. 1992;209:555–562. doi: 10.1111/j.1432-1033.1992.tb17320.x. [DOI] [PubMed] [Google Scholar]
- 37.Bryan J., Coluccio L. M. Kinetic analysis of F-actin depolymerization in the presence of platelet gelsolin and gelsolin–actin complexes. J. Cell Biol. 1985;101:1236–1244. doi: 10.1083/jcb.101.4.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 39.Dissmann E., Hinssen H. Immunocytochemical localization of gelsolin in fibroblasts, myogenic cells, and isolated myofibrils. Eur. J. Cell Biol. 1994;63:336–344. [PubMed] [Google Scholar]
- 40.Gimona M., Kaverina I., Resch G. P., Vignal E., Burgstaller G. Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol. Biol. Cell. 2003;14:2482–2491. doi: 10.1091/mbc.E02-11-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leinweber B., Tang Z. X., Stafford W. F., Chalovich J. M. Calponin interaction with α-actinin–actin: evidence for a structural role for calponin. Biophys. J. 1999;77:3208–3217. doi: 10.1016/S0006-3495(99)77151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mino T., Yuasa U., Nakamura F., Naka M., Tanaka T. Two distinct actin-binding sites of smooth muscle calponin. Eur. J. Biochem. 1998;251:262–268. doi: 10.1046/j.1432-1327.1998.2510262.x. [DOI] [PubMed] [Google Scholar]
- 43.Taniguchi S., Takeoka M., Ehara T., Hashimoto S., Shibuki H., Yoshimura N., Shigematsu H., Takahashi K., Katsuki M. Structural fragility of blood vessels and peritoneum in calponin h1-deficient mice, resulting in an increase in hematogenous metastasis and peritoneal dissemination of malignant tumor cells. Cancer Res. 2001;61:7627–7634. [PubMed] [Google Scholar]
- 44.Becker P. M., Kazi A. A., Wadgaonkar R., Pearse D. B., Kwiatkowski D., Garcia J. G. N. Pulmonary vascular permeability and ischemic injury in gelsolin-deficient mice. Am. J. Respir. Cell Mol. Biol. 2003;28:478–484. doi: 10.1165/rcmb.2002-0024OC. [DOI] [PubMed] [Google Scholar]
- 45.Chan M. W. C., El Sayegh T. Y., Arora P. D., Laschinger C. A., Overall C. M., Morrison C., McCulloch C. A. G. Regulation of intercellular adhesion strength in fibroblasts. J. Biol. Chem. 2004;279:41047–41057. doi: 10.1074/jbc.M406631200. [DOI] [PubMed] [Google Scholar]
- 46.Yoshikawa H., Taniguchi S.-i., Yamamura H., Mori S., Sugimoto M., Miyado K., Nakamura K., Nakao K., Katsuki M., Shibata N., Takahashi K. Mice lacking smooth muscle calponin display increased bone formation that is associated with enhancement of bone morphogenetic protein responses. Genes Cells. 1998;3:685–695. doi: 10.1046/j.1365-2443.1998.00214.x. [DOI] [PubMed] [Google Scholar]