Abstract
Chimeras created by fusing the monomeric red fluorescent protein (RFP) to a bacterial lipoprotein signal peptide (lipoRFPs) were visualized in the cell envelope by epifluorescence microscopy. Plasmolysis of the bacteria separated the inner and outer membranes, allowing the specific subcellular localization of lipoRFPs to be determined in situ. When equipped with the canonical inner membrane lipoprotein retention signal CDSR, lipoRFP was located in the inner membrane in Escherichia coli, whereas the outer membrane sorting signal CSSR caused lipoRFP to localize to the outer membrane. CFSR-RFP was also routed to the outer membrane, but CFNSR-RFP was located in the inner membrane, consistent with previous data showing that this sequence functions as an inner membrane retention signal. These four lipoproteins exhibited identical localization patterns in a panel of members of the family Enterobacteriaceae, showing that the lipoprotein sorting rules are conserved in these bacteria and validating the use of E. coli as a model system. Although most predicted inner membrane lipoproteins in these bacteria have an aspartate residue after the fatty acylated N-terminal cysteine residue, alternative signals such as CFN can and probably do function in parallel, as indicated by the existence of putative inner membrane lipoproteins with this sequence at their N termini.
Bacterial lipoproteins are a specialized class of membrane proteins that are anchored via fatty acids to the envelope of gram-negative and gram-positive bacteria. This subset of exported proteins performs a diverse set of functions such as maintaining cell envelope structural integrity (5, 31), delivering proteins to the outer membrane (15, 32, 39), antibiotic resistance (19), and adherence (16). Lipoproteins are often components of membrane-spanning machineries such as flagellar basal bodies and type III secretion complexes (2, 10).
In gram-negative bacteria, lipoproteins are exported from the cytoplasm by the Sec machinery and are processed and modified by three essential enzymes (38). Prolipoprotein diacylglycerol transferase (Lgt) adds a diacylglycerol to the cysteine residue of the L−3-A−2-G−1-C+1 lipobox motif, which allows cleavage between residues −1 and +1 by prolipoprotein signal peptidase (LspA). A third acyl chain is then added onto the free amino group of the diacylglycerocysteine by apolipoprotein N-acyltransferase (Lnt). The three fatty acid chains of the majority of mature lipoproteins are ultimately anchored to the periplasmic leaflet of either the inner or the outer membrane, although a few outer membrane lipoproteins are surface exposed (9, 22, 36).
The Lol machinery is required to sort bacterial lipoproteins to the inner face of the outer membrane, where most bacterial lipoproteins are found (35). The LolCDE complex is an inner membrane ABC transporter that recognizes and releases mature lipoproteins to the periplasmic chaperone LolA. The LolA-lipoprotein complex traverses the periplasm and delivers the lipoproteins to the outer membrane receptor LolB, itself an essential lipoprotein, which ultimately releases them into the outer membrane. The amino acids at positions +2 and +3 of the mature lipoprotein are critical in determining the final localization. Lipoproteins with an aspartate (D) at the +2 position are localized in the cytoplasmic membrane (20, 33, 41). The nature of the amino acid at the +3 position also influences the membrane localization. For example, amino acids D, glutamate (E), glutamine (Q), and asparagine (N) produce very strong inner membrane retention signals when D is in position +2 (33).
Inner membrane lipoproteins are fully fatty acylated but do not interact with LolCDE machinery (12, 27). The negative charge of the D residue at position +2 is critical for inner membrane retention (14). The distance between the Cα carbon and the negative charge is important and explains why glutamate at position +2 does not act as an inner membrane retention signal (14, 23, 33). The negative charge of D+2 is proposed to form an H bond with the amino group of phosphatidylethanolamine (PE), which creates a lipoprotein-PE complex with five fatty acids that cannot be accommodated by LolCDE (14). Negatively charged amino acids D and E at position +3 are proposed to reinforce the Lol avoidance signal by strengthening the interaction with PE (14, 33).
The amino acids tryptophan (W), glycine (G), proline (P), tyrosine (Y), and phenylalanine (F) at position +2 also act as inner membrane retention signals (29, 33). These alternative inner membrane retention signals are not present in any of the predicted or known Escherichia coli inner membrane lipoproteins. Interaction with PE cannot explain the inner membrane retention of lipoproteins with these amino acids at position +2 because they do not have a negative charge.
In vivo studies of the mechanism of lipoprotein localization are difficult and time-consuming because they require the isolation of bacterial membranes, sucrose gradient centrifugation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblot analysis (29). In an alternative in vitro method, the lipoprotein release assay (21, 40), radiolabeled spheroplasts or proteoliposomes reconstituted with LolCDE and lipoproteins are incubated with purified LolA, centrifuged, and monitored for lipoprotein release into the supernatant fraction. In this study, we established a simple, rapid in situ fluorescence microscopy method to visualize the inner or outer membrane localization of the monomeric red fluorescent protein (mRFP1) that was exported by fusing it to a lipoprotein signal peptide. Plasmid-encoded red fluorescent lipoprotein (lipoRFP) constructs with different amino acids at positions +2 and +3 were introduced into several species of the family Enterobacteriaceae in order to determine if the lipoprotein sorting rules are conserved, since most of the sorting studies done to date were performed with E. coli.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study are described in Table 1. All cultures were grown in Luria-Bertani medium at 30°C with ampicillin (50 μg/ml) to maintain plasmids.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Genotype or characteristics | Source or reference |
|---|---|---|
| E. coli PAP105 | Δ(lac-pro)/pAPIP501, [F′ lacIq ΔlacZM15 proAB+] | Laboratory strain |
| Yersinia pseudotuberculosis 32953 | Lacks virulence plasmid pYv | E. Carniel |
| Shigella flexneri | mxiD | 1 |
| Salmonella enterica serovar Typhimurium 14028 | Wild type | Laboratory strain |
| Erwinia carotovora | Wild type | Laboratory strain |
| Klebsiella oxytoca UNF5023 | Wild type | Laboratory strain |
| E. coli PAP9108 | PAP105 Δ(attL-lom)::bla lacIqP204-gfp-pulM | 3 |
| Plasmids | ||
| pDSW204 | Promoter down mutation in −35 of pTrc99A | 37 |
| pRSETb-mRFP1 | mRFP1 | 4 |
| pCHAP6614 | pDSW204, pulA CDSR signal peptide fusion vector | This study |
| pCHAP6615 | pDSW204, pulA CSSR signal peptide fusion vector | This study |
| pCHAP6617 | pDSW204, CDSR-RFP | This study |
| pCHAP6619 | pDSW204, CSSR-RFP | This study |
| pCHAP6636 | pDSW204, CFSR-RFP | This study |
| pCHAP6641 | pDSW204, CFNSR-RFP | This study |
| pCHAP7519 | pDSW204, PulM-RFP | N. Buddelmeijer |
Plasmid construction.
DNA encoding the signal peptide from Klebsiella oxytoca pullulanase PulA with either CD or CS after the signal peptidase cleavage site (29) was amplified with primer pairs CS_for-CS_rev and CS_for-CD_rev, respectively (Table 2 shows the primers used). PCR products were purified, digested with BspHI and XbaI, and cloned into NcoI (BspHI compatible) and XbaI sites in expression vector pDSW204 (37) to create signal peptide fusion vectors pCHAP6614 and pCHAP6615, respectively. The red fluorescent protein (mRFP1) lacking the native Met codon was amplified from pRSETb-mRFP1 with primers RFP_for and RFP_rev, digested with XbaI and HindIII, and cloned in frame with the DNA encoding lipoprotein signal peptides to create pCHAP6617 (CDSR-RFP) and pCHAP6619 (CSSR-RFP). The gene fusions are under the control of a modified trc promoter and can be repressed by vector-encoded lacIq (37). PCR amplifications were performed in 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min with 0.4 μM primers and the JumpStart REDTaq ReadyMix kit (Sigma).
TABLE 2.
DNA primers used in this study
| Primer | 5′-3′ sequence (restriction endonuclease)a |
|---|---|
| CS_for | GGCTCATGAGCCTCAGATATACCCGTAATGC (BspHI) |
| CS_rev | GCTCTAGAAGAGCAGCCGCTAAGTAAAACCAG (XbaI) |
| CD_rev | GCTCTAGAATCGCAGCCGCTAAGTAAAAC (XbaI) |
| RFP_for | GCTCTAGAGCCTCCTCCGAGGACGTCATC (XbaI) |
| RFP_rev | CCCAAGCTTAGGCGCCGGTGGAGTGG (HindIII) |
| F_mut_for | GGAAAAGCAGCCGCTAAGTAAAACCAG |
| Mut_rev | AGAGCCTCCTCCGAGGACGTC |
| FN_mut_for | GATCGCTGGTTTTACTTAGCGGCTGCTTTAACTCC |
| Xba-RFP-5 | CTAGTCTAGAATGGCCTCCTCC (XbaI) |
| HindIIIRFP-3 | CCTGAAGCTTTTAGGCGCCGGTGGAGTGGCGG (HindIII) |
Restriction endonuclease cleavage sites are underlined.
The CFSR-RFP and CFNSR-RFP constructs were generated by site-directed mutagenesis of the +2 codon. For mutagenic PCRs, the mutagenized codon was added to the 5′ end of the forward primer. The D+2 residue of CDSR-RFP was changed to CFSR-RFP with primers F_mut_for and Mut_rev or to CFNSR-RFP with primers FN_mut_for and Mut_rev. An inverse PCR with nonoverlapping, phosphorylated oligonucleotides was performed to introduce the desired mutation. PCR amplifications were performed in 15 cycles of 98°C for 10 s, 52°C for 20 s, and 72°C for 3 min with Phusion High-Fidelity DNA polymerase (Finnzymes).
PCR products were subsequently digested with DpnI to remove all circular template plasmid from the PCR product, ligated to circularize the linear PCR product, and transformed into E. coli. Plasmids were isolated from transformants and screened for loss of the XbaI site that encodes the S+3-R+4 linker between the signal peptide and mRFP1. A silent mutation in the S+3 codon was designed in the mutagenic primer to preserve the linker but not the XbaI site. All plasmids were sequenced to confirm the various signal peptide fusion constructions.
Live-cell imaging.
Overnight cultures were subcultured 1/100 and grown for 3 h (optical density at 600 nm [OD600] = 0.5). Cells were grown in the absence of the inducer or occasionally with 25 μM isopropyl-β-d thiogalactopyranoside (IPTG) for 1 h. To prepare cells for fluorescence microscopy, 0.5 ml of culture was pelleted and resuspended in 10 μl of Luria-Bertani medium or 10 μl of plasmolysis solution (15% sucrose, 25 mM HEPES [pH 7.4], 20 mM NaN3). One microliter of control cells or plasmolyzed cells was immobilized on a thin layer of 1% agarose in water or of 1% agarose in 15% sucrose (to maintain plasmolysis). For control experiments, cells were membrane stained by adding the fluorescent lipophilic dye FM 4-64 or FM 1-63 (Molecular Probes) to a final concentration of 5 μg/ml. Live cells were visualized by epifluorescence microscopy within 15 min of slide preparation with a Zeiss Axioplan 2 microscope equipped with a Hamamatsu charge-coupled device camera. Images were collected with OpenLab and processed with Photoshop. Red fluorescence was detected with rhodamine filters with exposure times of 1 to 10 s for mRFP1 and less than 1 s for FM 4-64. Green fluorescence was detected with fluorescein isothiocyanate filters.
Membrane fractionation.
Two-hundred-milliliter cultures were grown for 2.5 h (OD600 = 0.5). Membranes were prepared from cells disrupted in a French press and were separated by flotation sucrose gradient centrifugation as previously described (26, 27). Twenty fractions (250 μl) were collected from the top of the gradients, separated by 12% SDS-PAGE, and transferred to nitrocellulose membranes. LipoRFPs were detected by immunoblotting with primary antibodies raised in rabbits against affinity-purified, His-tagged mRFP1 and secondary anti-rabbit immunoglobulin G antibodies coupled to horseradish peroxidase (Amersham Biosciences). The primary antibodies were purified by adsorption to a soluble extract of E. coli DH5αF′ proteins to reduce nonspecific antibodies.
Osmotic shock and spheroplast preparation.
Osmotic shock was performed on 10-ml cultures grown for 2 h and then induced with 25 μM IPTG for 2 h until the OD600 was 0.5. Cells were pelleted, resuspended in 5 ml of ice-cold 15% sucrose in 25 mM HEPES (pH 7.4). The divalent cation chelator EDTA was slowly added to a final concentration of 1 mM. After 5 min on ice, cells were pelleted and resuspended in 0.5 ml of 5 mM MgSO4. Cells were centrifuged again, and the supernatant was kept as the osmotic shock fluid.
Five-milliliter cultures grown as described above were converted to spheroplasts as previously described by Randall and Hardy (25), with minor modifications. Cells from 3 ml of the culture were pelleted, resuspended in 75 μl of cold TSE buffer (0.1 M Tris acetate, 16% sucrose, 5 mM EDTA, pH 8.2) to which lysozyme (150 μg/ml) and 75 μl of cold water were added and incubated on ice for 5 min. Spheroplasts were stabilized with MgSO4 at a final concentration of 15 mM and pelleted, and the supernatant was kept as the periplasmic fraction. The spheroplast pellet was washed once in TSM buffer (0.05 M Tris acetate, 8% sucrose, 10 mM MgSO4, pH 8.2) and resuspended in sample buffer.
RESULTS
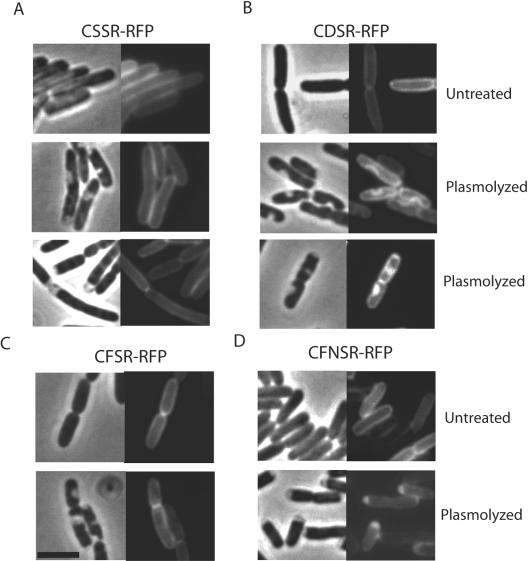
LipoRFPs localize to the cytoplasmic and outer membranes. In order to construct an improved synthetic lipoprotein for in situ localization studies, we created an in-frame chimera comprising mRFP1 and the signal peptide of the lipoprotein pullulanase (PulA) from K. oxytoca (18). mRFP1 was selected because, unlike the green fluorescent protein (GFP), it is fluorescent when exported to the periplasm by the Sec machinery (6). mRFP1 lacking its initiator methionine was fused to the lipoprotein signal peptide including the L−4-L−3-S−2-G−1 lipobox, followed by C+1-D+2 or C+1-S+2 and a two-amino-acid linker (S+3-R+4). Both CSSR-RFP and CDSR-RFP were shown to localize to the cell envelope by fluorescence microscopy (Fig. 1A and B).
FIG. 1.
Inner and outer membrane localization of lipoRFPs in E. coli. Live cells that were left untreated or plasmolyzed in 15% sucrose were visualized on agarose beds. Each panel consists of a phase-contrast image (left) and the corresponding fluorescence image. A to D, strains producing lipoRFPs with the N-terminal sequences CSSR, CDSR, CFSR, and CFNSR, respectively. The black bar is 3 μm in length.
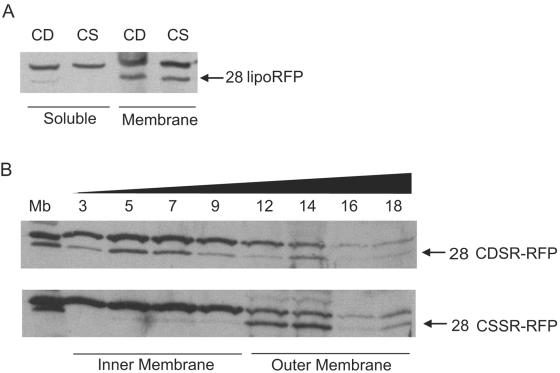
Soluble material from cells producing CSSR-RFP or CDSR-RFP was separated from membranes, which were subjected to flotation sucrose gradient centrifugation (27, 29). Both chimeras were located in the membrane fraction, with a very small amount of CDSR-RFP in the soluble fraction (Fig. 2A). CDSR-RFP was found primarily in the inner membrane fraction, and CSSR-RFP was found primarily in the outer membrane fraction after sucrose gradient analysis (Fig. 2B). The anti-RFP antibodies also recognized an unidentified inner membrane protein that is slightly larger than the lipoRFPs and serves as in internal control (Fig. 2). These profiles are similar to those observed previously for other synthetic lipoproteins (29). Thus, lipoprotein sorting signals function correctly when fused to mRFP1.
FIG. 2.
Localization of lipoRFPs CDSR-RFP and CSSR-RFP. (A) Analysis of membrane and soluble protein fractions. Samples were derived from identical starting cell suspension volumes. (B) Membranes were separated by flotation sucrose density gradient centrifugation. Total membranes were used as a control (Mb), and the sucrose gradient fraction number is indicated above. The black triangle indicates the increasing concentration of sucrose in the gradient fractions. Proteins in samples were separated by SDS-PAGE and immunoblotted with antibodies against mRFP1. The identities and molecular sizes (kilodaltons) of the lipoRFPs are indicated.
Since lipoproteins can be anchored to either membrane but are localized in the periplasm, visualization of untreated cells does not distinguish between lipoRFPs in the inner and outer membranes. To visualize the lipoRFPs directly in either the inner or the outer membrane, cells were plasmolyzed in 15% sucrose. This treatment results in the formation of plasmolysis bays where the cytoplasmic membrane retracts from the rigid peptidoglycan-outer membrane layer in response to hyperosmotic sucrose shock (8). The bays, which occur at the cell poles and along the cell length, were clearly visible in phase-contrast images (Fig. 1 and 3). In plasmolyzed cells with outer membrane-localized CSSR-RFP, the fluorescence pattern resembled the membrane fluorescence of untreated cells (Fig. 1A); i.e., the fluorescent contour was unaltered by plasmolysis. In plasmolyzed cells producing CDSR-RFP, fluorescence was detected along the edges of the periplasmic bay or throughout the area within the periplasmic bay (Fig. 1B). In summary, this fluorescence microscopy approach has sufficient resolution to differentiate between lipoproteins localized in the inner membrane and those localized in the outer membrane.
FIG. 3.
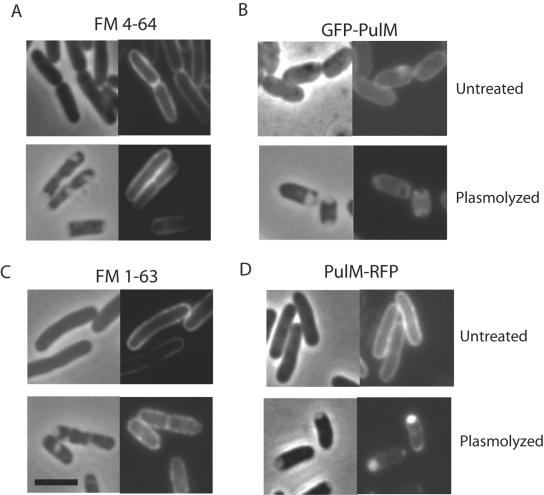
Outer membrane staining of lipophilic FM dyes and inner membrane localization of PulM. Live cells were left untreated or plasmolyzed in the presence of 5 μg/ml FM 4-64 (A) or FM 1-63 (C). Strains producing GFP-PulM (B) or PulM-RFP (D) were left untreated or plasmolyzed. Each panel consists of a phase-contrast image (left) and the corresponding fluorescence image. The black bar is 3 μm in length.
The fluorescent lipophilic dye FM 4-64 was used as a membrane-specific stain in control experiments. Fishov and Woldringh reported that this dye stains only the inner membrane of a plasmolyzed, filament-forming E. coli pbpB mutant (11). In our study, however, FM 4-64 and another lipophilic dye, FM 1-63, specifically stained the outer membrane of plasmolyzed wild-type E. coli (Fig. 3A and C). This fluorescence pattern was identical to that of outer membrane-localized CSSR-RFP (Fig. 1A), and therefore, the lipophilic dyes serve as controls for outer membrane localization patterns.
As a control for inner membrane fluorescence, we plasmolyzed cells producing the GFP-PulM or PulM-RFP fluorescent chimera. PulM is a component of the K. oxytoca type II secretion system that is anchored in the inner membrane by a single N-terminal transmembrane segment and has a periplasmic domain of approximately 120 amino acids (24). GFP-PulM was previously shown to exhibit uniform peripheral fluorescence, consistent with its location throughout the inner membrane (3). In plasmolyzed cells, fluorescent GFP-PulM was visualized along the edges of the periplasmic bay but did not fill the bay area, which is consistent with its known inner membrane localization (Fig. 3B). In contrast, the fluorescence image of PulM-RFP resembled the CDSR-RFP localization pattern, with intense fluorescence in the plasmolysis bays (Fig. 3D).
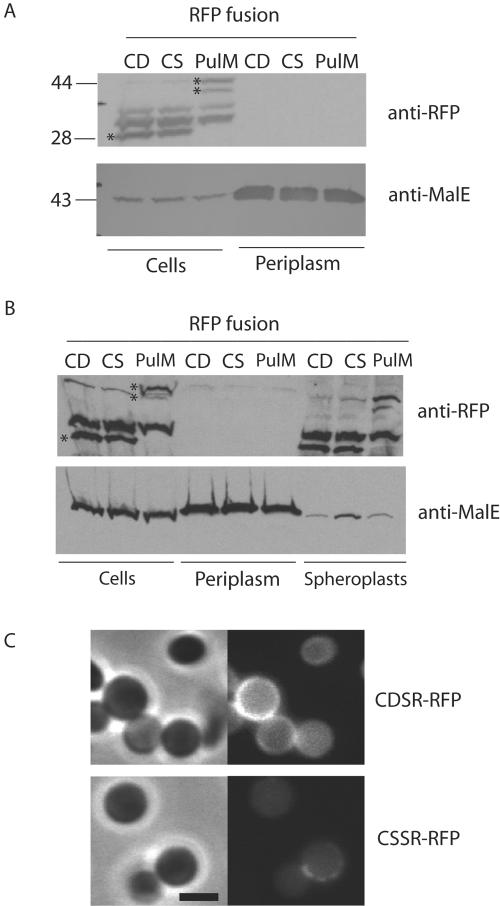
The bright fluorescence patches in the bays of plasmolyzed cells producing CDSR-RFP and PulM-RFP suggested that these chimeras might be released into the periplasm because of proteolysis or because of contraction of the cytoplasmic membrane. Proteolysis of CDSR-RFP is unlikely to explain the fluorescence in the plasmolysis bays since it was engineered to have only four amino acids after the lipid anchor in order to minimize proteolytic cleavage. To determine if the fluorescence in the periplasmic bays was due to the release of CDSR-RFP into the periplasm, periplasmic proteins were extracted by osmotic shock after plasmolysis with 15% sucrose. CDSR-RFP, CSSR-RFP, and PulM-RFP were not found in the extracted material, whereas periplasmic MalE was quantitatively released (Fig. 4A). The PulM-RFP protein was detected as two protein bands that likely result from proteolysis of the chimera (Fig. 4).
FIG. 4.
Plasmolyzed cells do not release fluorescent lipoproteins into the periplasm. (A) Proteins released by osmotic shock of cells producing CDSR-RFP, CSSR-RFP, or PulM-RFP after plasmolysis with 15% sucrose. (B) Proteins released by converting cells to spheroplasts by digesting the peptidoglycan layer with lysozyme following sucrose plasmolysis. In panels A and B, the samples were derived from the same volume of initial cell suspension. Proteins were separated by SDS-PAGE and immunoblotted with antibodies against mRFP1 or MalE. Asterisks indicate the lipoRFPs and PulM-RFP, and their molecular sizes (kilodaltons) are shown at the left of panel A. (C) Spheroplasts of cells producing CDSR-RFP and CSSR-RFP examined by phase-contrast and fluorescence microscopy. The black bar is 2 μm in length.
Since the peptidoglycan layer might impede the release of aggregated proteins upon osmotic shock, bacteria were converted to spheroplasts by lysozyme treatment (to degrade the peptidoglycan) and separated into periplasmic and spheroplast fractions. This treatment also failed to release either form of lipoRFP into the periplasm; i.e., both CSSR-RFP and CDSR-RFP remained in the spheroplast fraction (Fig. 4B). The spheroplasts from CDSR-RFP-producing cells showed uniform membrane fluorescence, whereas the fluorescence of CSSR-RFP-producing spheroplasts was weak except for occasional patchy fluorescence (Fig. 4C). The weak fluorescence probably results from large-scale release of outer membrane fragments during spheroplast preparation, while the patchy fluorescence probably corresponds to regions on the spheroplast where the outer membrane remains attached.
F+2-N+3 also targets lipoRFP to the inner membrane.
We reported previously that amino acids F, W, Y, P, and G, as well as D, at the +2 position cause inner membrane retention of fatty acylated MalE reporter proteins (29). To test whether this phenomenon is context dependent, the D+2 in CDSR-RFP was converted to F+2, Y+2, W+2, or G+2 and the cells were plasmolyzed and examined by fluorescence microscopy. In every case, the fluorescence pattern was identical to that of CSSR-RFP; i.e., the chimeras were located in the outer membrane (Fig. 1A and C and data not shown).
In the studies that led us to propose F+2 as an inner membrane lipoprotein retention signal (29), the amino acid at position +2 was followed by N+3, rather than S+3 as in the chimeras described above. Subsequent in vitro analyses showed that N+3 is essential for W+2 and F+2 to operate as inner membrane (Lol avoidance) signals (33). Thus, the outer membrane localization of CFSR-RFP, CWSR-RFP, CYSR-RFP, and CGSR-RFP was not unexpected. Site-directed mutagenesis was used to insert an N in front of the S+3 in CFSR-RFP to create CFNSR-RFP. The pattern of CFNSR-RFP fluorescence in plasmolyzed cells was identical to the inner membrane fluorescence of CDSR-RFP (Fig. 1B and D). Therefore, the lipoRFPs with the inner membrane retention signal D+2-S+3 or F+2-N+3 appear to localize specifically to the inner membrane.
Lipoprotein sorting in the family Enterobacteriaceae.
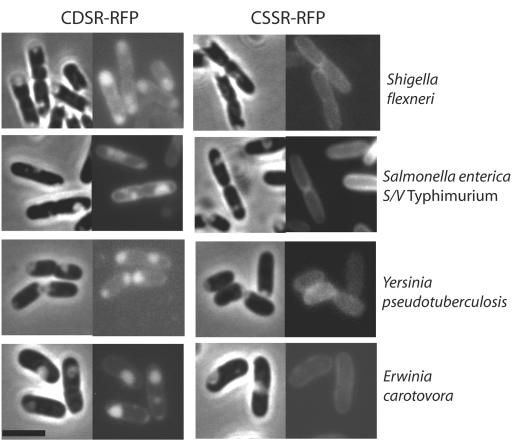
The validated in situ method for determining the membrane localization of lipoproteins was next extended to other gram-negative bacteria. We introduced the plasmids encoding the lipoRFPs into Salmonella enterica serovar Typhimurium, Shigella flexneri, Yersinia pseudotuberculosis, Erwinia carotovora, and K. oxytoca and examined the fluorescence of plasmolyzed cells. All species were readily plasmolyzed by 15% sucrose (Fig. 5). In all cases (Fig. 5 and data not shown), the lipoRFPs exhibited fluorescence patterns identical to those seen in E. coli; i.e., CDSR-RFP and CFNSR-RFP localized specifically to the periplasmic bays while CSSR-RFP and CFSR-RFP localized to the outer membrane. These results indicate that the rules determining lipoprotein localization are conserved among members of the family Enterobacteriaceae.
FIG. 5.
Inner and outer membrane localization of lipoRFPs in various gram-negative bacteria. CDSR-RFP and CSSR-RFP were visualized after plasmolysis. Each panel consists of a phase-contrast image (left) and the corresponding fluorescence image (right). The black bar is 3 μm in length. S/V, serovar.
The F+2-N+3 inner membrane retention signal is present in three predicted lipoproteins.
The LipoP algorithm (17) was used to analyze the genomes of 29 species of gram-negative bacteria, including the members of the family Enterobacteriaceae tested in this study, for predicted lipoproteins. Three predicted lipoproteins were found that contain C+1-F+2-N+3 at the N terminus. These closely related proteins encode a putative substrate-binding protein of an ABC transporter in E. carotovora (gi 50120484), in Klebsiella pneumoniae CG43 (gi 38016755), and in four sequenced strains of Salmonella (e.g., gi 56412308), although the Salmonella homologs are considerably shorter. Except for Salmonella, the gene encoding the putative binding protein is adjacent to genes encoding other components of the ABC transporter. The K. pneumoniae gene cluster is on a virulence plasmid and is flanked by insertion sequences (7). The full-length putative substrate-binding proteins are 37 to 47% identical to homologs in the gram-positive bacteria Corynebacterium diphtheriae (gi 38232945) and Streptomyces coelicolor (gi 3483044). These proteins are also predicted to be lipoproteins and have the sequences CFT and CFA at their N termini. The genes adjacent to the putative lipoprotein gene in these bacteria encode other components of an ABC transporter that is homologous to that in E. carotovora and K pneumoniae.
DISCUSSION
This report describes the use of fatty acylated mRFP1 to visualize the membrane localization and sorting of model bacterial lipoproteins in situ. When applied to plasmolyzed cells, this simple method allowed direct detection of fluorescent outer or inner membrane lipoproteins and enabled us to demonstrate the conservation of lipoprotein sorting signal function in members of the family Enterobacteriaceae; i.e., CDS and CFN at the N terminus cause inner membrane retention, and the sequence CSS or CFS permits sorting to the outer membrane.
LipoRFPs predicted to be targeted to the outer membrane were detected only in this membrane by epifluorescence microscopy of plasmolyzed E. coli. Two lipophilic FM dyes, FM 4-64 and FM 1-63, reproducibly produced the same outer membrane staining pattern. The former was previously reported to stain only the inner membrane (11, 13). In the original study, FM 4-64 at concentrations close to those used here appeared to stain the inner membrane of an E. coli pbpB mutant grown at 42°C to induce filament formation (11). We did not observe any staining of the inner membrane of wild-type bacteria grown at 42°C or when cells were stained for longer periods (data not shown). Thus, the aberrant staining pattern observed by others might be due to defects in cell wall assembly caused by mutations present in the strains used.
Inner membrane-retained lipoRFPs were observed along the edges of the plasmolysis bay and within the bay by epifluorescence microscopy. The formation of brightly fluorescent periplasmic bays was not due to the release of lipoRFPs after plasmolysis but was more likely due to the molecular crowding of RFP on the convex surface of the inner membrane that faces the periplasm, which makes it difficult to detect the contours of the inner membrane in this area of plasmolyzed cells because of the limited resolution capacity of the microscope.
CDX (where X is virtually any amino acid) appears to be the only inner membrane lipoprotein retention signal used in E. coli, as indicated by the absence of any predicted lipoprotein with the sequence CFN (or CWN, CYN, or CPN). Since the canonical sorting signal (D+2) is thought to act by interacting with positively charged PE (14), the unconventional signals must operate by an entirely different mechanism. Furthermore, these alternative sorting signals are rarely found in lipoproteins from other members of the family Enterobacteriaceae. Indeed, the only example found so far is a putative substrate-binding protein in Klebsiella, Salmonella, and Erwinia with the sequence CFN at its N terminus. Previous studies with a fatty acylated version of the periplasmic maltose-binding protein (lipoMalE) indicated that it can function when tethered to the inner membrane but not when tethered to the outer membrane (29). Therefore, the putative substrate-binding lipoproteins identified by genome screening are also likely to be tethered to the inner membrane, consistent with the presence of the CFN sorting signal.
We speculate that the genes for these proteins (and other components of the putative ABC transporter of which they are part) were acquired from the gram-positive bacterium C. diphtheriae or S coelicolor. In gram-positive bacteria, the substrate-binding proteins of ABC transporters are frequently tethered to the cytoplasmic membrane as lipoproteins to prevent their release from the cell surface (30). Evidence for acquisition of these genes by Erwinia bacteria from Streptomyces bacteria by horizontal transfer is provided by their GC content, which, at ca. 57%, is intermediate between the global GC content of Erwinia bacteria (ca. 51%) and both the global GC content of Streptomyces bacteria (ca. 70%) and the specific GC content of the putative gene (73%). The GC content of the gene in C diphtheriae (61%) is also higher than the overall GC content of the C. diphtheriae genome (ca. 54%). The sequences at the N termini of the proteins encoded by the genes in C. diphtheriae and S. coelicolor (CFT and CFA, respectively) would not function as inner membrane lipoprotein retention signals in Erwinia or Klebsiella bacteria, necessitating their conversion to CDT or CDA or to CFN for the proteins to become functional. The latter change is the one that seems to have occurred.
The functionality of the D+2 and F+2-N+3 inner membrane lipoprotein retention signals in the Enterobacteriaceae branch of gram-negative bacteria is consistent with the high-level conservation of the LolCDEAB machinery in these bacteria (40). These data also suggest that lipoprotein processing and modification proteins Lgt, LspA, and Lnt are functionally conserved. Despite the conservation of the lipoprotein sorting signals and machinery, there are apparent exceptions to these rules. For example, the +2 rule for lipoprotein sorting does not apply to the spirochete Borrelia burgdorferi, which produces many surface lipoproteins that are important for pathogenesis (28). Likewise, Neisseria meningitidis lipoprotein DsbA1 is an inner membrane-tethered disulfide oxidoreductase with serine at position +2 (34).
The method that we describe here should be useful to examine lipoprotein localization in all gram-negative bacteria that can be plasmolyzed and in which lipoRFPs can be produced at detectable levels. In preliminary studies, we have shown that N. meningitidis cannot be plasmolyzed and that lipoRFP fluorescence is weak in Vibrio cholerae and Pseudomonas aeruginosa. We are currently trying to adapt the methods reported here to these and other gram-negative bacteria with the aim of determining whether non-aspartate inner membrane retention signals are used outside of the family Enterobacteriaceae and, if so, to dissect the mechanisms of lipoprotein sorting that they use.
Acknowledgments
We thank Roger Tsien for pRSETb-mRFP1, Nienke Buddelmeijer for pCHAP7519, Olivera Francetic for antibodies against mRFP1, Elisabeth Carniel and Jost Enninga for strains, Benoît Arcangioli for use of microscope facilities, and all members of the Molecular Genetics Unit for support.
S.L. is supported by a postdoctoral fellowship from the Canadian Louis Pasteur Foundation. This work was supported in part by the Programme Microbiologie Fondamentale of the French Ministère Délégué de la Recherche et aux Nouvelles Technologies.
REFERENCES
- 1.Allaoui, A., P. Sansonetti, and C. Parsot. 1993. MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri Ipa invasins. Mol. Microbiol. 7:59-68. [DOI] [PubMed] [Google Scholar]
- 2.Allaoui, A., P. J. Sansonetti, and C. Parsot. 1992. MxiJ, a lipoprotein involved in secretion of Shigella Ipa invasins, is homologous to YscJ, a secretion factor of the Yersinia Yop proteins. J. Bacteriol. 174:7661-7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buddelmeijer, N., O. Francetic, and A. P. Pugsley. 2006. Green fluorescent chimeras indicate nonpolar localization of pullulanase secretion components PulL and PulM. J. Bacteriol. 188:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J. C., P. H. Viollier, and L. Shapiro. 2005. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 55:1085-1103. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y. T., H. Y. Chang, Y. C. Lai, C. C. Pan, S. F. Tsai, and H. L. Peng. 2004. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337:189-198. [DOI] [PubMed] [Google Scholar]
- 8.Cook, W. R., T. J. MacAlister, and L. I. Rothfield. 1986. Compartmentalization of the periplasmic space at division sites in gram-negative bacteria. J. Bacteriol. 168:1430-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coutte, L., E. Willery, R. Antoine, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2003. Surface anchoring of bacterial subtilisin important for maturation function. Mol. Microbiol. 49:529-539. [DOI] [PubMed] [Google Scholar]
- 10.Dailey, F. E., and R. M. Macnab. 2002. Effects of lipoprotein biogenesis mutations on flagellar assembly in Salmonella. J. Bacteriol. 184:771-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fishov, I., and C. L. Woldringh. 1999. Visualization of membrane domains in Escherichia coli. Mol. Microbiol. 32:1166-1172. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda, A., S.-I. Matsuyama, T. Hara, J. Nakayama, H. Nagasawa, and H. Tokuda. 2002. Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J. Biol. Chem. 277:43512-43518. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh, A. S., and K. D. Young. 2005. Helical disposition of proteins and lipopolysaccharide in the outer membrane of Escherichia coli. J. Bacteriol. 187:1913-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara, T., S. Matsuyama, and H. Tokuda. 2003. Mechanism underlying the inner membrane retention of lipoproteins caused by Lol avoidance signals. J. Biol. Chem. 279:40408-40414. [DOI] [PubMed] [Google Scholar]
- 15.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15:978-988. [PMC free article] [PubMed] [Google Scholar]
- 16.Jin, S., A. Joe, J. Lynett, E. K. Hani, P. Sherman, and V. L. Chan. 2001. JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol. Microbiol. 39:1225-1236. [DOI] [PubMed] [Google Scholar]
- 17.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornacker, M. G., and A. P. Pugsley. 1989. Molecular characterization of pulA and its product, pullulanase, a secreted enzyme of Klebsiella pneumoniae UNF5023. Mol. Microbiol. 4:73-85. [DOI] [PubMed] [Google Scholar]
- 19.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175:6299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda, K., S.-I. Matsuyama, and H. Tokuda. 2002. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl. Acad. Sci. USA 99:7390-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuyama, S., T. Tajima, and H. Tokuda. 1995. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO. J. 14:3365-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugsley, A. P., C. Chapon, and M. Schwartz. 1986. Extracellular pullulanase of Klebsiella pneumoniae is a lipoprotein. J. Bacteriol. 166:1083-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugsley, A. P., and M. G. Kornacker. 1991. Secretion of the cell surface lipoprotein pullulanase by Escherichia coli. Collaboration or competition between the specific secretion pathway and the lipoprotein sorting pathway. J. Biol. Chem. 266:13640-13645. [PubMed] [Google Scholar]
- 24.Pugsley, A. P., and I. Reyss. 1990. Five genes at the 3′ end of the Klebsiella pneumoniae pulC operon are required for pullulanase secretion. Mol. Microbiol. 4:365-379. [DOI] [PubMed] [Google Scholar]
- 25.Randall, L. L., and S. J. S. Hardy. 1986. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell 46:921-928. [DOI] [PubMed] [Google Scholar]
- 26.Robichon, C., M. Bonhivers, and A. P. Pugsley. 2003. An intramolecular disulphide bond reduces the efficacy of a lipoprotein plasma membrane sorting signal. Mol. Microbiol. 49:1145-1154. [DOI] [PubMed] [Google Scholar]
- 27.Robichon, C., D. Vidal-Ingigliardi, and A. P. Pugsley. 2005. Depletion of apolipoprotein N-acyltransferase causes mislocalization of outer membrane lipoproteins in Escherichia coli. J. Biol. Chem. 280:974-983. [DOI] [PubMed] [Google Scholar]
- 28.Schulze, R., and W. R. Zückert. 2006. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol. Microbiol. 59:1473-1484. [DOI] [PubMed] [Google Scholar]
- 29.Seydel, A., P. Gounon, and A. P. Pugsley. 1999. Testing the “+2 rule” for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34:810-821. [DOI] [PubMed] [Google Scholar]
- 30.Sutcliffe, I. C., and R. R. B. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki, H., Y. Nishimura, S. Yasuda, A. Nishimura, M. Yamada, and Y. Hirota. 1978. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol. Gen. Genet. 167:1-9. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka, K., S.-I. Matsuyama, and H. Tokuda. 2001. Deletion of lolB, encoding an outer membrane lipoprotein, is lethal for Escherichia coli and causes accumulation of lipoprotein localization intermediates in the periplasm. J. Bacteriol. 183:6538-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terada, M., T. Kuroda, S.-I. Matsuyama, and H. Tokuda. 2001. Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 276:47690-47694. [DOI] [PubMed] [Google Scholar]
- 34.Tinsley, C. R., R. Voulhoux, J. L. Beretti, J. Tommassen, and X. Nassif. 2004. Three homologues, including two membrane-bound proteins, of the disulfide oxidoreductase DsbA in Neisseria meningitidis: effects on bacterial growth and biogenesis of functional type IV pili. J. Biol. Chem. 279:27078-27087. [DOI] [PubMed] [Google Scholar]
- 35.Tokuda, H., and S. Matsuyama. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 1694:1-9. [PubMed] [Google Scholar]
- 36.van Ulsen, P., L. van Alphen, J. ten Hove, F. Fransen, P. van der Ley, and J. Tommassen. 2003. A neisserial autotransporter NalP modulating the processing of other autotransporters. Mol. Microbiol. 50:1017-1030. [DOI] [PubMed] [Google Scholar]
- 37.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, H. C. 1996. Biosynthesis of lipoproteins, p. 1005-1014. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington D.C. [Google Scholar]
- 39.Wu, T., J. Malinverni, N. Ruiz, S. Kim, T. J. Silhavy, and D. Kahne. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235-245. [DOI] [PubMed] [Google Scholar]
- 40.Yakushi, T., K. Masuda, S.-I. Narita, S.-I. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2:212-218. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi, K., F. Yu, and M. Inouye. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53:423-432. [DOI] [PubMed] [Google Scholar]