Abstract
In the present study, we report the identification and characterization of MEX (MEKK1-related protein X), a protein with homology to MEKK1 that is expressed uniquely in the testis. MEX is comprises four putative zinc-binding domains including an N-terminal SWIM (SWI2/SNF2 and MuDR) domain of unknown function and two RING (really interesting new gene) fingers separated by a ZZ zinc finger domain. Biochemical analyses revealed that MEX is self-ubiquitinated and targeted for degradation through the proteasome pathway. MEX can act as an E3, Ub (ubiquitin) ligase, through the E2, Ub-conjugating enzymes UbcH5a, UbcH5c or UbcH6. A region of MEX that contains the RING fingers and the ZZ zinc finger was required for interaction with UbcH5a and MEX self-association, whereas the SWIM domain was critical for MEX ubiquitination. The expression of MEX promoted apoptosis that was induced through Fas, DR (death receptor) 3 and DR4 signalling, but not that mediated by the BH3 (Bcl-2 homology 3)-only protein BimEL or the chemotherapeutic drug adriamycin. The enhancement of apoptosis by MEX required a functional SWIM domain, suggesting that MEX ubiquitination is critical for the enhancement of apoptosis. These results indicate that MEX acts as an E3 Ub ligase, an activity that is dependent on the SWIM domain and suggest a role for MEX in the regulation of death receptor-induced apoptosis in the testes.
Keywords: MEKK1-related protein X (MEX), really interesting new gene (RING) finger, SWI2/SNF2 and MuDR (SWIM) domain, testis, ubiquitination
Abbreviations: β-gal, β-galactosidase; Ab, antibody; BH3, Bcl-2 homology 3; DR, death receptor; DTT, dithiothreitol; E1, Ub-activating enzyme; E2, Ub-conjugating enzyme; E3, Ub ligase; EST, expressed sequence tag; HECT, homologous to E6-AP (associated protein) COOH terminus; HEK-293T, human embryonic kidney-293 where T stands for large T antigen; IKKβ, I-κB kinase subunit β; MEKK1, MAPK (mitogen-activated protein kinase)/ERK (extracellular-signal-regulated kinase) kinase kinase 1; MEX, MEKK1-related protein X; ORF, open reading frame; RING, really interesting new gene; SWIM, SWI2/SNF2 and MuDR; Ub, ubiquitin; U-box, UFD2 (Ub fusion degradation protein 2) homology box
INTRODUCTION
The Ub (ubiquitin)-proteasome pathway is a major pathway for targeted proteolysis in eukaryotic cells and plays a key role in diverse cellular functions including signal transduction, cell cycle progression, differentiation and apoptosis. This proteolytic pathway involves the covalent attachment of a polyUb chain to a protein substrate by the concerted action of the Ub-activating enzyme, E1, the Ub-conjugating enzyme, E2, and the Ub-protein ligase, E3 [1]. The first step in the Ub conjugation cascade is catalysed by E1 and involves the formation of a thioester bond between the C-terminus of Ub and the active site cysteine on E1 [1]. The activated Ub is then transferred from the E1 cysteine residue to a conserved cysteine residue of an E2 enzyme [1]. Finally, the modified Ub molecules are transferred from E2 to the target protein, a process that is mediated by an E3 ligase [1]. The resulting polyUb chain formed on the protein substrate is recognized and targeted for degradation by the 26 S proteasome [2,3].
The substrate specificity of the Ub-proteasome pathway is conferred by an E3 ligase. Two distinct classes of E3 ligases have been identified: the adaptor type that contains a RING (really interesting new gene) finger domain or a U-box [UFD2 (Ub fusion degradation protein 2) homology box] and the enzymatic HECT [homologous to E6-AP (associated protein) C-terminus]-type E3s. The E3 ligases that contain the RING finger or U-box include most of the Ub-ligases identified to date and function as adaptors that bridge the interaction between the E2–Ub intermediate and the lysine substrate in the target protein [4]. By contrast, HECT-type E3s possess both docking and catalytic activity and mediate the transfer of Ub to protein substrates [5–7]. The RING finger is a domain defined by the presence of eight cysteine and histidine residues in a conserved spacing that co-ordinates two Zn2+ ions [8–10].
Many RING finger domains have been shown to directly bind E2s [9–11]. RING finger E3 ligases are present in both single polypeptides such as Cbl and multi-subunit complexes, such as that found in the SCF (Skp1p-cullin-F-box protein) complex [12–15]. The SWIM (SWI2/SNF2 and MuDR) domain is a recently identified motif with predicted zinc-binding cysteine and histidine residues the signature of which is C-x-C-xn-C-x-H (where n=6–25) [16]. Many proteins that contain the SWIM domain have been identified in prokaryotic and eukaryotic cells [16]. Certain proteins such as MEKK1 [MAPK (mitogen-activated protein kinase)/ERK (extracellular-signal-regulated kinase) kinase kinase 1] contain both a RING finger and a SWIM domain [16]. However, the function of SWIM domains is currently unknown. In the present study, we describe MEX (MEKK1-related protein X), a protein expressed uniquely in the testis, and with homology to MEKK1, that is composed of two RING fingers and one SWIM domain. We provide evidence that MEX is self-ubiquitinated and targeted for degradation, an event that is mediated by the N-terminal RING finger, and SWIM domains. Moreover, MEX promotes apoptosis induced via DR (death receptor) signalling and this activity is dependent upon the SWIM domain.
EXPERIMENTAL
Isolation of the MEX cDNAs and preparation of expression plasmids
nt cDNA sequences encoding polypeptides with homology to rat MEKK1 (National Center for Biotechnology Information, protein database accession number NP_446339) were identified in the GenBank EST (expressed sequence tag) database using TBLASTN (NCBI). The nt sequences of the EST clones, 515103 and 603042, were determined by dideoxy sequencing. The EST clone, 603042, that contains a partial ORF (open reading frame) of mouse MEX was cloned into pcDNA3-FLAG [17] to generate pcDNA3-FLAG–MEX-T1 that contains an alternatively spliced exon, which causes premature termination of the ORF by insertion of a stop codon. A construct expressing full-length MEX, pcDNA3-FLAG–MEX, was generated by replacement of the region containing the alternatively spliced exon with that of the EST clone, 515103. The entire ORF from pcDNA3-FLAG–MEX was cloned into pcDNA3-Myc [18] to generate the expression plasmid, pcDNA3-Myc-MEX. Deletion and site-directed mutants of MEX (amino acids 1–398, 1–138 and 139–398; mutations C66S, C168S, C253S and C382S) were constructed by a PCR method and cloned into pcDNA3-FLAG and/or pcDNA3-Myc. All mutant MEX constructs were verified by DNA sequencing. pCGN-HA-Ub and pcDNA3-Myc-UbcH5a were provided by Dr R. Takahashi (Department of Neurology, Kyoto University Graduate School of Medicine, Japan). pcDNA3-Fas was from Dr V. M. Dixit (Genentech, Inc., San Francisco, CA, U.S.A.), and trimerized Fas ligand (LZ-FasL) was from Dr M. E. Peter (The Ben May Institute for Cancer Research, University of Chicago, IL, U.S.A.). FLAG–BimEL was a gift from Dr S. Cory (The Walter and Eliza Hall Institute, Melbourne, Australia). Plasmids expressing FLAG–IKKβ (I-κB kinase subunit β)-K44A, caspase-8-C377S–HA, caspase-9-C287S–HA, FLAG–DR3, and FLAG–DR4 were described previously [19].
Dot-blot and Northern blot hybridization
The full-length coding region of MEX was used to generate a probe of 1.9 Kb. The DNA fragment was labelled with [α-32P]- dCTP by random priming using a commercial kit (Roche Molecular Biochemicals, Indianapolis, IN, U.S.A.). The radiolabelled probe was used hybridized to Multiple Tissue Northern blots or a Multiple Tissue Expression array (BD Clontech, Palo Alto, CA, U.S.A.) according to the manufacturer's instructions.
Expression, immunoprecipitation and purification of recombinant proteins
HEK (human embryonic kidney)-293T (where T stands for large T antigen) cells were transiently co-transfected with pcDNA3-FLAG–MEX and various expression plasmids by the calcium phosphate method as described previously [19]. After transfection for 30 h, cells were treated with or without a 10 μM proteasome inhibitor, MG-132, (Calbiochem, San Diego, CA, U.S.A.), for 24 h, subsequently suspended in lysis buffer [50 mM Tris/HCl (pH 8.0), 400 mM NaCl, 1% Nonidet P40, 1mM DTT (dithiothreitol), 10% glycerol, 30 μM MG-132 and protease inhibitors], and sonicated for 30 s. After centrifugation (15000 g for 10 min at 4 °C), FLAG-tagged proteins were immunoprecipitated using an anti-FLAG monoclonal Ab (antibody, M2) (Sigma–Aldrich, St Louis, MO, U.S.A.) and Protein G-Sepharose (Zymed Laboratories, South San Francisco, CA, U.S.A.) as described [19]. The proteins were eluted from the resin by the addition of excess FLAG peptide (Sigma–Aldrich) and immunodetected with an anti-FLAG M2 Ab.
In vitro ubiquitination assay
Reaction mixtures were prepared with 50 ng of rabbit E1 (Sigma–Aldrich), 400 ng of various recombinant E2s (Boston Biochem, Cambridge, MA, U.S.A.), purified FLAG–MEX–Sepharose beads from 1.5 mg of HEK-293T cell lysate, and 400 ng of yeast Ub (Sigma–Aldrich) in 50 mM Tris/HCl (pH 7.4), 5 mM MgCl2, 2 mM ATP and 2 mM DTT. The reaction mixtures were incubated for 90 min at 30 °C, and the beads were washed. The MEX proteins were eluted with a FLAG peptide and analysed by immunoblotting with anti-FLAG or anti-Ub Abs (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.).
Cell death assay
HEK-293T cells (1×105) were co-transfected with 83 ng of the indicated expression plasmid and 33 ng of DR3, 7 ng of DR4, 17 ng of Fas, or 8 ng of the BimEL expression plasmid in the presence of 37 ng of pEF-BOS-β-gal (β-galactosidase). The total amount of transfected plasmid DNA was adjusted with pcDNA3. At 24 h post-transfection, the cells were fixed and stained for β-gal as described previously [17]. The percentage of apoptotic cells in triplicate cultures was determined by calculating the fraction of membrane-blebbed blue cells from the total population of blue cells [17].
RESULTS AND DISCUSSION
MEX is a RING finger and SWIM domain-containing protein with homology to MEKK1
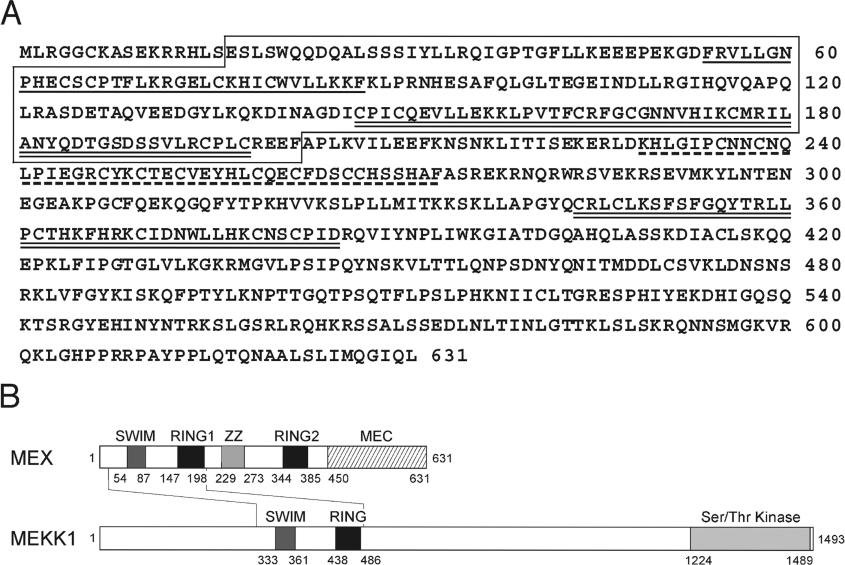
To identify novel MEKK1-like molecules, public nt databases were searched for DNA sequences encoding polypeptides with homology to the rat MEKK1 sequence. The search identified several mouse cDNA sequences encoding a polypeptide with 48% amino acid similarity (27% identity) to the N-terminal region (residues 297–490) of mouse MEKK1. Sequence analysis of the EST clones revealed an ORF encoding a 631 amino acid protein with a predicted molecular mass of approx. 72 kDa (Figure 1A). This protein was designated MEX (MEKK1-related protein X). MEX is identical to a Zswim2, an uncharacterized protein the sequence of which has been recently deposited in public databases (NCBI, accession number NP_082240). A search for a human orthologue revealed several overlapping cDNAs that could be constructed to form a single ORF encoding a 633 amino acid protein with 67% identity to the mouse sequence. The human MEX gene mapped to chromosome 2q32.1 and the corresponding mouse gene mapped to chromosome 2 E1.
Figure 1. Deduced amino acid sequence and domain structure of MEX.
(A) Amino acid sequence of mouse MEX. SWIM, RING1 and RING2, and ZZ are indicated by underlining, double-underlining, and a broken line respectively. The region homologous to MEKK1 is boxed. (B) Domain structure of MEX. Numbers correspond to amino acid residues are shown in (A). The structure of MEKK1 is shown as a comparison.
Analysis of the amino acid sequence of MEX revealed that its structure comprises four putative zinc-binding domains, an N-terminal SWIM domain, two RING fingers (RING1 and RING2) and a ZZ zinc finger domain, and an unique C-terminal domain (Figures 1A and 1B). Further analysis of protein sequences revealed that the homology between MEKK1 and MEX was restricted to the region that contains the SWIM and N-terminal RING domain (Figure 1B). The C-terminal region of MEX referred to in the present study as the MEC (MEX C-terminus) domain lacked a kinase domain and possessed no significant homology to any other protein found in public databases.
The expression of MEX is restricted to testis
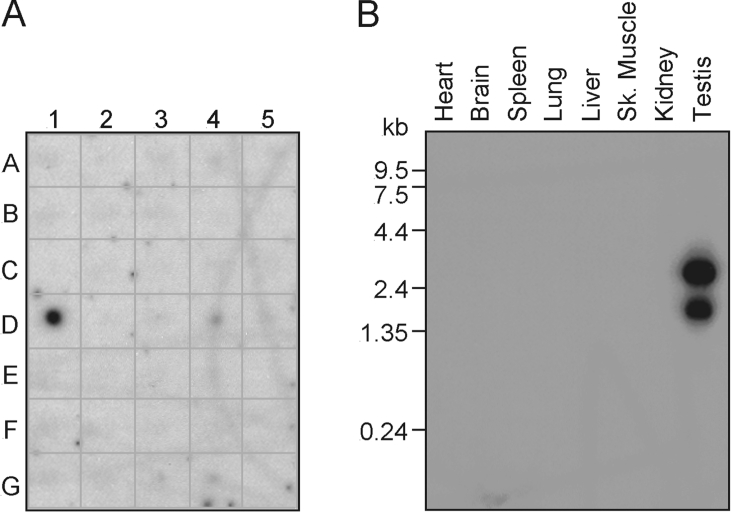
We performed dot-blot analysis of total RNA isolated from multiple mouse tissues to assess the expression of MEX. Hybridization with a MEX-specific labelled probe showed expression of MEX in testis, but not in any other adult tissue including brain, eye, liver, lung, kidney, heart, skeletal muscle, smooth muscle, pancreas, thyroid, thymus, spleen, ovary, prostate, uterus or mouse embryos at different stages of development (Figure 2A). To verify these results, mRNA samples from a panel of mouse tissues were analysed by Northern blot analysis with a labelled MEX probe. Consistent with the dot-blot results, MEX expression was detected as approx. 2.8 and 1.9 Kb transcripts in mouse testis but not in any other tissue analysed (Figure 2B).
Figure 2. Tissue distribution of MEX mRNA.
(A) Dot-blot analysis of MEX expression in mouse tissues. A1–A5: brain, eye, liver, lung and kidney respectively. B1–B5: heart, skeletal muscle, smooth muscle, blank and blank. C1–C5: pancreas, thyroid, thymus, submaxillary gland and spleen. D1–D5: testis, ovary, prostate, epididymus and uterus. E1–E5: 7-day embryo, 11-day embryo, 15-day embryo, 17-day embryo and blank. F1–F5: yeast total RNA, yeast tRNA, Escherichia coli rRNA, E. coli DNA and blank. G1–G5: blank, Cot 1 DNA, mouse DNA (100 ng), mouse DNA (500 ng) and blank. (B) Northern blot analysis of mouse tissues. The blots were analysed by autoradiography.
MEX is ubiquitinated and degraded via the proteasome
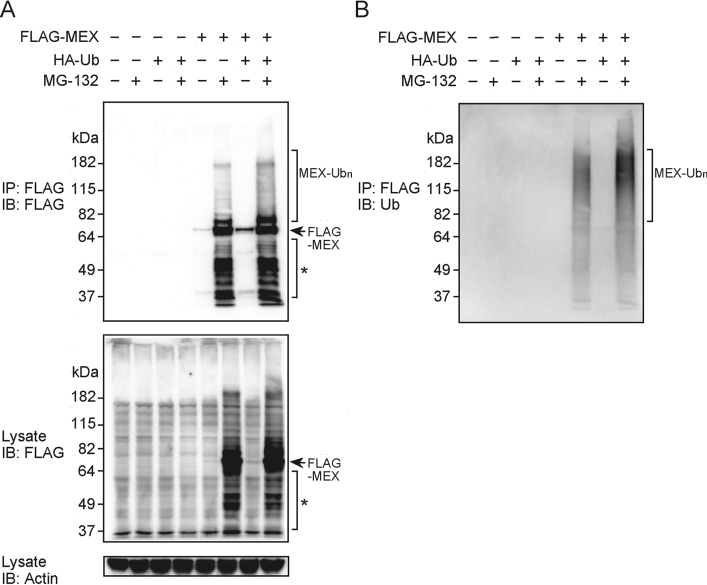
MEX contains several putative zinc-binding motifs that have been linked to ubiquitination. To test whether MEX can be ubiquitinated, we transiently co-transfected constructs producing FLAG-tagged full-length MEX and HA-tagged Ub into HEK-293T cells in the presence or absence of MG-132, an inhibitor of the proteasome. The levels of total and ubiquitinated MEX were monitored by immunoprecipitation with an anti-FLAG Ab and immunoblotting with anti-FLAG and anti-Ub Abs. In the absence of MG-132, the expression of MEX was minimal in the presence or absence of exogenous Ub (Figure 3A). By contrast, the level of MEX expression was greatly enhanced when MG-132 was added to the cultured cells (Figure 3A), suggesting that MEX is targeted for degradation via the Ub-proteasome pathway. Immunoblotting of MEX immunoprecipitates with anti-FLAG and anti-Ub Abs revealed the presence of ubiquitinated full-length and degraded MEX proteins (Figures 3A and 3B). Furthermore, ubiquitination was detected in cells that were untransfected with the Ub construct (Figure 3B), although MEX ubiquitination was enhanced by the exogenous expression of Ub (Figure 3B). We detected degraded MEX in the presence of MG-132, suggesting that there is a pathway of MEX degradation that is not sensitive to MG-132 or, alternatively, that under our experimental conditions the drug cannot totally inhibit proteasome-dependent degradation.
Figure 3. Ubiquitination of MEX in vivo.
(A) HEK-293T cells were co-transfected with HA-tagged ubiquitin (HA–Ub) and/or FLAG-tagged MEX (FLAG–MEX). Cells were treated with 10 μM MG-132 or left untreated for 24 h. The lysates were subjected to immunoprecipitation (IP) with an anti-FLAG Ab, and immunocomplexes were eluted with excess FLAG peptide. The eluted proteins were analysed by immunoblotting (IB) with an anti-FLAG Ab (top panel). Total lysates were blotted with an anti-FLAG Ab (middle panel) or an anti-actin Ab (bottom panel). (B) After stripping, the membrane used in (A) was immunoblotted with an anti-Ub Ab. Ubiquitinated full-length, and degraded (*) MEX proteins are indicated by brackets.
MEX exhibits E3 Ub ligase activity through UbcH5a, UbcH5c and UbcH6
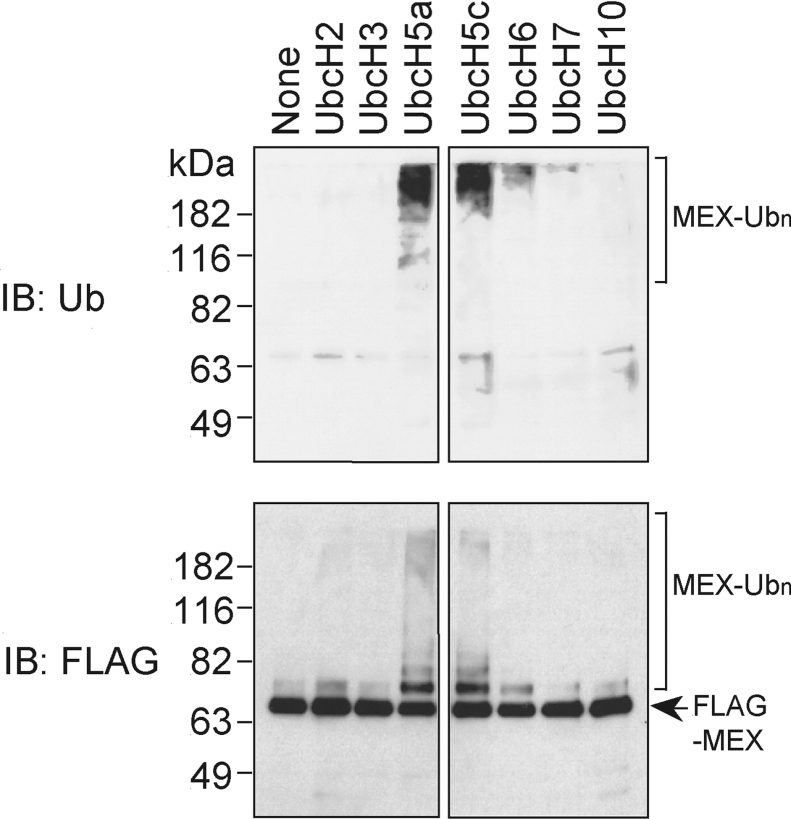
We next assessed the E3 Ub ligase activity of MEX using a panel of recombinant E2s and purified MEX. Using this in vitro assay, MEX was ubiquitinated in the presence of the E2 enzymes, UbcH5a and UbcH5c, and to a lesser degree with UbcH6, but not in the presence of UbcH2, UbcH3, UbcH7 or UbcH10 (Figure 4, upper panel). Immunoblotting revealed equal loading of purified MEX in the assay (Figure 4, lower panel).
Figure 4. MEX acts as a E3 Ub ligase in vitro.
Immunopurified MEX was incubated with purified E1 and various E2s for 90 min at 30 °C. The MEX-containing beads were washed and immunocomplexes were eluted with a FLAG peptide. The eluted proteins were immunoblotted with an anti-FLAG Ab (lower panel), and an anti-Ub Ab (upper panel).
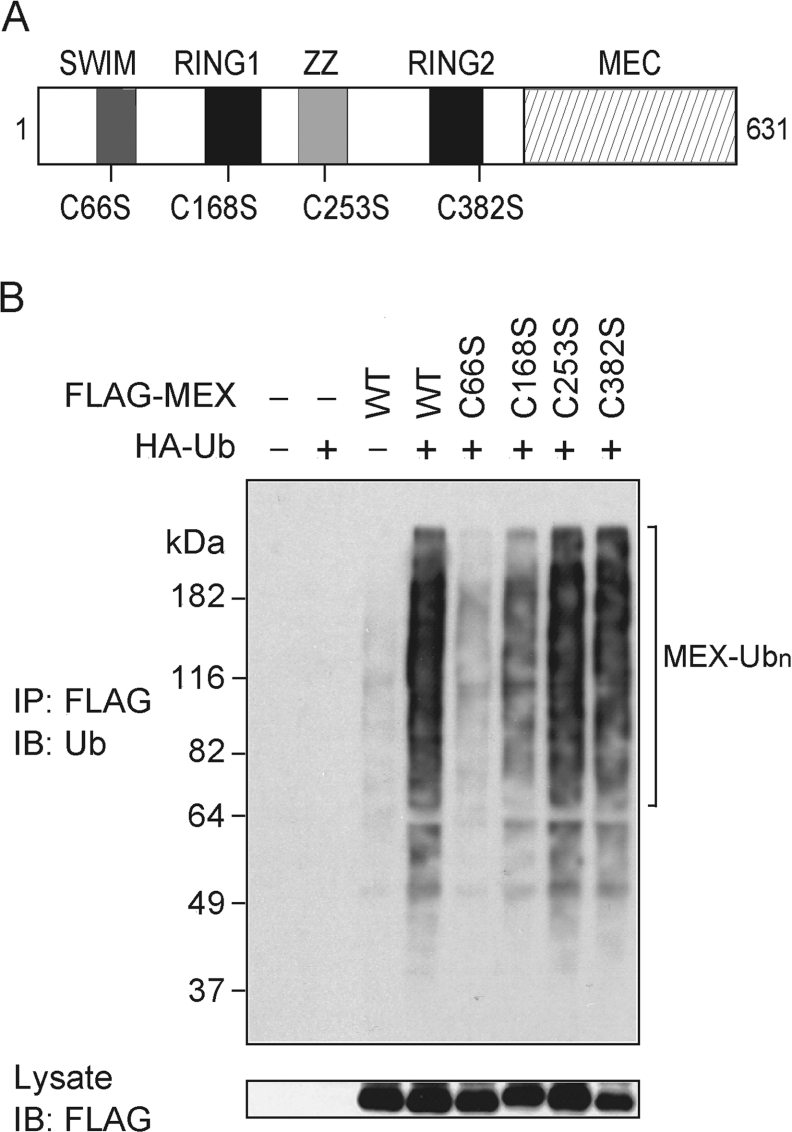
The SWIM domain is essential for MEX ubiquitination
MEX comprises four putative zinc-finger-like domains that could mediate self-ubiquitination of MEX. The RING finger domains are known to act as an E3 ligase but there is no evidence that SWIM domains can promote protein ubiquitination [16]. To determine the role of the zinc-binding motifs in MEX ubiquitination, we generated point mutants in which an essential cysteine residue in each of the zinc-finger domains was replaced by a serine residue (Figure 5A). Immunoblot analysis of cell extracts showed similar levels of the MEX mutants after transfection of each construct into HEK-293T cells (Figure 5B, lower panel). Immunoprecipitation of MEX followed by immunoblotting with an anti-Ub Ab revealed that the C66S mutation in the SWIM domain almost abolished the ubiquitination of MEX (Figure 5B, upper panel). The C168S mutation located in the N-terminal RING domain of MEX also exhibited decreased ubiquitination, whereas mutation of the ZZ domain (C253S) and C-terminal RING domain (C382S) had little or no effect on MEX ubiquitination (Figure 5B).
Figure 5. Mutational analysis of MEX ubiquitination.
(A) Schematic diagram of mutant MEX proteins. The positions of point mutations introduced into the MEX gene are shown. (B) HEK-293T cells were co-transfected with HA-tagged Ub and/or the indicated FLAG-tagged MEX constructs. Cells were treated as described in Figure 3(A). Immunocomplexes were analysed by immunoblotting (IB) with an anti-Ub Ab (upper panel). Total lysates were blotted with an anti-FLAG Ab (lower panel). IP, immunoprecipitate.
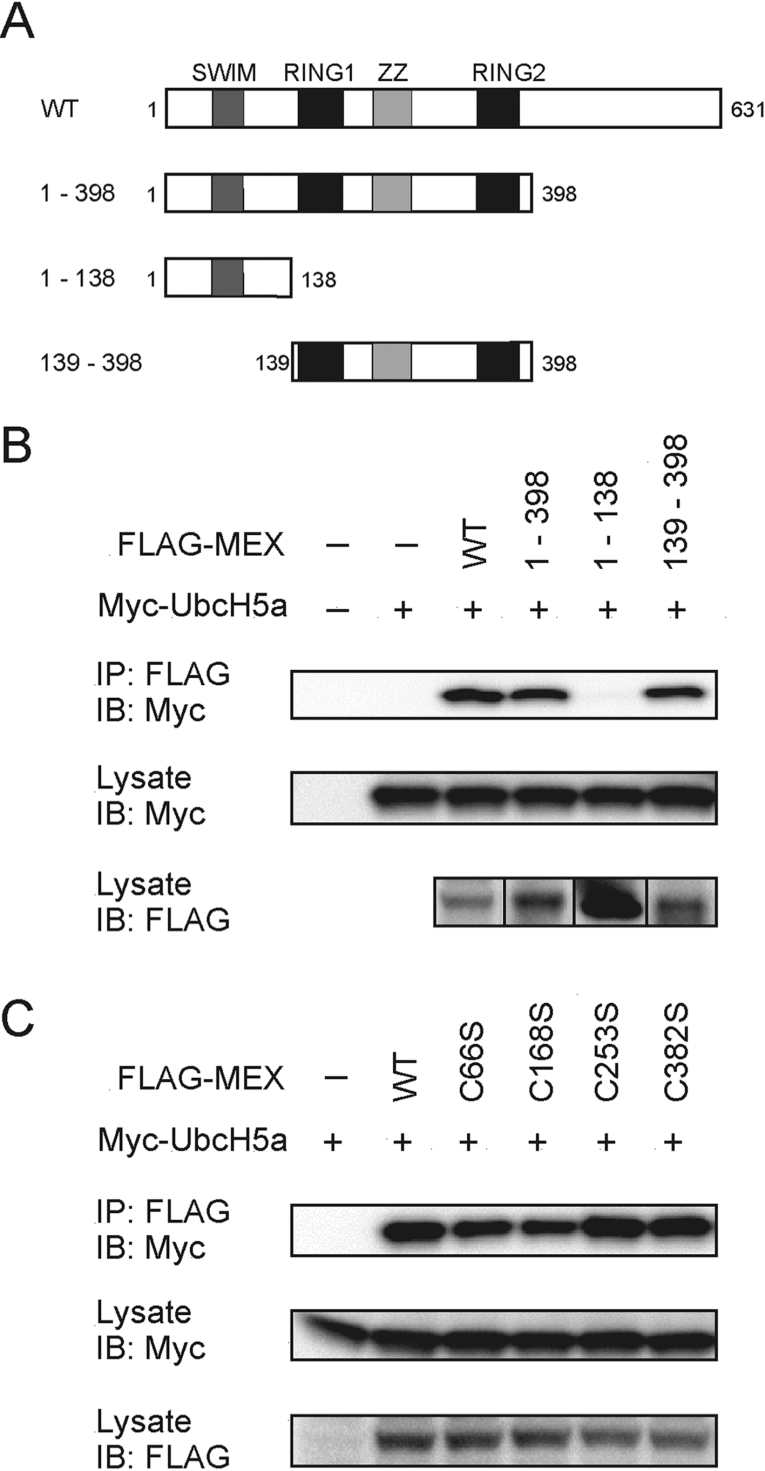
The region of MEX containing residues 139–398 interacts with UbcH5a
We next assessed the region of MEX involved in the interaction with UbcH5a. Analysis of MEX truncation mutants revealed that residues 139–398 that contain the RING1, RING2 and ZZ-zinc finger domains mediate the interaction with UbcH5a (Figures 6A and 6B). Further analysis showed that the critical cysteine residues that are required for MEX ubiquitination were dispensable for the association with UbcH5a, although the association of the mutant, C168S, with UbcH5a was slighted decreased when compared with that observed using wild-type MEX (Figure 6C).
Figure 6. MEX binds with UbcH5a as an E2.
(A) Wild-type (WT) and deletion mutants of MEX proteins. (B) HEK-293T cells were co-transfected with the indicated FLAG-tagged MEX and the Myc-tagged UbcH5a constructs. Cells were treated as described in Figure 3(A) using an anti-Myc Ab (top panel) for immunoblotting (IB). Total lysates were blotted with an anti-Myc (middle panel) or an anti-FLAG Ab (bottom panel). (C) HEK-293T cells were co-transfected with the indicated constructs and protein interactions were analysed as described in (B). IP, immunoprecipitate.
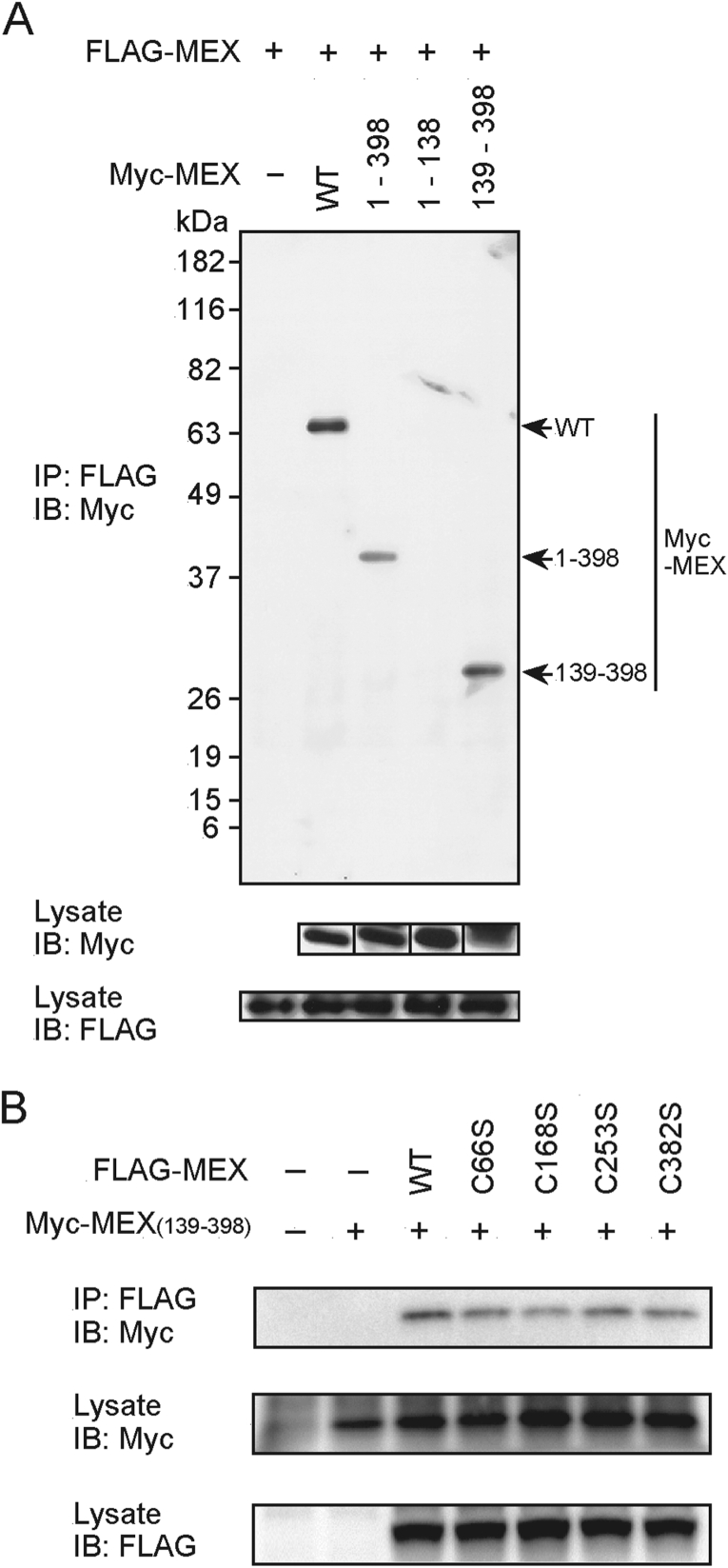
MEX self-associates
We next assessed the ability of MEX to dimerize, by immunoprecipitation followed by immunoblotting. This analysis revealed that MEX associated with itself (Figure 7A) and that this activity was independent of the SWIM domain and was mediated through the region containing residues 139–398 (Figures 7A and 7B). Further analysis revealed that the critical cysteine residues that are required for MEX ubiquitination were not essential for MEX self-association, although the interaction of C168S with MEX (amino acids 139–398) was slightly decreased when compared with that observed for wild-type MEX (Figure 7B). These results indicate that the region containing the RING1, RING2 and ZZ domains is involved in both the interaction with UbcH5a and MEX self-association.
Figure 7. MEX self-association is independent of the SWIM domain.
(A) HEK-293T cells were co-transfected with FLAG-tagged MEX and the indicated Myc-tagged MEX constructs. Cells were treated as described in Figure 3(A) using an anti-Myc Ab (top panel) for immunoblotting (IB). Total lysates were blotted with an anti-Myc (middle panel) or an anti-FLAG Ab (bottom panel). (B) HEK-293T cells were co-transfected with the Myc-tagged MEX mutant (amino acids 139–398) construct and the indicated FLAG-tagged MEX constructs. Protein interactions were analysed as described in (A). IP, immunoprecipitate.
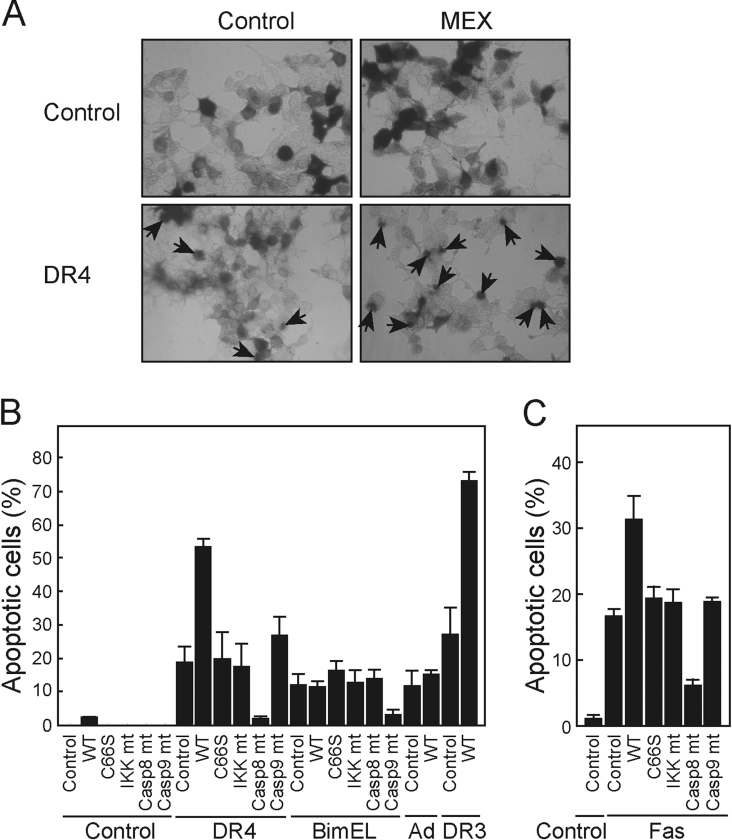
MEX regulates Fas-, DR3- and DR4-mediated apoptosis
The Ub-proteasome pathway is known to regulate a diverse array of cellular processes including cell cycle, apoptosis and stress responses [1]. We tested whether the expression of MEX regulates apoptosis in HEK-293T cells. In these experiments, we co-expressed MEX and proteins that activate the DR or mitochondrial pathways of apoptosis. The expression of MEX alone did not induce significant apoptosis in HEK-293T cells above that found after transfection with the control plasmid (Figure 8A). As expected, expression of DR4 induced apoptosis in HEK-293T cells (Figure 8A). Notably, DR4-induced apoptosis was significantly increased by the expression of MEX but not by a dominant negative form of IKKβ that was used as a control (Figures 8A and 8B). Consistent with this finding, MEX also enhanced apoptosis induced by DR3 (Figure 8B) and Fas (Figure 8C), but not that mediated by BimEL, a BH3-only family member, or the chemotherapeutic drug adriamycin, two pro-apoptotic stimuli that activate the mitochondrial pathway [20,21]. Importantly, the ability of MEX to promote DR4- and Fas-induced induced apoptosis was abrogated by mutation of the critical cysteine residue at position 66 in the SWIM domain (Figures 8B and 8C). These results indicate that a functional SWIM domain is critical for MEX to promote DR-induced apoptosis. DR4- and Fas-induced apoptosis were inhibited by the expression of dominant negative forms of caspase-8, a caspase that is essential for apoptosis mediated through DR signalling, but not dominant negative forms of caspase-9 or IKKβ (Figure 8B). Conversely, BimEL-induced apoptosis was suppressed by a dominant interfering mutant of caspase-9 but not by dominant negative caspase-8 (Figure 8B).
Figure 8. Enhancement of DR-induced apoptosis by MEX.
(A) HEK-293T cells were co-transfected with DR4 and/or MEX expression plasmids in the presence of pEF-BOS-β-gal. After 24 h of transfection, the cells were fixed and stained for β-gal activity. Arrows show round-up apoptotic cells with membrane blebbing. (B) and (C) HEK-293T cells were co-transfected with control vector, DR3, DR4, BimEL, or Fas expression plasmids and wild-type MEX (WT), MEX mutant with replacement of critical cysteine residue in the SWIM domain (C66S), IKKβ K44A (IKK mt), caspase-8-C377S (Casp8 mt), caspase-9-C287S (Casp9mt), or vector control in the presence of pEF-BOS-β-gal. The cells transfected with Fas were treated with 1 μg/ml trimerized Fas ligand (C). As an additional control, cells transfected with wild-type MEX or vector control were treated with 0.1 μg/ml adriamycin (Ad). After 24 h of transfection, the percentage of apoptotic cells±S.D. was calculated in triplicate cultures.
In the present study, we report the initial characterization of MEX, a novel protein with multiple zinc-finger domains. We provide evidence that MEX is ubiquitinated and degraded through the proteasome pathway. A main finding is the identification of the SWIM domain of MEX as a critical zinc-finger motif required for ubiquitination. The SWIM domain was originally identified as a zinc-finger-like module with a C-x-C-xn-C-x-H signature distinct from that found in other zinc-binding domains including RING fingers and ZZ motifs [16], but its function was unknown. To our knowledge, this is the first report demonstrating that the SWIM domain functions in the regulation of protein ubiquination. Because the SWIM domain is found in a variety of prokaryotic and eukaryotic proteins, it is likely that this zinc-finger domain regulates multiple cellular processes through protein ubiquitination. MEKK1 contains a SWIM domain and a RING finger or PHD (plant homoeodomain) [22]. In the case of MEKK1, its RING finger domain mediates ubiquitination and degradation of ERK1 and ERK2 [22]. The role of the SWIM domain in the regulation of MEKK1 is unknown. Our studies suggest that the SWIM domain of MEKK1 regulates self-ubiquitination or that of MEKK1-interacting proteins. The expression of MEX is restricted to the testis but its physiological function remains unclear. Our studies indicate that MEX promotes apoptosis induced by Fas, DR3 and DR4 but not that mediated by BimEL or adriamycin. These results suggest that MEX targets a component(s) of DR pathways but not factors such as caspase-3 that are shared with the mitochondrial apoptotic pathway. Previous studies have shown that Fas, DR4 and their ligands are expressed in germ cells of human and rat testis [23,24]. Thus MEX could be involved in the regulation of paracrine or autocrine germ cell apoptosis that is induced during germ cell differentiation or hormonal withdrawal. Future studies should address these possibilities.
Acknowledgments
This work was supported by grants CA84064 (to G.N.) and GM60421 (to N.I.) from the National Institutes of Health.
References
- 1.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Coux O., Tanaka K., Goldber A. L. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 3.Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 4.Cyr D. M., Hohfeld J., Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 5.Scheffner M., Huibregtse J. M., Vierstra R. D., Howley P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 6.Scheffner M., Nuber U., Huibregtse J. M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature (London) 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 7.Huibregtse J. M., Scheffner M., Beaudenon S., Howley P. M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freemont P. S. The RING finger. A novel protein sequence motif related to the zinc finger. Ann. N.Y. Acad. Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 9.Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freemont P. S. Ubiquitination: RING for destruction? Curr. Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 11.Hu G., Zhang S., Vidal M., Baer J. L., Xu T., Fearon E. R. Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:2701–2714. doi: 10.1101/gad.11.20.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joazeiro C. A., Wing S. S., Huang H., Leverson J. D., Hunter T., Liu Y. C. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science (Washington, DC) 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 13.Kamura T., Koepp D. M., Conrad M. N., Skowyra D., Moreland R. J., Iliopoulos O., Lane W. S., Kaelin W. G., Jr, Elledge S. J., Conaway R. C., et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science (Washington, DC) 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 14.Skowyra D., Koepp D. M., Kamura T., Conrad M. N., Conaway R. C., Conaway J. W., Elledge S. J., Harper J. W. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science (Washington, DC) 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 15.Jackson P. K., Eldridge A. G. The SCF ubiquitin ligase: an extended look. Mol. Cell. 2002;9:923–925. doi: 10.1016/s1097-2765(02)00538-5. [DOI] [PubMed] [Google Scholar]
- 16.Makarova K. S., Aravind L., Koonin E. V. SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem. Sci. 2002;27:384–386. doi: 10.1016/s0968-0004(02)02140-0. [DOI] [PubMed] [Google Scholar]
- 17.Inohara N., Koseki T., Chen S., Wu X., Nunez G. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 1998;274:2526–2533. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inohara N., Koseki T., Chen S., Benedict M. A., Nunez G. Identification of regulatory and catalytic domains in the apoptosis nuclease DFF40/CAD. J. Biol. Chem. 1999;274:270–274. doi: 10.1074/jbc.274.1.270. [DOI] [PubMed] [Google Scholar]
- 19.Inohara N., Koseki T., del Peso L., Hu Y., Yee C., Chen S., Carrio R., Merino J., Liu D., Ni J., Nunez G. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-κB. J. Biol. Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi H., Wang H. G. Bcl-XL protects BimEL-induced Bax conformational change and cytochrome c release independent of interacting with Bax or BimEL. J. Biol. Chem. 2002;277:41604–41612. doi: 10.1074/jbc.M207516200. [DOI] [PubMed] [Google Scholar]
- 21.Kotamraju S., Kalivendi S. V., Konorev E., Chitambar C. R., Joseph J., Kalyanaraman B. Oxidant-induced iron signaling in Doxorubicin-mediated apoptosis. Methods Enzymol. 2004;378:362–382. doi: 10.1016/S0076-6879(04)78026-X. [DOI] [PubMed] [Google Scholar]
- 22.Lu Z., Xu S., Joazeiro C., Cobb M. H., Hunter T. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell. 2002;9:945–956. doi: 10.1016/s1097-2765(02)00519-1. [DOI] [PubMed] [Google Scholar]
- 23.Grataroli R., Vindrieux D., Gougeon A., Benahmed M. Expression of tumor necrosis factor-α-related apoptosis-inducing ligand and its receptors in rat testis during development. Biol. Reprod. 2002;66:1707–1715. doi: 10.1095/biolreprod66.6.1707. [DOI] [PubMed] [Google Scholar]
- 24.Grataroli R., Vindrieux D., Selva J., Felsenheld C., Ruffion A., Decaussin M., Benahmed M. Characterization of tumour necrosis factor-α-related apoptosis-inducing ligand and its receptors in the adult human testis. Mol. Hum. Reprod. 2004;10:123–128. doi: 10.1093/molehr/gah016. [DOI] [PubMed] [Google Scholar]