Abstract
The Bacillus subtilis ferric uptake regulator (Fur) protein mediates the iron-dependent repression of at least 20 operons encoding ∼40 genes. We investigated the physiological roles of Fur-regulated genes by the construction of null mutations in 14 transcription units known or predicted to function in siderophore biosynthesis or iron uptake. We demonstrate that ywbLMN, encoding an elemental iron uptake system orthologous to the copper oxidase-dependent Fe(III) uptake system of Saccharomyces cerevisiae, is essential for growth in low iron minimal medium lacking citric acid. 2,3-Dihydroxybenzoyl-glycine (Itoic acid), the siderophore precursor produced by laboratory strains of B. subtilis, is of secondary importance. In the presence of citrate, the YfmCDEF ABC transporter is required for optimal growth. B. subtilis is unable to grow in minimal medium containing the iron chelator EDDHA unless the ability to synthesize the intact bacillibactin siderophore is restored (by the introduction of a functional sfp gene) or exogenous siderophores are provided. Utilization of the catecholate siderophores bacillibactin and enterobactin requires the FeuABC importer and the YusV ATPase. Utilization of hydroxamate siderophores requires the FhuBGC ABC transporter together with the FhuD (ferrichrome) or YxeB (ferrioxamine) substrate-binding proteins. Growth with schizokinen or arthrobactin is at least partially dependent on the YfhA YfiYZ importer and the YusV ATPase. We have also investigated the effects of a fur mutation on the proteome and documented the derepression of 11 Fur-regulated proteins, including a newly identified thioredoxin reductase homolog, YcgT.
Iron is an essential nutrient for nearly all living organisms (1). Transport systems for the uptake of iron and iron-chelation complexes are therefore critical for growth (64). In many bacteria, the ferric uptake regulator (Fur) protein coordinates the expression of iron uptake and homeostasis pathways in response to available iron (1). For example, in Escherichia coli K-12 the Fur regulon includes genes for the biosynthesis and transport of the catechol siderophore enterobactin, the uptake of various hydroxamate siderophores, and a substrate-inducible ferric citrate uptake system (25, 45). In addition, Fur represses a small RNA, RyhB, that in turn negatively regulates the expression of iron-rich enzymes such as succinate dehydrogenase, fumarase, and aconitase (36). This allows the production of these enzymes to be activated in response to available iron.
The production of low-molecular-weight Fe(III) chelators (siderophores) enables microorganisms to efficiently scavenge iron even in aerobic environments where iron exists primarily as insoluble hydroxides (64). The first bacterial siderophore to be structurally characterized was Itoic acid, isolated from low-iron fermentation cultures of Bacillus subtilis (31). Itoic acid (DHBG) is the glycine conjugate of 2,3-dihydroxybenzoic acid (DHBA) (30, 47). In retrospect, DHBA and DHBG [collectively referred to as DHB(G)] are precursors of the catecholate siderophore bacillibactin, the cyclic trimeric lactone of 2,3-dihydroxybenzoyl-Gly-Thr (37, 55). A siderophore of this apparent structure was first described as a product of Corynebacterium glutamicum and named corynebactin (12), but recent genome sequence and biochemical analyses suggest that this organism is unlikely to produce this compound (16). Thus, bacillibactin is now the preferred name for the B. subtilis siderophore (16, 37), although several papers refer to this siderophore as corynebactin (8, 55).
In most laboratory strains of B. subtilis (derived from strain 168) DHB(G) accumulates in the medium due to the presence of the sfp0 mutation (52). The Sfp phosphopantetheinyl transferase is required for the posttranslational activation of DhbB and DhbF, which are required for the latter stages of bacillibactin biosynthesis (37). Due to its relatively weak iron-chelating ability, DHB(G) is unlikely to have physiological relevance as a siderophore under natural conditions (15), but it can facilitate iron uptake under carefully controlled laboratory conditions.
Early studies established many of the essential features of iron uptake in B. subtilis (47-49, 63). The production of DHB(G) was shown to be repressed by iron, and transport was shown to be both temperature and energy dependent (49, 63). A variety of related catechol compounds were also shown to mediate iron uptake and to suppress siderophore production (48). These physiology studies suggested the presence of either a very broad selectivity transport system (or a hand-off of the iron to a common carrier) or the presence of multiple uptake systems.
Schneider and Hantke reported the first detailed molecular analysis of iron uptake pathways in B. subtilis (57). Selection for mutants resistant to antibiotics that structurally mimic hydroxamate siderophores (albomycin and ferrimycin) allowed the isolation of loci encoding ABC transporter subunits needed for the uptake of ferric hydroxamate compounds. The FhuD lipoprotein functions as a substrate-binding protein (SBP) for ferrichrome (and the related siderophores ferrichrysin and ferricrocin), and transport requires the FhuBG transmembrane proteins and the FhuC ATPase. A mutation (foxD) specifically defective in ferrioxamine B and E uptake was postulated to define an SBP for this siderophore (57). Their genetic mapping data, together with the subsequent delineation of the Fur regulon, suggested that the foxD mutation was likely within the Fur-regulated yxeB gene (5). This hypothesis was generated independently by an analysis of the protein similarities for all of the ABC transport systems, and corresponding SBPs, in B. subtilis (53). The identification of candidate Fur box elements in the regulatory region for the divergently transcribed fhuD and fhuB genes also provided an early indication that B. subtilis iron uptake functions were likely regulated by a Fur homolog (57).
B. subtilis provides a model system for the investigation of metal ion homeostasis in gram-positive bacteria (40). We have identified metalloregulatory proteins controlling iron (Fur), zinc (Zur), and manganese (MntR) homeostasis and characterized the corresponding regulons by using transcriptional profiling and DNA-binding studies (5, 11, 20, 22). B. subtilis Fur represses ∼40 genes in response to the available iron by binding to suitably positioned Fur box elements (4, 5). The largest class of Fur-regulated genes encodes ABC-transport systems, suggesting the presence of multiple iron uptake pathways. However, the contributions of these Fur-regulated operons to cell physiology have not been systematically assessed.
In the present study we have defined the effects of a fur mutation on the cytoplasmic and extracytoplasmic proteomes, identified a putative thioredoxin reductase as a novel member of the Fur regulon, and investigated the role of various Fur-regulated operons in enabling growth under conditions of iron limitation. In B. subtilis 168 strains, unable to synthesize functional siderophore, the YwbLMN proteins provide a dominant pathway for iron uptake in minimal medium lacking citric acid. YwbLMN is a predicted elemental iron uptake system orthologous to the copper oxidase-dependent Fe(III) uptake system of Saccharomyces cerevisiae. In the presence of citrate, the YfmCDEF ABC transporter is required for optimal growth. Mutational studies provide insights into the transport specificities for the multiple ferric-siderophore uptake systems encoded in the B. subtilis genome and demonstrate the important role that bacillibactin can play in iron physiology.
MATERIALS AND METHODS
Bacterial strains.
The B. subtilis strains used in the present study are derivatives of CU1065 and are listed in Table 1. Like other 168 strains, CU1065 (trpC2 attSPβ) is an sfp0 strain and carries a frameshift mutation in the sfp gene encoding a phosphopantetheinyl transferase required for the synthesis of bacillibactin and other secondary metabolites, including surfactin (52). In order to restore bacillibactin synthesis, we replaced the sfp0 allele with an intact gene from Bacillus subtilis OKB105 (43) by congression. Transformation of CU1065 cells with OKB105 DNA, followed by selection for prototrophy identified transformants that had acquired an intact trpC gene. These transformants were screened to identify those that had also acquired a copy of the unlinked sfp gene by halo formation on Difco Sporulation Medium. Surfactin-producing strains produce a Ca2+-dependent halo and were selected for further characterization. One such isolate, HB5800, was checked by DNA sequencing to verify the presence of an intact sfp gene.
TABLE 1.
Strains used in this study
| Strain | Genotype (siderophore)a | Source or referenceb |
|---|---|---|
| CU1065 | trpC2 attSPβ sfp0 [DHB(G)+] | Lab stock |
| NCIB3610 | sfp+ (bacillibactin+) | 10 |
| HB2501 | CU1065 fur::kan [DHB(G)+] | 19 |
| HB5600 | CU1065 dhbA::spc | LFH-PCR → CU1065 |
| HB5601 | CU1065 dhbA::mls | LFH-PCR → CU1065 |
| HB5602 | CU1065 fhuD::spc [DHB(G)+] | LFH-PCR → CU1065 |
| HB5604 | CU1065 yclN::mls [DHB(G)+] | LFH-PCR → CU1065 |
| HB5605 | CU1065 yclN::mls dhbA::spc | HB5600 chr DNA → HB5604 |
| HB5606 | CU1065 yhfQ::cat [DHB(G)+] | LFH-PCR → CU1065 |
| HB5607 | CU1065 yhfQ::cat dhbA::spc | HB5600 chr DNA → HB5606 |
| HB5608 | CU1065 fhuB::mls [DHB(G)+] | LFH-PCR → CU1065 |
| HB5609 | CU1065 fhuB::mls dhbA::spc | HB5600 chr DNA → HB5608 |
| HB5610 | CU1065 yusV::mls [DHB(G)+] | LFH-PCR → CU1065 |
| HB5611 | CU1065 yusV::mls dhbA::spc | HB5600 chr DNA → HB5610 |
| HB5612 | CU1065 yfmC::mls [DHB(G)+] | LFH-PCR → CU1065 |
| HB5613 | CU1065 yfmC::mls dhbA::spc | HB5600 chr DNA → HB5612 |
| HB5614 | CU1065 yuiI::spc [DHB(G)+] | LFH-PCR → CU1065 |
| HB5615 | CU1065 yuiI::spc dhbA::mls | HB5601 chr DNA → HB5614 |
| HB5616 | CU1065 yfiY::spc [DHB(G)+] | LFH-PCR → CU1065 |
| HB5617 | CU1065 yfiY::spc dhbA::mls | HB5601 chr DNA → HB5616 |
| HB5618 | CU1065 yfiZ::cat [DHB(G)+] | LFH-PCR → CU1065 |
| HB5619 | CU1065 yfiZ::cat dhbA::mls | HB5601 chr DNA → HB5618 |
| HB5620 | CU1065 ywjA::spc [DHB(G)+] | LFH-PCR → CU1065 |
| HB5621 | CU1065 ywjA::spc dhbA::mls | HB5601 chr DNA → HB5620 |
| HB5622 | CU1065 ywbL::cat [DHB(G)+] | LFH-PCR → CU1065 |
| HB5623 | CU1065 ywbL::cat dhbA::spc | HB5600 chr DNA → HB5622 |
| HB5624 | CU1065 feuA::spc [DHB(G)+] | LFH-PCR → CU1065 |
| HB5625 | CU1065 feuA::spc dhbA::mls | HB5601 chr DNA → HB5624 |
| HB5626 | CU1065 yxeB::spc [DHB(G)+] | LFH-PCR → CU1065 |
| HB5628 | CU1065 yfhA::spc [DHB(G)+] | LFH-PCR → CU1065 |
| HB5630 | CU1065 yusV::mls feuA::spc [DHB(G)+] | HB5610 chr DNA → HB5624 |
| HB5800 | sfp+ (prototroph) (bacillibactin+) | OKB105 chr DNA → CU1065 |
| HB5700 | HB5800 dhbA::spc | HB5600 chr DNA → HB5800 |
| HB5701 | HB5800 dhbA::mls | HB5601 chr DNA → HB5800 |
| HB5712 | HB5800 yfmC::mls (bacillibactin+) | HB5612 chr DNA → HB5800 |
| HB5722 | HB5800 ywbL::cm (bacillibactin+) | HB5622 chr DNA → HB5800 |
| HB5710 | HB5800 yusV::mls | HB5610 chr DNA → HB5800 |
| HB5714 | HB5800 yuiI::spc | HB5614 chr DNA → HB5800 |
| HB5724 | HB5800 feuA::spc | HB5624 chr DNA → HB5800 |
| HB5730 | CU1065 amyE::PycgT-lacZ(cat) | See Materials and Methods |
| HB5731 | CU1065 fur::kan amyE::PycgT-lacZ(cat) | See Materials and Methods |
The siderophores produced by each strain are indicated. Only sfp+ strains produce bacillibactin. DHB(G) refers to the mixture of 2,3-dihydroxybenzoic acid and its glycine conjugate (Itoic acid) produced by 168 (Sfp0) strains that are not mutated at the dhb locus. Except where indicated, all gene disruptions were double-crossover, allelic replacements using the indicated antibiotic cassettes: kan, kanamycin resistance; spc, spectinomycin resistance; mls, macrolide-lincomycin streptogramin B resistance; cat, chloramphenicol resistance.
Some strains were constructed by long-flanking homology PCR (LFH-PCR) or by transformation of chromosomal DNA (chr DNA) from one donor strain into the indicated recipient with selection for the antibiotic marker associated with the donor DNA.
Mutations were introduced into various Fur-regulated genes by using long-flanking homology PCR (62). Briefly, oligonucleotide primers were used to amplify ∼1-kb DNA segments upstream and downstream of the gene(s) to be deleted (primer sequences available upon request) using high-fidelity Pfu DNA polymerase. These two DNA segments were mixed with the desired antibiotic resistance cassettes which had been amplified with primers containing appropriate sequence overlaps with the flanking cassettes. Antibiotic resistance cassettes were amplified from pGEM-cat(cat) (66), pDG647(mls), or pDG1727(spc) (24). The three DNA fragments were then combined by sequence overlap PCR, using the Expand High FidelityPLUS PCR system (Roche), to generate the desired allelic replacement products. These PCR products were used for direct transformation into CU1065 to generate the strains shown in Table 1. In several cases, mutations were made by using more than one resistance cassette to facilitate the construction of double mutant strains. Multiply mutant strains were constructed by chromosomal DNA transformation as summarized in Table 1. Mutants defective in predicted iron uptake functions were checked in multiple strain backgrounds including CU1065(sfp0), CU1065(sfp+), and CU1065dhbA (Table 1 and data not shown).
For construction of a ycgT-lacZ promoter fusion, the putative promoter region of ycgT was amplified by using a forward primer ∼350 bp upstream of the putative fur box (with restriction site EcoRI) and a reverse primer immediately upstream of the ycgT start codon (with restriction site BamHI). Chromosomal DNA of CU1065 was amplified by using the Expand High FidelityPLUS PCR system (Roche), and the resulting PCR fragment was cloned into pDG1661 (23). The ycgT promoter-pDG1661 clone was linearized by digestion with ScaI and integrated into the amyE locus of B. subtilis CU1065 and the isogenic fur mutant (HB2501) by double crossover recombination with selection for chloramphenicol resistance.
Chemical reagents and siderophores.
Unless otherwise indicated, all chemical reagents were obtained from Sigma Chemical Co. (St. Louis, MO) in the highest available purity. The siderophores and chelators used in the present study are summarized in Table 2. Ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDHA) was obtained from Sigma-Aldrich, but we note that it is no longer commercially available. 2,3-Dihydroxybenzoic acid (DHBA) was used as a standard when monitoring the production of siderophore from B. subtilis cultures. Ferrichrome and desferrioxamine were obtained from Sigma-Aldrich in both iron-free and iron-loaded forms. Enterobactin (iron-free and iron-loaded), and iron-loaded rhizoferrin and coprogen were obtained from EMC Microcollections GmbH Tuebingen. Iron-loaded schizokinen and arthrobactin were obtained from K. Hantke (Tuebingen, Germany). Note that growth stimulation was obtained with iron-free siderophores due to their ability to solubilize and scavenge trace iron in the growth medium.
TABLE 2.
Siderophores and chelators used in this study
| Siderophore or chelator | Chemical class | Representative producing strain(s)a | pFeb |
|---|---|---|---|
| DHBA | Catechol | Bacillus subtilis 168 strains | ∼15 |
| Bacillibactin | Catecholate | Bacillus subtilis sfp+ strains | 33.1 |
| Enterobactin | Catecholate | Escherichia coli, Salmonella enterica, and other enterobacteria | 34.3 |
| Ferrioxamine b | Hydroxamate | Streptomyces spp., Arthrobacter simplex, Pseudomonas stuzeri, Chromobacterium violaceum, and Hafnia alvei | 26.6 |
| Ferrichrome | Hydroxamate | Ustilago spp., Aspergillus spp., Penicillium parvum, Neovossia indica, and Trichophyton mentagrophytes | 25.2 |
| Coprogen | Hydroxamate | Neurospora crassa, Penicillium spp., curvularia lunata, and Epicoccum purpurascens | 27.5 |
| Schizokinen | Carboxylate/hydroxamate | B. megaterium and Ralstonia solanacearum | NA |
| Arthrobactin | Carboxylate/hydroxamate | Arthrobacter pascens | NA |
| Citric acid | Carboxylate | Bradyrhizobium japonicum | 17.7 |
| EDDHA | Carboxylate | (Ferric chelator) | 26.9 |
Adapted from Stintzi and Raymond (60).
pFe = (−log[Fe3+] at pH 7.4, [Fe]tot = 1 μM, [L]tot = 10 μM). Values for pFe are from DHBA (estimated from measurements of 2,3-dihydroxy-N,N-dimethylbenzamide [26]), enterobactin and bacillibactin (16), other siderophores (65), citric acid (50), and EDDHA (58; K. Raymond, unpublished data). NA, values not available in the literature.
Proteome analysis.
Cells were grown in Luria-Bertani (LB) medium until the transition into the stationary phase and harvested 1 h after the transition phase. To prepare extracellular proteins, the proteins in the medium were precipitated with ice-cold 10% (wt/vol) trichloroacetic acid and prepared for proteome analysis as described previously (2). To prepare cytoplasmic proteins, cells were washed and disrupted by using a French pressure cell, and the soluble protein fraction was separated from the cell debris by centrifugation as described previously (12). The protein content was determined by using the Bradford dye-binding assay (9), and 200 μg of the protein extract was separated by two-dimensional gel electrophoresis (2D-PAGE) using the nonlinear immobilized pH gradients (Amersham Biosciences) in the pH range 4 to 7 or 3 to 10 for cytoplasmic or extracellular proteins, respectively, as described previously (6, 7). The resulting 2D gels were fixed in 40% (vol/vol) ethanol-10% (vol/vol) acidic acid and stained with colloidal Coomassie brilliant blue (Amersham Biosciences). The image analysis was performed with the DECODON Delta 2D software. Spot cutting, tryptic digestion of the proteins, and spotting of the resulting peptides onto the matrix-assisted laser desorption ionization (MALDI) targets (Voyager DE-STR; PerSeptive Biosystems) was performed by using the Ettan Spot Handling Workstation (Amersham-Biosciences, Uppsala, Sweden) as described previously (17). The MALDI-time of flight (TOF)-TOF measurement of spotted peptide solutions was carried out on a Proteome-Analyzer 4700 (Applied Biosystems, Foster City, CA) as described previously (17).
Electrophoretic mobility shift assay.
A 316-bp ycgT promoter fragment containing a PvuI restriction site was generated by PCR with B. subtilis CU1065 genomic DNA as a template and the primers 2606 (5′-GGAATTCCTATTATAGAGTTTTCAGC-3′) and 2607 (5′-TATCGCGAATGATTTTCTCAGG-3′). The fragment was gel purified and end labeled by using T4 polynucleotide kinase and [γ-32P]dATP, followed by restriction digestion with PvuI to give a 196-bp specific and a 121-bp nonspecific fragment. The Fur protein and labeled DNA were incubated in binding buffer (20 μl) consisting of 20 mM Tris-HCl (pH 8.0), 5% (vol/vol) glycerol, 5 μg of salmon sperm DNA/ml, 50 μg of bovine serum albumin/ml, and 50 mM KCl for 10 min. The reaction was then subjected to electrophoresis on a 6% (29:1) nondenaturing gel with 40 mM Tris-borate buffer (pH 8.0) at 60 V for 1.5 h using Mini-PROTEIN 3 Cell (Bio-Rad). After drying, the gel was visualized by using a STORM PhosphorImager, and the DNA was quantified by using ImageQuant software (Molecular Dynamics).
Growth media.
Precultures were grown by overnight cultivation in LB medium at 37°C. LB medium has, in general, between 8 and 10 μM iron, and the Fur regulon is repressed under these conditions. Overnight precultures were diluted 1:100 into Fe starvation minimal medium (FS-MM). FS-MM was prepared by using Milli-Q purified water and contains 40 mM potassium morpholinepropanesulfonic acid (MOPS) (adjusted to pH 7.4 with KOH), 2 mM potassium phosphate buffer (pH 7.0), glucose (2% [wt/vol]), (NH4)2SO4 (2 g/liter), MgSO4 · 7H2O (0.2 g/liter), trisodium citrate · 2H2O (1 g/liter), potassium glutamate (1 g/liter), tryptophan (10 mg/liter), 3 nM (NH4)6Mo7O24, 400 nM H3BO3, 30 nM CoCl2, 10 nM CuSO4, 10 nM ZnSO4, and 80 nM MnCl2. To prepare FS-MM, a fivefold-concentrated MM salts solution (omitting metal ions) was prepared and treated with Chelex to help eliminate adventious iron contamination. Where indicated, EDDHA was added to the FS-MM during growth of cells in liquid culture. Cells were cultured in 15-ml borosilicate glass culture tubes that were pretreated by washing with 1 N HCl and extensive rinsing with Milli-Q water. For preparation of the FS-MM agarose plates, 1.5% Bacto-Agar was used for solidification and Fe-deficient conditions were maintained by the inclusion of 10 μM EDDHA.
Assay of DHB(G) production.
Production of DHBA and DHBG by iron-starved B. subtilis 168 derivatives was monitored in 1 ml of cell-free supernatant by the addition of 50 μl of 10 mM FeCl3 (in 100 mM HCl), followed by neutralization with 100 μl of 1 M Tris-HCl buffer (pH 8.0). The resulting purple DHB(G)-Fe complex was measured spectrophotometrically. An optical density at 510 nm (OD510) of 0.5 corresponds to ∼80 μg of DHB(G) per ml.
β-Galactosidase assays.
Each strain was grown overnight in LB medium containing appropriate antibiotics and diluted 100-fold into fresh LB medium without antibiotics or into FS-MM as indicated. β-Galactosidase activity was measured in triplicate by using the method of Miller (38).
Disk diffusion assays.
B. subtilis strains were grown in 5 ml of FS-MM at 37°C until cells reached middle logarithmic phase (OD600 ∼ 0.5) and assayed for growth stimulation by siderophores using a modification of the assay of Schneider and Hantke (57). Then, 100 μl of the culture was added to 3 ml of soft agar plus 10 μM EDDHA and immediately poured onto plates containing 15 ml of FS-MM. After solidification, a filter paper disk (6 mm) containing 10 μl of a 1-mg/ml solution of the siderophore to be tested was placed in the center of the petri dish. The plates were incubated for 24 h at 37°C. Those strains able to obtain iron in the presence of the added siderophore formed a zone of growth surrounding the disk. To test for growth stimulation by exogenous bacillibactin, 10 μl of filter-sterilized supernatant from iron-starved cultures of B. subtilis NCBI3610 or CU1065sfp+ was used as the source of siderophore. Note that this growth stimulation assay is useful only for those siderophores that have an Fe3+ affinity comparable to or greater than that of EDDHA (Table 2).
Liquid culture growth experiments in the presence of exogenous siderophores.
The ability of siderophores to support growth in liquid culture was assessed by monitoring the growth rate and cell yield after overnight growth in the presence of various iron sources. For preliminary screening of all mutants, cultures were grown in 96-well microtiter plates with FS-MM containing 1 μM EDDHA to restrict growth (instead of the 10 μM used in the disk diffusion assays on agar plates). Siderophores were added to a final concentration of 1 μM. Growth was monitored by determining the OD600 using a Tecan Rainbow microplate reader. For more accurate quantitation of growth, cells were cultured in 15 ml of acid-washed borosilicate glass culture tubes in a volume of 5 ml, and the OD600 was determined as a function of time (see, for example, Fig. 4 and 5).
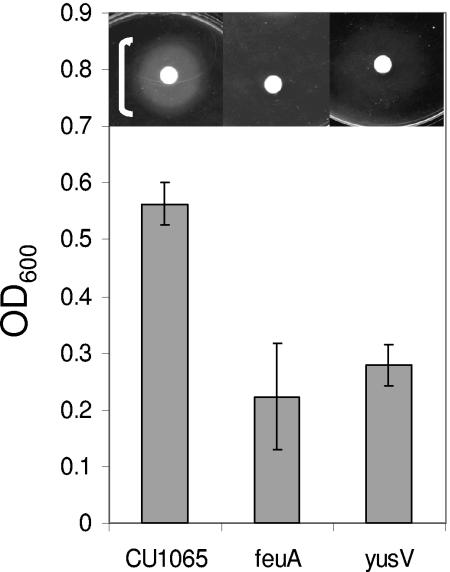
FIG. 4.
Genetic requirements for growth stimulation by enterobactin. Filter disks (inset) containing enterobactin were added to iron starvation minimal medium (FS-MM) plates containing EDDHA to impose Fe limitation. White brackets indicate the extent of observed growth stimulation. A thin bracket indicates weak growth. For growth in liquid culture, the indicated strains were grown in FS-MM in triplicate, and the final cell density (OD600) was measured. The results shown are mean ± the standard deviation.
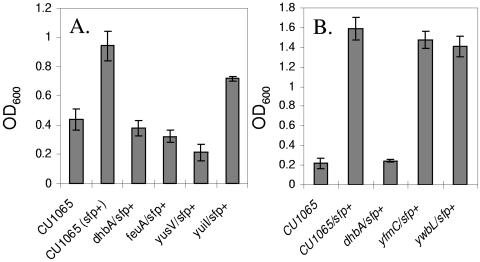
FIG. 5.
Effects of bacillibactin synthesis on growth under conditions of iron limitation. Cells were grown in FS-MM+Cit containing 5 μM EDDHA and 1 μM ferric ammonium citrate. (A) Under these conditions, growth to high cell density is blocked by mutants deficient in bacillibactin synthesis (dhb and sfp0 in CU1065) or transport (feuA and yusV). (B) Bacillibactin-dependent growth is independent of the yfmC and ywbL loci. All strains were cultured in triplicate, and the final cell density (OD600) was measured after overnight growth. The results shown are mean ± the standard deviation. Note that the background level of growth differs between panels A and B, presumably due to differences in the medium and carryover of iron from the inoculum.
Liquid culture growth experiments in the absence of exogenous siderophores.
B. subtilis cells were grown in LB medium for 24 h at 37°C, and 100 μl was used to inoculate 5 ml of LB medium to mid-logarithmic growth phase (OD600 ∼ 0.4). Cells were harvested and washed once with 1 ml of FS-MM without citrate. Cells were resuspended to a final OD600 of 0.8, and 5 μl was used to inoculate 200 μl of medium in a 96-well microtiter plate. The cell density was measured as a function of time of incubation at 37°C by using a Tecan Rainbow microplate reader. The results shown (see Fig. 6) are representative of growth experiments performed at least three times. Similar results were seen in cultures grown in glass tubes.
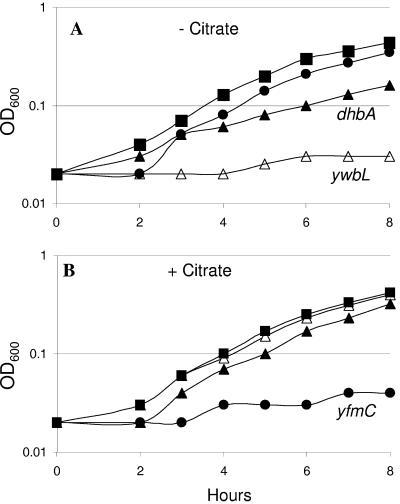
FIG. 6.
Genetic requirements for growth of B. subtilis 168 in FS-MM with or without citrate. (A) In FS-MM lacking citrate, good growth was observed for CU1065 (▪) and the isogenic yfmC mutant (•). The dhbA mutant (▴) grows somewhat more slowly, and the ywbL mutant (▵) fails to grow. (B) In the presence of citrate, all strains grow well with the exception of the yfmC mutant. Growth was assessed by using 96-well microtiter plates as described in Materials and Methods.
RESULTS AND DISCUSSION
In previous work, Fur was shown to regulate the transcription of about 20 operons (∼40 genes) in response to iron availability, many of which encode putative transporters (5). The Fur regulon includes at least four different ABC transporters (transmembrane and ATPase components), together with five substrate-binding lipoproteins (5). In addition, the ywbLMN operon encodes a protein (YwbL) with similarity to the elemental iron uptake channel in the yeast Saccharomyces cerevisiae, as noted originally by Dancis and coworkers (59) (Table 3). Here, we have used proteomics to monitor and identify Fur-regulated proteins and assessed the functional role of the various transport systems by mutational analysis.
TABLE 3.
Fur regulon: comparison of transcriptome and proteomics results
| Operon(s)a | Derepression by DP/furb | Proteins detected in fur mutantc | Proteins detected in previous proteome analysesd | Functione |
|---|---|---|---|---|
| dhbACEBF | 107/150 | DhbACEB (c) | DhbACEB (NaCl, ox) | Bacillibactin biosynthesis (37) |
| ybbB | 8/9 | AraC regulator fused to FeuA domain (adjacent to feuABCybbA) | ||
| feuABCybbA | 7/23 | FeuA (c) | FeuA (c, s*) | ABC-transporter/esterase: bacillibactin and enterobactin |
| yxeB | 26/17 | YxeB (c, s) | YxeB (m, s*) | SBP ferrioxamine: works with FhuBCG |
| fhuD | 5/5 | FhuD (m, s*) | SBP ferrichrome: works with FhuBCG | |
| fhuBCG | 8/7 | FhuC (m) | ABC-transporter: ferric hydroxamates | |
| yfiY | 6/11 | YfiY (s) | YfiY (c, s) | SBP schizokinin, arthrobactin |
| yfiZ yfhA | 3/2 | ABC-transporter: schizokinin, arthrobactin | ||
| yusV | 2/2 | ATPase component for YfiYZYfhA and FeuABC | ||
| yhfQ | 5/4 | YhfQ (m) | SBP: unkown siderophore uptake | |
| yclNOPQ | 5/2 | YclQ (c, s) | YclQ (s, m) | ABC transporter: unknown siderophore uptake |
| ywbLMN | 4/3 | YwbM (c) | YwbM (m) | Elemental Fe transport (yeast FTS3 homolog) |
| YwbN (Tat) | Predicted iron-dependent peroxidase | |||
| yuiI | 20/25 | Predicted esterase (adjacent to dhb operon) | ||
| yfmCDEF | 3/2 | YfmC (s) | YfmC (m, s*) | Fe-citrate uptake |
| yoaJ | 3/4 | YoaJ (s) | Secreted endoglucanasef | |
| ywjAB | 5/5 | ABC transporter homolog, DHFR homolog | ||
| ycgT | 9/10 | YcgT (c) | Thioredoxin reductase homolog |
Insertional inactivation mutants of the 14 transcription units shown in boldface were analyzed in this study (Table 1). A total of 17 operons, encoding 36 proteins, are included here. Other Fur-regulated transcription units include the fliDST operon, ykuNOP, and ydbN, and two small RNAs (4, 5; unpublished results).
Fold derepression (average for all genes in operon) in cells treated with 2,2′-dipyridyl (DP) or in a fur-null mutant (from previously published results [5], although ycgT was not included in the previous tabulation of the Fur regulon; see the text).
That is, proteins detected in the present study. Proteins detected for the first time in proteomics studies are in boldface. A “c” in parentheses indicates proteins detected in the cytoplasmic fraction, and “s” indicates proteins detected in the extracellular (secreted) proteome of wild-type cells and includes proteins normally tethered to the cell membrane or cell wall.
That is, proteins detected in previous proteome studies of B. subtilis. “NaCl, ox” refers to the detection of the Dhb biosynthetic enzymes under conditions of high-salt or oxidative stress known to induce the Fur regulon (29, 42). “s” refers to proteins detected in the extracellular proteome, and “s*” indicates lipoproteins that are shed to the medium in a Δlgt strain deficient in the diacylglycerol transferase (Lgt) involved in achoring proteins to the extracellular surface of the membrane (3). “m” refers to proteins identified using sodium dodecyl sulfate-PAGE, followed by ESI-MS/MS (17), or by washing purified membranes with the nondetergent sulfobetaine 256 (13). “Tat” refers to the fact that YwbN was only detected after epitope tagging and was identified as a specific substrate of the tatAy tatCy twin-arginine dependent secretion system (32).
Functional assignments inferred from the genetic and physiological studies reported here are in boldface. SBP, substrate-binding protein.
Proteome analysis of fur mutant cells.
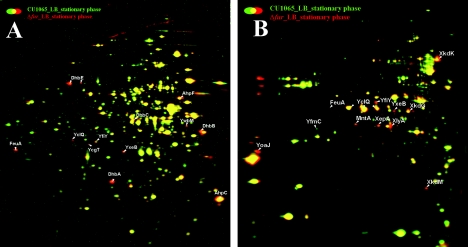
To monitor the effects of Fur on the proteome, we compared wild-type and fur mutant cell fractions by using 2D-PAGE (Fig. 1). The fur mutant displayed elevated levels of the Dhb proteins involved in the synthesis of the B. subtilis bacillibactin siderophore (8, 37, 56), YwbM, and four predicted siderophore-binding proteins (FeuA, YfiY, YxeB, and YclQ [53]). These and other Fur-regulated proteins have been detected in previous proteome studies: derepression of the Dhb biosynthetic enzymes has been noted under conditions of salt stress (29) and oxidative stress (42), and many of the substrate-binding proteins have been detected in analyses of membrane-associated (13, 17) or secreted proteins (3) (Table 3). Analysis of the extracellular proteome also revealed derepression of the Fur-regulated YoaJ protein, a predicted endoglucanase. This protein had not been previously detected in proteome analyses. In addition, the fur mutant displayed an increased synthesis of proteins encoded by the PBSX defective prophage. It is not clear whether this is a direct or an indirect effect of the fur deletion.
FIG. 1.
Effects of a fur mutation on the B. subtilis proteome. The dual-channel image of the cytoplasmic (A) and extracellular proteomes (B) of B. subtilis wild type (CU1065) grown in complete medium (green image) compared to the isogenic fur mutant (red image). Cells grown in LB medium were harvested 1 h after the transition phase. Cytoplasmic and extracytoplasmic proteins were prepared and separated by 2D-PAGE as described in Materials and Methods. The image analysis was performed by using Decodon Delta 2D software. Proteins (red) that displayed elevated levels in the fur mutant in comparison to the wild type are labeled. Note that FeuA, YfiY, YxeB and YclQ were found in both cellular fractions.
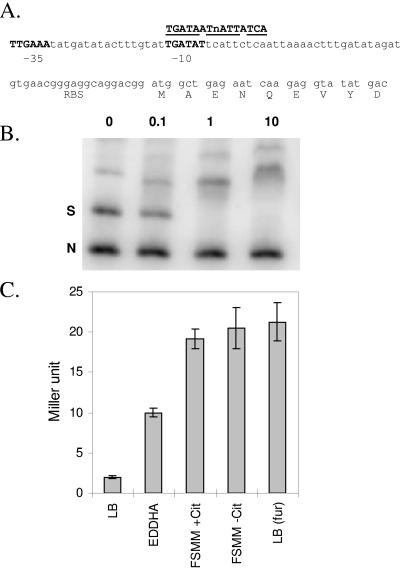
Analysis of the cytoplasmic proteome in the fur mutant also revealed an ∼3-fold derepression of a putative thioredoxin reductase, YcgT. In our previous transcriptome studies (4, 5), this gene was detected as an ∼10-fold derepressed in iron-starved (dipyridyl-treated) cells and was similarly derepressed in a fur mutant strain. However, because of variability in the data (due largely to one aberrant datum point), this gene was filtered out during analysis and therefore not included in our tabulation of Fur-regulated genes. Inspection of the ycgT promoter region revealed the presence of a candidate σA-dependent promoter and a 12/14 match to the minimal Fur-binding site (4) (Fig. 2A). Therefore, we hypothesized that ycgT is a previously unrecognized member of the Fur regulon. Consistent with this hypothesis, in electrophoretic mobility shift assays Fur protein bound with high affinity to the ycgT regulatory region (Fig. 2B), and a ycgT::lacZ fusion was ∼10-fold derepressed in a fur mutant strain and when grown under iron-limiting conditions imposed either by the addition of EDDHA or the use of iron-starvation minimal medium (FS-MM) (Fig. 2C). Coregulation of ycgT with Fur-regulated genes is also apparent in other transcriptome studies under conditions that perturb expression of the Fur regulon (39, 41).
FIG. 2.
Identification of YcgT as a new member of the Fur regulon. (A) DNA sequence of the ycgT regulatory region showing candidate −35 and −10 elements for σA-holoenzyme and an overlapping Fur box with a 12/14 match to the 7-1-7 consensus for Fur protein (4). The ribosome-binding site (RBS) and the DNA encoding the first nine codons are shown. (B) Binding of purified Fur protein to the ycgT regulatory region. A 316-bp DNA fragment spanning the ycgT regulatory region was amplified using PCR and digested with PvuI to generate a specific fragment (S; containing the candidate Fur box element) and a nonspecific fragment (N). Concentrations of added Fur protein (nM monomer) are indicated. Complete binding of the specific fragment is observed with 1 nM Fur monomer. (C) Expression of a ycgT::lacZ transcriptional fusion in wild-type cells (CU1065) is repressed when growing in iron-rich medium (LB medium) but induced 30 min after treatment with 10 μM EDDHA or during growth in FS-MM (with or without citrate [+Cit or −Cit]). Derepression is also noted in LB medium-grown cells of the isogenic fur mutant (right column).
Identification of genes required for siderophore-dependent growth stimulation.
To address the substrate specificity of the various Fur-regulated transporters, we generated gene disruption mutants by using allelic replacement mutagenesis (Table 1). Mutant strains were then tested for their ability to use various siderophores (Table 2) as a source of iron under iron-deficient conditions (see Materials and Methods). For our initial studies, we took advantage of the fact that B. subtilis 168 derivatives carry the sfp0 mutation and produce DHB(G) under iron limitation. Since DHB(G) binds Fe(III) with relatively low affinity (Table 2), sfp0 strains were unable to grow in Fe-starvation minimal medium (FS-MM) amended with the iron-chelator EDDHA. Where noted, we have also used derivatives of our wild-type strain that produce bacillibactin (CU1065 sfp+) or strains deficient for the production of both DHB(G) and bacillibactin due to the presence of a dhb mutation.
Utilization of hydroxamate siderophores.
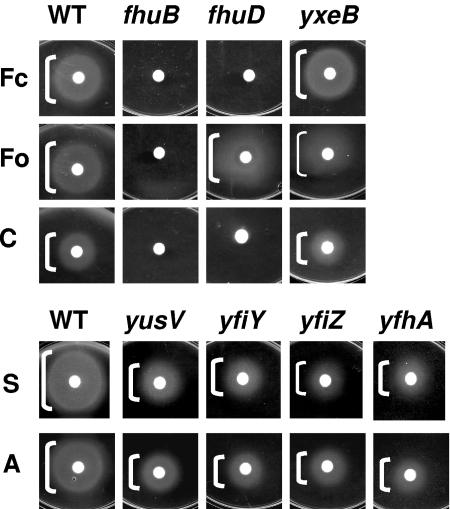
Like many bacteria, B. subtilis can utilize a range of siderophores made by other organisms. Significant growth stimulation was observed with the hydroxamate siderophores ferrichrome, ferrioxamine, coprogen, schizokinen, and arthrobactin. As noted previously (57), growth stimulation by ferrichrome and ferrioxamine required the fhuBGC ABC transporter (Fig. 3A). In addition, growth with ferrichrome required the FhuD substrate-binding protein, whereas stimulation by ferrioxamine was at least partially dependent on YxeB. This is consistent with the previous isolation of mutants (foxD) defective in ferrioxamine uptake (57) that map very near the yxeB locus (5). Note that in these and subsequent experiments, only mutants with significant growth defects are shown. All growth effects seen in this plate assay have also been confirmed by using growth experiments in liquid medium (data not shown).
FIG. 3.
Genetic requirements for growth stimulation by hydroxamate siderophores. Filter disks containing ferrichrome (Fc), ferrioxamine (Fo), coprogen (C), schizokinen (S), or arthrobactin (A) were added to iron starvation minimal medium (FS-MM) plates containing EDDHA to impose Fe limitation. The strains used were CU1065 (wild type [WT]) or the indicated isogenic mutants. Only mutations leading to a growth defect are shown. White brackets indicate the extent of observed growth stimulation. Thin brackets indicate weak growth.
Growth stimulation in the presence of schizokinen and arthrobactin was significantly reduced in the yfiY, yfiZ, yfhA, and yusV single mutants: in each case the diameter of growth stimulation on plates was reduced by ∼2-fold (Fig. 3B). We note that since yfiZyfhA are in an operon (Table 3), the yfiZ mutant was likely polar on yfhA expression. We conclude that YfiY, YfhA, and the YusV ATPase are all required for efficient growth stimulation and, in light of the observed genetic organization, it is likely that YfiZ functions as part of the same ABC transporter. The growth defects in this experiment were not as dramatic as those observed with ferrichrome and ferrioxamine (Fig. 3A). This is likely due to the fact that schizokinen and arthrobactin were only available in their iron-loaded forms. Studies performed with apo-siderophores (when available) more clearly revealed the genetic requirements for ferri-siderophore uptake, presumably due to contaminating free iron in the iron-loaded siderophores. For example, iron-free ferrichrome was unable to stimulate growth of the fhuB or fhuD mutants (Fig. 3A), whereas Fe-loaded ferrichrome gave a small but significant zone of growth stimulation (data not shown).
Utilization of catecholate siderophores.
B. subtilis 168 strains contain a single siderophore biosynthesis operon (dhbACEBF) that encodes the biosynthetic enzymes for the catecholate siderophore bacillibactin: the trimeric cyclic ester of 2,3-dihydroxybenzoyl-Gly-Thr (37). Since B. subtilis 168 derivatives cannot synthesize bacillibactin (due to the sfp0 mutation), growth in the presence of the strong iron chelator EDDHA requires transport of an exogenously provided siderophore. Growth stimulation by enterobactin required the feuABC-encoded ABC transporter and the yusV-encoded ATPase (Fig. 4). Growth stimulation by cell-free supernatants containing bacillibactin (from iron-starved B. subtilis sfp+ cells) also required the FeuABC transporter and YusV ATPase but was not affected by other mutations (data not shown).
The use of a common transporter for both enterobactin and bacillibactin is consistent with recent transport studies that demonstrated effective inhibition of radiolabeled bacillibactin uptake in the presence of excess, nonlabeled enterobactin (16). Similar results have been reported in Salmonella enterica serovar Typhimurium, where the IroN outer membrane protein can recognize both ferric-siderophores (54). In contrast, E. coli FepA recognizes ferric enterobactin but interacts very poorly with ferric bacillibactin (2). Despite the fact that both enterobactin and bacillibactin are trimeric, cyclic lactones with dihydroxybenzoate chelating groups, the two molecules form ferric complexes with opposite chirality (8), and this distinction may account for the discrimination noted for some siderophore-binding proteins.
As expected, sfp+ derivatives of B. subtilis did not require exogenous siderophore for growth even in the presence of up to 50 μM EDDHA, a finding consistent with their ability to synthesize bacillibactin. Mutants defective either for bacillibactin synthesis (dhbA) or for uptake (feuA or yusV) grew poorly under these conditions (Fig. 5A). Other tested mutations (e.g., yuiI, yfmC, and ywbL) had little or no effect on growth (Fig. 5). Together, these results indicate that the uptake of bacillibactin requires the FeuABC transporter and YusV ATPase regardless of whether the siderophore is provided exogenously or produced by the cells.
Bacillibactin, like enterobactin, binds Fe(III) with extremely high affinity (16). Indeed, enterobactin is the strongest iron-chelator characterized to date with an estimated Kd of ∼10−49 M (∼10−35 M at physiological pH; see, for example, Table 2) (8, 55). The release of iron from enterobactin requires hydrolysis of the ferric-enterobactin complex inside the cell, although reduction of bound Fe(III) to Fe(II) may also play a role. Enterobactin hydrolysis requires the Fes protein, a member of the α,β-hydrolase family of enzymes. Sequence comparisons suggest that YbbA, encoded by the feuABCybbA operon, may play a similar role in B. subtilis (67). B. subtilis also encodes an additional predicted bacillibactin esterase, YuiI. The yuiI gene is immediately upstream of the dhbACEBF biosynthetic operon and encodes an esterase most closely related to IroE. Since IroE functions to linearize apo-enterobactin during export (34), and yuiI is located next to the biosynthetic gene cluster, it is reasonable to suggest that yuiI may also encode an esterase active during export. As noted above, a mutation in yuiI had only a small effect on the ability of cells to utilize bacillibactin for iron uptake (Fig. 5A). Further studies will be needed to define the roles of YuiI and YbbA in bacillibactin processing during export or subsequent to import.
Identification of an iron-citrate uptake system.
In the absence of added exogenous siderophore, CU1065 was unable to grow in liquid medium amended with >1 μM EDDHA. However, good growth was routinely observed in FS-MM in the absence of EDDHA. To gain insight into the pathway(s) responsible for iron uptake in FS-MM, we again tested our panel of mutant strains for effects on growth. The only strain that was dramatically impaired in growth contained a mutation in the yfmCDEF operon (Fig. 6B). Since this operon encodes an ABC-transporter, we hypothesized that this system transports ferric citrate into the cell. Consistent with this hypothesis, a yfmC mutant was unaffected in its ability to grow in FS-MM in the absence of citrate (Fig. 6A). Operons similar to yfmCDEF are widely distributed in bacterial genomes and are typically annotated as encoding ABC transporters for ferrichrome or, more generally, iron uptake. An operon orthologous to yfmCDEF is encoded in E. coli O157/H7 (Z4385-Z4382) and is a predicted member of the Fur regulon in this pathogen (45). A distantly related operon in B. cereus, one of several annotated as encoding ferrichrome-ABC uptake systems, has recently been shown to allow growth on ferric citrate as sole iron source (27), which is consistent with our findings here.
The inability of the yfmC mutant to grow in FS-MM containing citrate suggests that DHB(G) is unable to compete effectively with citrate for the trace iron in this medium. This is consistent with the greater stability of the ferric citrate complex compared to the DHB(G) complexes at physiological pH (Table 2) and with the previous observation that Salmonella mutants that produce DHBA are unable to obtain iron from citrate-supplemented medium (51). We reasoned that the production of bacillibactin, a much stronger Fe(III) chelator than DHB(G), should be able to restore growth to the yfmC mutant strain. Consistent with this expectation, a yfmC sfp+ strain grew well even in the presence of 5 μM EDDHA (Fig. 5B). However, in FS-MM containing citrate, growth of the yfmC sfp+ strain was variable and dependent on the growth phase of the inoculum. This is surprising since bacillibactin should, at equilibrium, bind iron in the presence of citrate (Table 2). However, it is known that the kinetics of iron transfer from ferric citrate complexes to siderophores are highly sensitive to solution conditions (18, 21), and this likely accounts for the variable results observed.
Role of elemental Fe uptake.
B. subtilis CU1065 was still able to grow in FS-MM lacking citrate. Thus, under these conditions another high-affinity pathway of iron uptake must be active. Mutant analyses revealed that the ywbLMN operon was required for growth under these conditions (Fig. 6A). Bioinformatic analyses suggest that this operon encodes an elemental iron uptake system (61). By analogy with the well-characterized Saccharomyces cerevisiae Ftr1p/Fet3p system, we suggest that YwbLMN functions as an Fe(III) permease. It has been previously shown that YwbN is unique among secreted B. subtilis proteins in that it requires the TatAyCy twin-arginine translocase for secretion (32). Secretion via a dedicated Tat system presumably reflects the requirement for a bound cofactor assembled into the YwbN protein: a member of the dye-decolorizing family of predicted iron-dependent peroxidases (COG2837). Although widely conserved among the bacteria (61), these genes have not, to our knowledge, been previously demonstrated to function in iron homeostasis. Under these conditions, the dhb operon also contributes to growth. Presumably, DHB(G) contributes to the solubilization of trace iron in the medium which may then be transported into the cell using the YwbLMN permease. The role of the YwbLMN system in the uptake of iron from ferric-DHB(G) is consistent with recent transport studies demonstrating that ferric-bacillibactin competes poorly with radiolabeled 55Fe-DHBG for uptake (16).
Iron uptake in other bacilli.
Genome sequence analysis has facilitated recent insights into the iron uptake pathways for several gram-positive pathogens, including B. anthracis and B. cereus. B. anthracis, for example, contains biosynthesis operons for two distinct siderophores (14). The B. anthracis anthrabactin (bac) genes are similar to the B. subtilis bacillibactin biosynthesis genes (dhb operon), while the asb operon is similar to genes for the synthesis of the hydroxamate siderophores (14). Analysis of culture supernatant demonstrates the presence of a catecholate siderophore of the predicted molecular mass for bacillibactin and a second, more abundant species containing 3,4-catechol moieties and assigned as petrobactin (33). B. anthracis can also obtain iron from heme, hemoproteins, and transferrin. B. cereus has been shown to express catecholate siderophores under the regulation of Fur (28, 46), and the fecABC operon encoding ferric citrate uptake functions was found to be required for virulence in an insect infection model (27).
Summary and conclusions.
The major outlines of the B. subtilis Fur regulon were previously defined using a combination of transcriptome analyses and searches of the genome for Fur box like-regulatory elements (4, 5). Here we have complemented that work with a proteome analysis comparing wild-type and fur mutant strains. This confirms the Fur-dependent repression and subcellular localization of several regulon members and additionally identifies YcgT as a new member of the Fur regulon. The ycgT gene was missed during our previous work due to the conservative statistical filtering algorithm used and the fact that the ycgT Fur box closely matches the 7-1-7 consensus sequence (4) but not the longer 19-bp consensus sequence. The function of YcgT, encoding a thioredoxin reductase homolog, is not obvious, and proteins of this family are not commonly observed as members of Fur regulons.
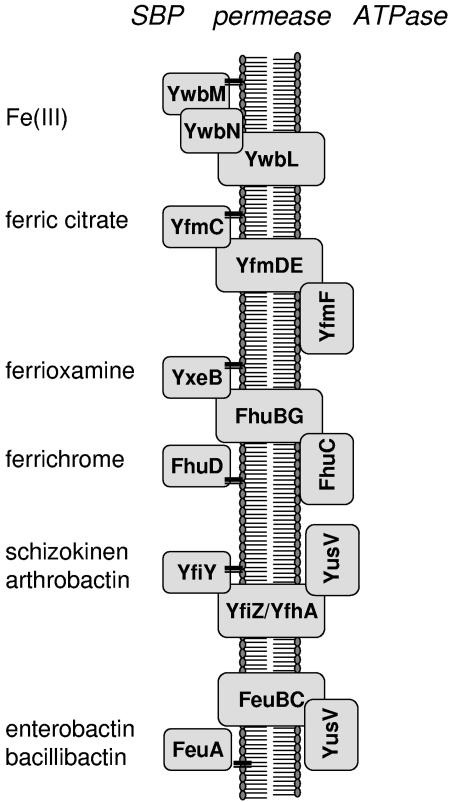
We have also determined the roles of various Fur-regulated ABC transporters in supporting growth in the presence of various iron sources (Fig. 7). These results significantly extend and refine previous studies of iron uptake in B. subtilis 168, which focused primarily on hydroxamate-class siderophores (57). Our results reveal an important role of iron-citrate transport, mediated by the YfmCDEF system, in mediating growth in the absence of siderophores and demonstrate that the predicted YwbLMN elemental iron uptake system is required for growth in iron-limited medium lacking citrate. Orthologs of the YwbLMN system are widely distributed in the bacteria, and bioinformatic analyses had suggested a likely role in iron uptake. The physiological studies presented here provide experimental support for this inferred role.
FIG. 7.
Summary of major pathways for iron uptake in B. subtilis. The four ABC transporters shown contain two predicted permease subunits, siderophore-specific substrate-binding proteins (SBPs; predicted lipoproteins are indicated as tethered to the membrane), and cytoplasmic ATPase subunits. The FeuA SBP mediates uptake of both enterobactin and bacillibactin, while YfiY allows growth with the hydroxamate siderophores schizokinen, arthrobactin, and corprogen. The FhuBG permease is needed for growth with ferrichrome and ferrioxamine, but these siderophores use different SBPs. Note that the YusV ATPase apparently functions as part of two different ABC uptake systems. YwbM, like the SBPs, is a predicted lipoprotein and YwbN, a predicted iron peroxidase, requires a dedicated Tat export pathway for secretion (32).
Acknowledgments
We thank K. Hantke and G. Winkelmann for providing siderophores and advice on their use, C. M. Moore for helpful advice, and K. Raymond and his students for advice on pM calculations.
This study was supported by a grant from the National Institutes of Health (GM59323 to J.D.H.).
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 2.Annamalai, R., B. Jin, Z. Cao, S. M. Newton, and P. E. Klebba. 2004. Recognition of ferric catecholates by FepA. J. Bacteriol. 186:3578-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484-1502. [DOI] [PubMed] [Google Scholar]
- 4.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 6.Bernhardt, J., K. Buttner, C. Scharf, and M. Hecker. 1999. Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20:2225-2240. [DOI] [PubMed] [Google Scholar]
- 7.Bernhardt, J., U. Volker, A. Volker, H. Antelmann, R. Schmid, H. Mach, and M. Hecker. 1997. Specific and general stress proteins in Bacillus subtilis: a two-dimensional protein electrophoresis study. Microbiology 143:999-1017. [DOI] [PubMed] [Google Scholar]
- 8.Bluhm, M. E., S. S. Kim, E. A. Dertz, and K. N. Raymond. 2002. Corynebactin and enterobactin: related siderophores of opposite chirality. J. Am. Chem. Soc. 124:2436-2437. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 12.Budzikiewicz, H., A. Bossenkamp, K. Taraz, A. Pandey, and J. M. Meyer. 1997. Bacterial constituents. 72. Corynebactin, a cyclic catecholate siderophore from Corynebacterium glutamicum ATCC 14067 (Brevibacterium sp. DSM 20411). Z. Naturforsch. C-a J. Biosci. 52:551-554. [Google Scholar]
- 13.Bunai, K., M. Ariga, T. Inoue, M. Nozaki, S. Ogane, H. Kakeshita, T. Nemoto, H. Nakanishi, and K. Yamane. 2004. Profiling and comprehensive expression analysis of ABC transporter solute-binding proteins of Bacillus subtilis membrane based on a proteomic approach. Electrophoresis 25:141-155. [DOI] [PubMed] [Google Scholar]
- 14.Cendrowski, S., W. MacArthur, and P. Hanna. 2004. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol. Microbiol. 51:407-417. [DOI] [PubMed] [Google Scholar]
- 15.Chipperfield, J. R., and C. Ratledge. 2000. Salicylic acid is not a bacterial siderophore: a theoretical study. Biometals 13:165-168. [DOI] [PubMed] [Google Scholar]
- 16.Dertz, E. A., J. Xu, A. Stintzi, and K. N. Raymond. 2006. Bacillibactin-mediated iron transport in Bacillus subtilis. J. Am. Chem. Soc. 128:22-23. [DOI] [PubMed] [Google Scholar]
- 17.Eymann, C., A. Dreisbach, D. Albrecht, J. Bernhardt, D. Becher, S. Gentner, T. Tam le, K. Buttner, G. Buurman, C. Scharf, S. Venz, U. Volker, and M. Hecker. 2004. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 4:2849-2876. [DOI] [PubMed] [Google Scholar]
- 18.Faller, B., and H. Nick. 1994. Kinetics and mechanism of iron(III) removal from citrate by desferrioxamine B and 3-hydroxy-1,2-dimethyl-4-pyridone. J. Am. Chem. Soc. 116:3860-3865. [Google Scholar]
- 19.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaballa, A., T. Wang, R. W. Ye, and J. D. Helmann. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 184:6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautier-Luneau, I., C. Merle, D. Phanon, C. Lebrun, F. Biaso, G. Serratrice, and J. L. Pierre. 2005. New trends in the chemistry of iron(III) citrate complexes: correlations between X-ray structures and solution species probed by electrospray mass spectrometry and kinetics of iron uptake from citrate by iron chelators. Chemistry 11:2207-2219. [DOI] [PubMed] [Google Scholar]
- 22.Guedon, E., C. M. Moore, Q. Que, T. Wang, R. W. Ye, and J. D. Helmann. 2003. The global transcriptional response of Bacillus subtilis to manganese involves the MntR, Fur, TnrA, and sigmaB regulons. Mol. Microbiol. 49:1477-1491. [DOI] [PubMed] [Google Scholar]
- 23.Guérout-Fleury, A.-M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 24.Guérout-Fleury, A.-M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 25.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 26.Harris, W. R., C. J. Carrano, S. R. Cooper, S. R. Sofen, A. E. Avdeef, J. V. MacArdle, and K. N. Raymond. 1979. Coordination chemistry of microbial iron transport compounds. 19. Stability constants and electrochemical behavior of ferric enterobactin and model complexes. J. Am. Chem. Soc. 101:6097-6104. [Google Scholar]
- 27.Harvie, D. R., and D. J. Ellar. 2005. A ferric dicitrate uptake system is required for the full virulence of Bacillus cereus. Curr. Microbiol. 50:246-250. [DOI] [PubMed] [Google Scholar]
- 28.Harvie, D. R., S. Vilchez, J. R. Steggles, and D. J. Ellar. 2005. Bacillus cereus Fur regulates iron metabolism and is required for full virulence. Microbiology 151:569-577. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann, T., A. Schutz, M. Brosius, A. Volker, U. Volker, and E. Bremer. 2002. High-salinity-induced iron limitation in Bacillus subtilis. J. Bacteriol. 184:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito, T. 1993. Enzymatic determination of itoic acid, a Bacillus subtilis siderophore, and 2,3-dihydroxybenzoic acid. Appl. Environ. Microbiol. 59:2343-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito, T., and J. B. Neilands. 1958. Products of “low-iron fermentation” with Bacillus subtilis: isolation, characterization, and synthesis of 2,3-dihydroxybenzoylglycine. J. Am. Chem. Soc. 80:4645-4647. [Google Scholar]
- 32.Jongbloed, J. D., U. Grieger, H. Antelmann, M. Hecker, R. Nijland, S. Bron, and J. M. van Dijl. 2004. Two minimal Tat translocases in Bacillus. Mol. Microbiol. 54:1319-1325. [DOI] [PubMed] [Google Scholar]
- 33.Koppisch, A. T., C. C. Browder, A. L. Moe, J. T. Shelley, B. A. Kinkel, L. E. Hersman, S. Iyer, and C. E. Ruggiero. 2005. Petrobactin is the primary siderophore synthesized by Bacillus anthracis Str. Sterne under conditions of iron starvation. Biometals 18:577-585. [DOI] [PubMed] [Google Scholar]
- 34.Lin, H., M. A. Fischbach, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 127:11075-11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mader, U., H. Antelmann, T. Buder, M. K. Dahl, M. Hecker, and G. Homuth. 2002. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol. Genet. Genomics 268:455-467. [DOI] [PubMed] [Google Scholar]
- 36.Masse, E., and M. Arguin. 2005. Ironing out the problem: new mechanisms of iron homeostasis. Trends Biochem. Sci. 30:462-468. [DOI] [PubMed] [Google Scholar]
- 37.May, J. J., T. M. Wendrich, and M. A. Marahiel. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276:7209-7217. [DOI] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Moore, C. M., A. Gaballa, M. Hui, R. W. Ye, and J. D. Helmann. 2005. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol. Microbiol. 57:27-40. [DOI] [PubMed] [Google Scholar]
- 40.Moore, C. M., and J. D. Helmann. 2005. Metal ion homeostasis in Bacillus subtilis. Curr. Opin. Microbiol. 8:188-195. [DOI] [PubMed] [Google Scholar]
- 41.Moore, C. M., M. M. Nakano, T. Wang, R. W. Ye, and J. D. Helmann. 2004. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J. Bacteriol. 186:4655-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 43.Nakano, M. M., M. A. Marahiel, and P. Zuber. 1988. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 170:5662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogura, M., H. Yamaguchi, K. Yoshida, Y. Fujita, and T. Tanaka. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA, and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2001. Comparative analysis of FUR regulons in gamma-proteobacteria. Nucleic Acids Res. 29:5195-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park, R. Y., M. H. Choi, H. Y. Sun, and S. H. Shin. 2005. Production of catechol-siderophore and utilization of transferrin-bound iron in Bacillus cereus. Biol. Pharm. Bull. 28:1132-1135. [DOI] [PubMed] [Google Scholar]
- 47.Peters, W. J., and R. A. Warren. 1968. Itoic acid synthesis in Bacillus subtilis. J. Bacteriol. 95:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peters, W. J., and R. A. J. Warren. 1970. The mechanism of iron uptake in Bacillus subtilis. Can. J. Microbiol. 16:1285-1291. [DOI] [PubMed] [Google Scholar]
- 49.Peters, W. J., and R. A. J. Warren. 1968. Phenolic acids and iron transport in Bacillus subtilis. Biochim. Biophys. Acta 165:225-232. [DOI] [PubMed] [Google Scholar]
- 50.Pierre, J. L., and I. Gautier-Luneau. 2000. Iron and citric acid: a fuzzy chemistry of ubiquitous biological relevance. Biometals 13:91-96. [DOI] [PubMed] [Google Scholar]
- 51.Pollack, J. R., B. N. Ames, and J. B. Neilands. 1970. Iron transport in Salmonella typhimurium: mutants blocked in the biosynthesis of enterobactin. J. Bacteriol. 104:635-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quadri, L. E., P. H. Weinreb, M. Lei, M. M. Nakano, P. Zuber, and C. T. Walsh. 1998. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37:1585-1595. [DOI] [PubMed] [Google Scholar]
- 53.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287:467-484. [DOI] [PubMed] [Google Scholar]
- 54.Rabsch, W., W. Voigt, R. Reissbrodt, R. M. Tsolis, and A. J. Baumler. 1999. Salmonella typhimurium IroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J. Bacteriol. 181:3610-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raymond, K. N., E. A. Dertz, and S. S. Kim. 2003. Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 100:3584-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rowland, B. M., T. H. Grossman, M. S. Osburne, and H. W. Taber. 1996. Sequence and genetic organization of a Bacillus subtilis operon encoding 2,3-dihydroxybenzoate biosynthetic enzymes. Gene 178:119-123. [DOI] [PubMed] [Google Scholar]
- 57.Schneider, R., and K. Hanke. 1993. Iron-hydroxamate uptake systems in Bacillus subtilis: identification of a lipoprotein as part of a binding protein-dependent transport system. Mol. Microbiol. 8:111-121. [DOI] [PubMed] [Google Scholar]
- 58.Sierra, M. A., M. Gomez-Gallego, R. Alcazar, J. J. Lucena, F. Yunta, and S. Garcia-Marco. 2004. Effect of the tether on the Mg(II), Ca(II), Cu(II), and Fe(III) stability constants and pM values of chelating agents related to EDDHA. Dalton Trans. 21:3741-3747. [DOI] [PubMed] [Google Scholar]
- 59.Stearman, R., D. S. Yuan, Y. Yamaguchi-Iwai, R. D. Klausner, and A. Dancis. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552-1557. [DOI] [PubMed] [Google Scholar]
- 60.Stintzi, A., and K. N. Raymond. 2002. Siderophore chemistry, p. 273-319. In D. M. Templeton (ed.), Molecular and cellular iron transport. Marcel Dekker, New York, N.Y.
- 61.van Bakel, H., M. Huynen, and C. Wijmenga. 2004. Prokaryotic diversity of the Saccharomyces cerevisiae Atx1p-mediated copper pathway. Bioinformatics 20:2644-2655. [DOI] [PubMed] [Google Scholar]
- 62.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in Saccharomyces cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 63.Walsh, B. L., W. J. Peters, and R. A. J. Warren. 1971. The regulation of phenolic acid synthesis in Bacillus subtilis. Can. J. Microbiol. 17:53-59. [DOI] [PubMed] [Google Scholar]
- 64.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 65.Wong, G. B., M. J. Kappel, B. Matzanke, and G. Winkelmann. 1983. Coordination chemistry of microbial iron transport compounds. 24. Characterization of coprogen and ferricrocin, two ferric hydroxamate siderophores. J. Am. Chem. Soc. 105:810-815. [Google Scholar]
- 66.Youngman, P. 1990. Use of transposons and integrational vectors for mutagenesis and construction of gene fusions in Bacillus species, p. 221-266. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Ltd., Chichester, United Kingdom.
- 67.Zhu, M., M. Valdebenito, G. Winkelmann, and K. Hantke. 2005. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology 151:2363-2372. [DOI] [PubMed] [Google Scholar]