Abstract
Helicobacter pylori was previously reported to lack a stringent response. In contrast, we show that after nutrient downshift, H. pylori produced abundant ppGpp and less total RNA. pH downshift also caused (p)ppGpp accumulation. Our observations indicate that nutrient deprivation and acid shock activate the stringent response in H. pylori.
The stringent response controls the ability to adapt to certain nutrient stress conditions, is associated with the onset of stationary phase in many bacteria, and directs a general decrease in metabolic activity. Hallmarks of the stringent response include rapid accumulation of guanosine tetraphosphate (ppGpp) and inhibition of stable RNA (sRNA [rRNA and tRNA]) synthesis.
Using model systems like Escherichia coli, it has been shown that upon amino acid starvation, the presence of uncharged tRNA molecules triggers ribosome-associated RelA to produce guanosine pentaphosphate (pppGpp), which is subsequently hydrolyzed to ppGpp (4). ppGpp is thought to bind to and alter the affinity of RNA polymerase for various promoters, such as those associated with adaptation to adverse conditions (4, 6). In E. coli, SpoT, a protein homologous to RelA, is responsible for ppGpp hydrolysis (4) but also has the capacity to synthesize pppGpp (25) under certain conditions (21).
All bacteria have either separate RelA and SpoT proteins, similar to E. coli, or dual-function enzymes (23, 24) that both synthesize pppGpp and hydrolyze ppGpp at different active sites (13). Since there is at least one relA/spoT homolog in all bacteria (16), the stringent response serves as a highly conserved means of fine-tuning metabolic activity in response to stresses.
Recent work has shown that persistence of pathogenic bacteria within specialized host niches requires the stringent response (9, 12, 17). For example, the stringent response allows the persistence of Mycobacterium tuberculosis within the host (17), the activation of pathogenesis-related genes in Legionella pneumophila (2, 12), and the interaction of the enteric pathogen Campylobacter jejuni with host cells (9). Such observations strongly suggest that changes brought about by ppGpp allow these bacteria to adjust to and thrive within a specialized host environment.
Although the genes responsible for ppGpp production are broadly conserved and required for the success of several pathogenic interactions, it was previously reported that Helicobacter pylori, a widespread gram-negative bacterium that colonizes the stomach and causes peptic ulcer disease and gastric carcinoma (7), lacks a stringent response (18). In that study, initiation of the stringent response in H. pylori was tested by addition of the amino acid biosynthesis inhibitors pseudomonic acid and serine hydroxamate (SH). After addition of these compounds, the authors assayed protein synthesis and the abundance of a single rRNA transcript. They observed that although translation was inhibited, rRNA continued to accumulate. The authors concluded that H. pylori does not initiate the stringent response during translational pausing and that this characteristic was unique among all eubacteria (18). Despite the fact that both sequenced H. pylori strains harbor relA/spoT homologs annotated as spoT (8), the authors further hypothesized that bacteria inhabiting “protected” niches (e.g., the epithelium of the stomach), in contrast to those found in the general environment, do not require and thereby have lost the ability to induce the stringent response (18).
We demonstrate that, in contrast to the previous conclusions (18), H. pylori has a pronounced stringent response, as evidenced by the production of significant amounts of ppGpp and a marked decrease in sRNA under stringent total nutrient starvation conditions. Three H. pylori strains were assayed for ppGpp production: the sequenced strains J99 and 26695 (1, 20) and strain G27, a strain commonly used to investigate H. pylori interactions with gastric epithelial cells (10, 11, 19). Strains were grown in rich medium (brucella broth plus 10% fetal bovine serum) under microaerobic conditions to an early log optical density at 600 nm of ∼0.2. The bacteria were then washed and transferred to either morpholinepropanesulfonic acid (MOPS)-MGS (14) lacking mannitol (22), a defined minimal medium that contains no carbon or phosphate, or fresh rich medium. ppGpp production was assayed by an established protocol (3). In brief, immediately after transfer to new medium, samples were labeled with 100 μCi/ml 32Pi (Amersham) for 45 to 60 min under microaerobic conditions and then treated with an equal volume of 2 M formic acid to lyse cells and extract nucleotides. Small volumes (typically 3 μl) were spotted onto polyethyleneimine-cellulose thin-layer chromatography (TLC) plates and developed in 1.5 M KH2PO4 to visualize intracellular nucleotide pools. Intensity was determined by autoradiograph, phosphorimager (Bio-Rad), and densitometry analyses. Figure 1 shows that upon nutrient downshift, all three H. pylori strains produced significant amounts of ppGpp, whereas the same strains produced little to no ppGpp when transferred to rich medium.
FIG. 1.
H. pylori produces ppGpp upon nutrient downshift. Three wild-type strains of H. pylori, J99, 26695, and G27, were grown to early log phase in rich medium (brucella broth plus 10% fetal bovine serum), shifted to either new rich medium or minimal medium lacking carbon sources and phosphate (MOPS-MGS without mannitol or phosphate: 50 mM MOPS [pH 7.4], 1 mM MgSO4, 0.25 mM CaCl2, 19 mM glutamic acid, and 0.004 mM biotin), and immediately labeled with 32Pi. Nucleotides were resolved by TLC.
Stringent control in organisms such as E. coli also results in inhibition of sRNA synthesis (4). To test this in H. pylori, we measured total RNA produced upon a nutrient downshift. As previously reported, H. pylori does not harbor uracil uptake machinery (18); thus, assays of RNA turnover involving [3H]uracil uptake cannot be executed in H. pylori. Nonetheless, as cessation of sRNA synthesis will result in a net decrease in total RNA (∼90% of which is sRNA) over time, total RNA was harvested from G27 grown to log phase and then either (i) maintained in rich medium, (ii) shifted to minimal medium (as in the ppGpp assays described above), or (iii) shifted to minimal medium plus chloramphenicol (25 μg/ml), a bacteriostatic agent that interferes with protein synthesis. It is known that, in bacteria like E. coli, halting protein synthesis by treatment with chloramphenicol causes a “relaxed” phenotype in which normal “stringent” responses do not occur, even under severe starvation conditions (4). Viability (CFU) and RNA levels were monitored for 3 h under microaerobic conditions. As expected, H. pylori did not grow in the minimal medium, although numbers of CFU in both nutrient-downshifted samples remained constant with no loss of viability over the entire 3 h (data not shown). However, the total amount of RNA per CFU declined dramatically for G27 shifted to minimal medium without chloramphenicol, in contrast to G27 maintained in rich medium or to G27 shifted to minimal medium plus chloramphenicol (Fig. 2A).
FIG. 2.
Nutrient downshift of H. pylori causes a protein synthesis-dependent decrease in sRNA (A) and a relative increase in spoT RNA versus 16S rRNA (B). (A) Strain G27 was grown to early log phase in rich medium and then transferred either to new rich medium, to minimal medium lacking carbon sources and phosphate (MOPS-MGS without mannitol or phosphate), or to minimal medium plus 25 μg/ml chloramphenicol and incubated microaerobically at 37°C for 3 h. Samples were harvested at the indicated times and assayed for total RNA (∼90% of which is sRNA) and viability (CFU counts). RNA was quantified by spectrophotometric analysis. Average amounts (femtograms) of RNA per CFU are shown. Samples from rich medium are solid black; samples from minimal medium are gray; samples from minimal medium plus chloramphenicol are hatched gray lines. CFU counts in minimal medium-shifted samples remained constant throughout the experimental time course (data not shown). (B) G27 was grown as described above and shifted to either rich or minimal medium for the indicated times. RNA was harvested, reverse transcribed, and subjected to quantitative PCR analyses using the TaqMan system from ABI. Primers used were 16S-fwd (5′-CAGCCATGTTGCGGTGAAT-3′), 16S-rev (5′-TGTGACGGGCGGTGAGTA-3′), 16S probe (5′-6FAM-CGTTCCCGGGTCTT-3′), spoT-fwd (5′-ACCTCGTTTCATTTGGATGGAT-3′), spoT-rev (5′-TGGATGCGCAAATGGTTTT-3′), and spoT probe (5′-6FAM-AGCTTAAAACTTCTAAGGCT-3′).
To further support these findings, precise levels of spoT RNA and 16S rRNA were assayed by reverse transcription-quantitative PCR analyses. We observed a significant (>3-fold) increase in the ratio of spoT RNA to 16S rRNA in the minimal medium samples over time compared to rich medium samples (Fig. 2B), suggesting either a relative increase in spoT RNA, a relative decrease in 16S rRNA, or a combination of both during starvation conditions. Together, the observations presented in Fig. 2 are consistent with the notion that H. pylori indeed mounts a stringent response that is induced during total nutrient deprivation.
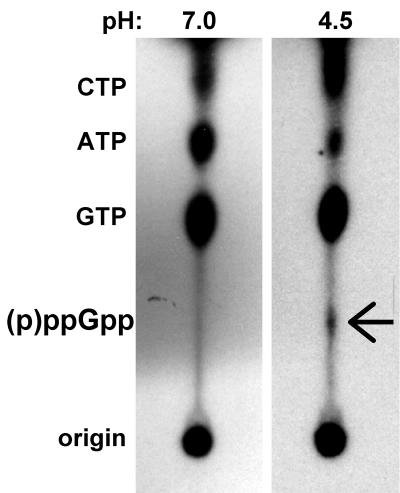
Recently, a pH downshift was shown to result in numerous gene expression changes in H. pylori (15). We therefore sought to determine whether H. pylori initiates the stringent response during a pH downshift. Strain G27 was grown to early log phase in rich medium at pH 7.0 and then either downshifted to pH 4.5 by a 1:1 dilution in rich medium at pH 3.0 (achieved by addition of HCl to the medium) or maintained at pH 7.0 by dilution in medium at pH 7.0 and labeled with 32Pi for 50 min. CFU stability in mock-labeled samples indicated that bacterial viability was unaffected by the incubation at pH 4.5 and that the pH of the media remained constant throughout the experiment (data not shown). We also note that a pH of 4.5 is slightly lower than that used in the above-mentioned work (15). We observed moderate but significant and reproducible (p)ppGpp accumulation during pH downshift compared to the nonshifted control (Fig. 3). By densitometer analysis, the amount of (p)ppGpp produced upon pH downshift was ∼4.5-fold higher than the nonshifted control. These data, consistent with the observation that the stringent response-related gene gppA is up-regulated upon acid induction (15), imply a role for the stringent response in the adjustment of H. pylori to survival within its specialized niche, the stomach.
FIG. 3.
H. pylori produces (p)ppGpp upon pH downshift. H. pylori strain G27 was grown to early log phase in rich medium at neutral pH and then diluted 1:1 either in rich medium at neutral pH (final pH 7.0) or in rich medium at pH 3.0 (final pH 4.5) and labeled with 32Pi. Nucleotides were resolved by TLC. (p)ppGpp corresponding to guanosine penta- and tetraphosphate in the pH 4.5 sample is indicated by the arrow. This spot was quantified by densitometer analysis and found to be ∼4.5-fold higher than in the pH 7.0 control.
It has recently been shown in E. coli that only very specific amino acid deprivations trigger the stringent response (5). Furthermore, stringent response regulation in organisms harboring a single dual-function RelA/SpoT protein is neither well understood nor necessarily directed by the same signals that activate RelA (i.e., SH, uncharged tRNA, etc.) in E. coli. In our assays for ppGpp production in bacteria such as Sinorhizobium meliloti, C. jejuni, and H. pylori, we have found that addition of amino acid analogs such as SH to rich medium is ineffective (data not shown). Therefore, signals that trigger the RelA-dependent stringent response in E. coli might fail to do so in these organisms. Although the authors of the prior publication observed an effect on protein synthesis, they also concluded that sRNA continued to accumulate (18). However, only a single rRNA transcript was analyzed. Thus, two alternative explanations are that (i) the particular rRNA transcript assayed was expressed from an unregulated rRNA promoter and (ii) as with ppGpp accumulation, addition of SH to rich medium fails to elicit a strong effect on sRNA levels in H. pylori. Finally, the authors suggested that the presence of only a spoT homolog (and not relA) in H. pylori could explain the lack of a traditional stringent response under the conditions observed. We and others have found that an increasing number of gram-negative bacteria, including S. meliloti and the close H. pylori relative C. jejuni, harbor a single, dual-function RelA/SpoT enzyme that mediates the stringent response in these bacteria (9, 22). Indeed, we have recently found that the H. pylori spoT gene fully mediates its stringent response, which in turn controls several important phenotypes (K. Mouery, B. Rader, E. Gaynor, and K. Guillemin, unpublished observations).
In summary, we have found that H. pylori has a clear and defined stringent response during a total nutrient downshift. Our work suggests that ppGpp accumulation may be important for survival of H. pylori in the acidic stomach environment, implying possible roles for the H. pylori stringent response not only in surviving general stresses but also in adaptation to a highly specialized niche.
Acknowledgments
We thank Sharon Long and Stanley Falkow for support and encouragement throughout the course of this work, Sarah Svensson and George Spiegelman for helpful discussions, Karen Guillemin and Kyle Mouery for helpful discussions and assistance with RNA experiments, and Hirofumi Hara and William Mohn for assistance with RT-qPCR experiments.
D.H.W. was supported by NIH grant GM30962 to Sharon Long. E.C.G. is supported by a Career Development Award from the Burroughs Wellcome Fund and a grant from the Canadian Institutes of Health Research.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 3.Cashel, M. 1969. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J. Biol. Chem. 244:3133-3141. [PubMed] [Google Scholar]
- 4.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 5.Chang, D. E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289-306. [DOI] [PubMed] [Google Scholar]
- 6.Chatterji, D., and A. Kumar Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 7.Cover, T. L., and S. R. Blanke. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 3:320-332. [DOI] [PubMed] [Google Scholar]
- 8.Doig, P., B. L. de Jonge, R. A. Alm, E. D. Brown, M. Uria-Nickelsen, B. Noonan, S. D. Mills, P. Tummino, G. Carmel, B. C. Guild, D. T. Moir, G. F. Vovis, and T. J. Trust. 1999. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol. Mol. Biol. Rev. 63:675-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaynor, E. C., D. H. Wells, J. K. MacKichan, and S. Falkow. 2005. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol. Microbiol. 56:8-27. [DOI] [PubMed] [Google Scholar]
- 10.Guillemin, K., N. R. Salama, L. S. Tompkins, and S. Falkow. 2002. Cag pathogenicity island-specific responses of gastric epithelial cells to Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 99:15136-15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, B. P., and J. J. Mekalanos. 2002. Rapid genetic analysis of Helicobacter pylori gastric mucosal colonization in suckling mice. Proc. Natl. Acad. Sci. USA 99:8354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 13.Hogg, T., U. Mechold, H. Malke, M. Cashel, and R. Hilgenfeld. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117:57-68. [DOI] [PubMed] [Google Scholar]
- 14.Mendrygal, K. E., and J. E. Gonzalez. 2000. Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 182:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittenhuber, G. 2001. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J. Mol. Microbiol. Biotechnol. 3:585-600. [PubMed] [Google Scholar]
- 17.Primm, T. P., S. J. Andersen, V. Mizrahi, D. Avarbock, H. Rubin, and C. E. Barry III. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182:4889-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scoarughi, G. L., C. Cimmino, and P. Donini. 1999. Helicobacter pylori: a eubacterium lacking the stringent response. J. Bacteriol. 181:552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA 96:14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 21.Vinella, D., C. Albrecht, M. Cashel, and R. D'Ari. 2005. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 56:958-970. [DOI] [PubMed] [Google Scholar]
- 22.Wells, D. H., and S. R. Long. 2002. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol. Microbiol. 43:1115-1127. [DOI] [PubMed] [Google Scholar]
- 23.Wendrich, T. M., C. L. Beckering, and M. A. Marahiel. 2000. Characterization of the relA/spoT gene from Bacillus stearothermophilus. FEMS Microbiol. Lett. 190:195-201. [DOI] [PubMed] [Google Scholar]
- 24.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 26:65-79. [DOI] [PubMed] [Google Scholar]
- 25.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]