Abstract
The Escherichia coli Aer protein contains an N-terminal PAS domain that binds flavin adenine dinucleotide (FAD), senses aerotactic stimuli, and communicates with the output signaling domain. To explore the roles of the intervening F1 and HAMP segments in Aer signaling, we isolated plasmid-borne aerotaxis-defective mutations in a host strain lacking all chemoreceptors of the methyl-accepting chemotaxis protein (MCP) family. Under these conditions, Aer alone established the cell's run/tumble swimming pattern and modulated that behavior in response to oxygen gradients. We found two classes of Aer mutants: null and clockwise (CW) biased. Most mutant proteins exhibited the null phenotype: failure to elicit CW flagellar rotation, no aerosensing behavior in MCP-containing hosts, and no apparent FAD-binding ability. However, null mutants had low Aer expression levels caused by rapid degradation of apparently nonnative subunits. Their functional defects probably reflect the absence of a protein product. In contrast, CW-biased mutant proteins exhibited normal expression levels, wild-type FAD binding, and robust aerosensing behavior in MCP-containing hosts. The CW lesions evidently shift unstimulated Aer output to the CW signaling state but do not block the Aer input-output pathway. The distribution and properties of null and CW-biased mutations suggest that the Aer PAS domain may engage in two different interactions with HAMP and the HAMP-proximal signaling domain: one needed for Aer maturation and another for promoting CW output from the Aer signaling domain. Most aerotaxis-defective null mutations in these regions seemed to affect maturation only, indicating that these two interactions involve structurally distinct determinants.
The Aer protein of Escherichia coli promotes movement toward optimal concentrations of oxygen and other electron acceptors (8, 20). Aer homologs have been found in many other species of bacteria, where they probably mediate similar aerotactic behaviors. Aer communicates with the flagellar motors through a cytoplasmic signaling domain that modulates the activity of the histidine kinase CheA to control the phosphorylation state of the CheY response regulator. The Aer signaling domain is closely related to that of transmembrane chemoreceptors of the methyl-accepting chemotaxis protein (MCP) family, but Aer differs from orthodox MCPs in several important respects. First, the Aer signaling domain does not employ reversible methylation changes for sensory adaptation. Second, the Aer molecule, although membrane associated, does not have a periplasmic sensing domain but rather a cytoplasmic PAS domain that is thought to sense oxygen stimuli by monitoring cellular electron transport activity. PAS domains are widespread in proteins that sense cellular energy levels, redox potential, light, and other stimuli (24). The PAS structural motif comprises a binding pocket that can be adapted for a variety of small-molecule ligands, such as heme (in FixL). In the case of Aer, the PAS ligand is flavin adenine dinucleotide (FAD), which probably serves as a redox-sensing moiety.
The domain organization of Aer is summarized in Fig. 1. Its PAS domain is located at the N terminus of the molecule, followed by a segment (F1) of unknown function (7). A 40-residue hydrophobic segment (M) in the middle of the molecule anchors Aer to the inner side of the cytoplasmic membrane (3). The M segment is followed by a 50-residue HAMP domain that is a common feature of many membrane-associated bacterial signaling proteins, including sensor histidine kinases and MCP chemoreceptors. HAMP domains appear to contain two amphipathic α-helices (AS-1 and AS-2) joined by a connector of unknown structure, but their roles in signal transduction are not well understood (5, 27). The AS-2 element of the Aer HAMP domain adjoins the C-terminal signaling domain and most likely regulates its activity. The PAS-HAMP portion of Aer can mediate aerotactic behavior when coupled to a heterologous MCP signaling domain, demonstrating that the aerosensing and output control functions of Aer lie in the N-terminal half of the molecule (7, 9, 21). We designate the PAS-HAMP segment the Aer input-output (I/O) module.
FIG. 1.
Aer domain organization. The scale at the top denotes residue numbers in the Aer protein. For this study, genetically silent restriction sites were created at aer codons 101, 210, and 270 in order to manipulate the F1 and HAMP portions of the coding region. The structure of the F1 segment of the Aer protein is unknown, whereas the HAMP region is predicted to consist of two amphipathic α-helices (AS-1 and AS-2) joined by a connector of unknown structure. The first and last residues of AS-1 and AS-2, based on HAMP sequence alignments (4, 5, 27), are indicated. The residue boundaries for the HAMP connector segment are slight modifications of those reported by Ma et al. (16).
Redox changes in the FAD prosthetic group of the Aer PAS domain are thought to trigger conformational changes that are somehow propagated to the Aer signaling domain to modulate flagellar-controlling output signals. The signaling route through the Aer molecule is not well understood. In conventional MCPs, the HAMP domain lies next to the cytoplasmic face of the inner membrane and communicates input signals from the ligand-binding periplasmic domain to the cytoplasmic output domain. The HAMP domain in Aer occupies an analogous position in the molecule, but because stimuli are sensed through the PAS domain, sensory signals in Aer must necessarily follow a different path to the output domain. Genetic evidence suggests that the Aer PAS domain could interact directly with the C terminus of the HAMP AS-2 segment to modulate activity of the adjoining signaling domain (26).
To explore the input-output signaling pathway through the Aer molecule, we isolated and characterized mutations in the F1 and HAMP segments of E. coli Aer. We found that most aerotaxis-defective mutations in these regions caused null phenotypes, owing to rapid degradation and low steady-state levels of the mutant Aer molecules. It appears that these null mutations perturb mainly the Aer maturation process and not native Aer function. In contrast, some mutations abrogated aerotaxis by shifting the Aer molecule into a clockwise (CW) signaling state with reduced sensitivity to aerotactic stimuli. The distribution of null and CW-biased lesions and their functional interactions with wild-type Aer and with heterologous MCPs provide new insights into the functional organization of the Aer molecule and its mechanism(s) of input-output control.
MATERIALS AND METHODS
Bacterial strains.
The following strains used are derivatives of E. coli K-12 RP437, our wild-type reference (19), and their genetic markers relevant to the current study are given in parentheses or brackets: UU1117 (Δaer-1) (8), UU1250 [Δaer-1 Δtsr-7028 Δ(tar-tap)5201 Δtrg-100] (9), and UU1446 [Δaer-1 Δtsr-7028 Δ(tar-tap)5201 Δtrg-100 recA srlC::Tn10] (this work).
Plasmids.
Aer expression plasmids were derived from two replication-compatible parent vectors: pCJ30 (ampicillin resistance, IPTG [isopropyl-β-d-thiogalactopyranoside]-inducible expression from the Ptac promoter) (8) and pKG116 (chloramphenicol resistance, salicylate-inducible expression from the PnahG promoter [1, 28]) (this work). Aer plasmids were pSB20 (pCJ30 aer+) (8), pDM17 (pSB20 with silent restriction sites EagI at codon 210 and BamHI at codon 270) (A. C. Miller, S. I. Bibikov, and J. S. Parkinson, unpublished results), pMB1 (pDM17 NcoI at codon 101) (this work), and pKG117 (pKG116 aer+) (this work).
Soft agar aerotaxis assays.
Aerotactic ability of plasmid-containing UU1250 and UU1446 strains was assessed at 30°C on tryptone soft agar plates (17) containing 0.27% Bacto agar (Difco), plasmid-selective antibiotics (50 μg/ml ampicillin [pSB20, pDM17, and pMB1] and/or 12.5 μg/ml chloramphenicol [pKG117]), and appropriate concentrations of the inducers IPTG and/or sodium salicylate. Aerotaxis of plasmid-containing UU1117 strains was assessed at 30°C on minimal succinate soft agar plates (7, 8) containing 0.25% agar and the same antibiotic and inducer concentrations used in tryptone plates. Note that these agar concentrations are approximations; agar potency was empirically calibrated with each new lot to ensure optimal performance.
Mutant isolation.
The HAMP and F1 coding regions of plasmid pSB20 were mutagenized by error-prone PCR, using Taq DNA polymerase (Stratagene, La Jolla, CA) at 8 mM MgCl2 to reduce proofreading activity. PCR machine settings were 30 s at a 94°C initial denaturation, followed by 6 cycles with 30 s at 94°C, 30 s at 55°C, and 90 s at 74°C and a final elongation for 5 min at 74°C. PCR products were cleaned with a QIAquick PCR purification kit (QIAGEN, Valencia, CA), followed by restriction enzyme digestion (see below) and another purification step. Oligonucleotide primers for HAMP introduced silent restriction sites (EagI at codon 210 and BamHI at codon 270) at the fragment ends for cloning PCR segments into the corresponding region of pDM17. Similarly, F1 primers carried NcoI and EagI sites at codons 101 and 210 for cloning into pMB1. Recombinant plasmids were introduced into strain UU1250, and transformant cells were plated (∼500 cells per plate) in tryptone miniswarm agar (0.35% agar, 50 μg/ml ampicillin, 50 μM IPTG). Plates were screened for nonaerotactic colonies after overnight incubation at 30°C. Candidate mutants were retested on tryptone soft agar plates, and then their plasmids were extracted and subjected to DNA sequence analysis. For all plasmid mutants described in this report, we sequenced the entire aer coding region in order to detect and discard any mutants with multiple mutations.
FAD-binding assay.
UU1250 cells carrying mutant Aer plasmids were grown at 30°C to mid-log phase in minimal Casamino Acids medium. IPTG (1 mM) was added to the cultures to induce high-level expression of the mutant protein, and the cells were allowed to grow for an additional 4 h, after which membrane-associated FAD was extracted and measured as previously described (7).
Cell tethering assay.
UU1250 cells carrying mutant Aer plasmids were grown at 30°C to mid-log phase in tryptone broth containing 100 μg/ml ampicillin and 50 μM IPTG. Cells were collected, tethered to microscope coverslips with antiflagellar antibodies, and analyzed for flagellar rotation pattern as previously described (2).
Steady-state protein levels.
UU1250 cells carrying mutant Aer plasmids were grown at 30°C to mid-log phase in tryptone broth containing 100 μg/ml of ampicillin and 50 μM IPTG. Cells were harvested by centrifugation, washed once in phosphate-buffered saline (pH 7.4), and lysed by boiling for 10 min in sodium dodecyl sulfate (SDS) sample buffer (15), followed by freezing and thawing. Samples were analyzed by electrophoresis in 10% SDS-containing polyacrylamide gels and immunoblotting with a polyclonal rabbit antiserum directed against Aer residues 1 to 263 (12). Aer bands were visualized using Cy5-labeled goat anti-rabbit immunoglobulin G (heavy plus light chains; Amersham) and quantified with a Molecular Dynamics Typhoon 8600 imager. We analyzed a series of dilutions for each sample and used those in the linear dose-response range to determine protein levels in the cell extracts.
Protein degradation assay.
Minor modifications of published procedures were followed (10, 22). In brief, UU1250 cells carrying mutant Aer plasmids were grown at 30°C to mid-log phase in tryptone broth containing 100 μg/ml of ampicillin and then induced with 50 μM IPTG and grown for an additional 2.5 h. Protein synthesis in the cultures was then blocked by addition of 500 μg/ml chloramphenicol, and incubation continued at 30°C. Samples were collected over the following 4 h and analyzed for full-length Aer protein as described above.
RESULTS
Isolation of F1 and HAMP mutations.
Functionally silent restriction sites were created at strategic points in the aer gene to facilitate manipulations of the F1 and HAMP portions of the coding region (Fig. 1). F1 mutations were induced by error-prone PCR of wild-type aer, using opposing primers bearing NcoI and EagI restriction sites, flanking the F1 coding region. Mutant PCR products were cloned into pMB1, which had been gapped between its NcoI and EagI sites. HAMP mutations were induced in a similar fashion, and mutant coding fragments were cloned into pMB1 gapped at its EagI and BamHI sites (Fig. 1). The F1 mutagenesis included portions of the adjacent PAS domain and membrane segment (M) (Fig. 1). The HAMP mutagenesis covered all but the first five residues of HAMP and extended through the proximal signaling domain (Fig. 1). Candidate mutant plasmids were tested for the ability to support aerotaxis in strain UU1250, which lacks aer and the four MCP genes (tar, tap, trg, and tsr). Transformants were screened for aerotaxis defects by colony morphology on tryptone soft agar miniswarm plates.
We obtained aerotaxis-defective mutations at only 6 of the 45 residues comprising F1 (see Fig. 5). Perhaps most F1 lesions cause other phenotypes, such as poor viability, that would have been overlooked in our screen. Alternatively, F1 is poorly conserved in primary structure among Aer homologs, so many of its residues could conceivably be resistant to inactivating mutations. The adjacent M segment is also poorly conserved and, in fact, yielded no missense mutations, suggesting that it, too, tolerates many amino acid replacements. We also obtained mutations at three PAS residues within the targeted region; these mutant plasmids were included in subsequent characterizations. By contrast, the HAMP region was much less refractory to mutation. We obtained mutations at 16 of the 44 targeted HAMP residues and at nine sites in the proximal signaling domain. Most of these mutant sites were hit multiple, independent times, indicative of an effectively saturated survey (data not shown). In all, 47 different mutations were subjected to additional characterization tests, whose results are summarized in Fig. 5 and described in more detail below.
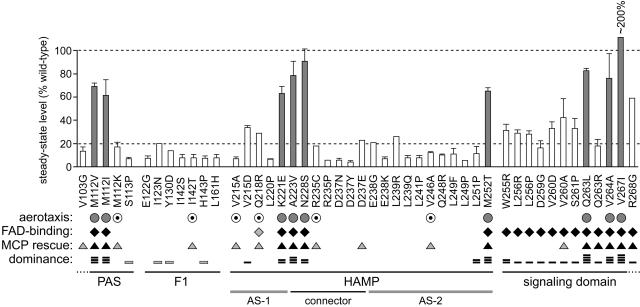
FIG. 5.
Summary of Aer mutant (Aer*) properties. The residue numbers and amino acid replacements of pMB1-Aer* characterized in this study are listed below the histogram, which shows the steady-state expression level of each mutant protein, determined as detailed in Materials and Methods. Shaded vertical bars indicate Aer proteins with CW-biased signal output, as determined by flagellar rotation pattern. Symbols: for aerotaxis (UU1250 colony phenotype on tryptone soft agar), shaded circles, fuzzy, and dotted circles, leaky; for FAD binding (ability to increase cellular FAD levels when highly induced), black diamonds, increases comparable to those produced by wild-type Aer, and shaded diamonds, intermediate-level increase; for MCP rescue (recovery of aerotactic ability in UU1117), black triangles, fully rescued, and shaded triangles, partial rescue or rescue at high induction; for dominance (ability to block aerotaxis in UU1250 carrying a wild-type aer plasmid), one bar reduced colony size on soft agar (shading denotes effects seen only at high induction); two bars, impaired ring formation; three bars, reduced size and impaired ring formation; and four bars, null phenotype.
Aerotaxis phenotypes.
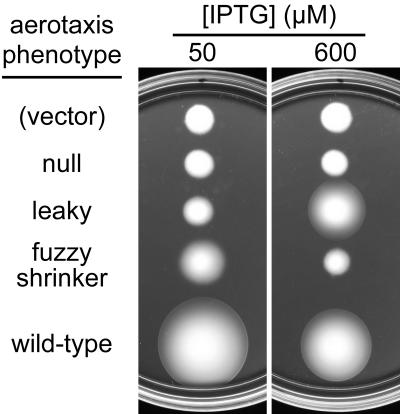
In UU1250, the mutant plasmids produced several distinct aerotaxis phenotypes in tryptone soft agar (Fig. 2). The majority of the mutant isolates (37/47) showed a null phenotype with a colony morphology indistinguishable from that of the vector control at 50 μM IPTG, the optimal inducer concentration for wild-type aerotaxis. Most (31/37) of these null mutants appeared to have a total loss of Aer function, because their colony morphology did not change at high levels (600 μM) of IPTG inducer. The remainder (6/37) of the null mutants produced larger colonies upon elevated expression, consistent with a partial loss-of-function defect. Other mutants (9/47) produced fuzzy, cylindrical colonies that lacked a distinct ring of aerotactic cells at their edge. At 600 μM IPTG, the fuzzy colonies became smaller in size, suggesting that the mutant gene product had some functional activity (Fig. 2). Finally, one mutant isolate (V103G, not shown in Fig. 2) exhibited a null phenotype at 50 μM IPTG and a fuzzy phenotype at 600 μM IPTG.
FIG. 2.
Phenotypes of aerotaxis mutants on tryptone soft agar plates. Mutant plasmids were tested in strain UU1250, which lacks Aer and the four conventional MCP family chemoreceptors (Tap, Tar, Trg, and Tsr). Colonies were transferred to tryptone soft agar plates containing 50 μg/ml ampicillin and either 50 or 600 μM IPTG to induce Aer expression. Plates were incubated at 30°C for 17 h. Plasmids were pCJ30 (vector), pMB1-L220P (null), pMB1-V246A (leaky), pMB1-N228S (fuzzy), and pSB20 (wild type).
Flagellar rotation patterns.
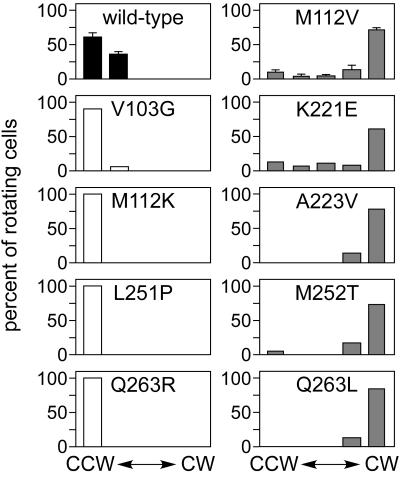
Strain UU1250, which lacks all chemoreceptors, shows exclusively counterclockwise (CCW) flagellar rotation because there are no transducers available to activate the signaling phosphorelay via CheA. Wild-type Aer alone, at an appropriate expression level, imparts sufficient CW bias to UU1250 to enable it to perform aerotaxis. To assess the steady-state signaling activity of mutant Aer proteins, we examined the flagellar rotation pattern of UU1250 strains carrying mutant plasmids that had been induced at 50 μM IPTG, which yields optimal expression levels for wild-type Aer function (Fig. 3). Null mutants exhibited exclusively CCW rotation, the UU1250 default behavior, consistent with a complete loss of function. However, the V103G null mutant (fuzzy colonies at 600 μM IPTG) exhibited a small fraction of reversing cells at 50 μM IPTG, indicative of some residual ability to activate the CheA phosphorelay. In contrast, cells containing wild-type Aer exhibited higher levels (∼36%) of reversing individuals, despite an overall CCW bias. Five representative fuzzy mutants were tested, and all exhibited a pronounced CW bias (Fig. 3). We conclude that fuzzy mutants represent Aer proteins with CW-biased or -locked signal outputs, a gain-of-function behavior.
FIG. 3.
Flagellar rotation profiles of Aer mutants. UU1250 strains carrying plasmids with the indicated Aer mutations were analyzed by cell tethering as detailed in Materials and Methods. The histograms show the percentages of rotating cells assigned to each of five behavioral categories, from exclusively CCW to exclusively CW, during the observation period. The shading of the data bars denotes colony phenotype in soft agar assays: white, null mutants; gray, fuzzy mutants; and black, wild type.
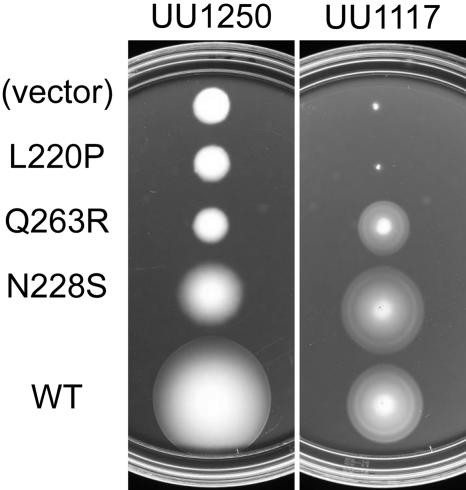
Functional rescue by MCPs.
Aer mutants with biased rather than locked signal outputs might be able to mediate aerotactic responses in cells with a normal flagellar rotation pattern. To test this idea, we transferred pMB1 mutant plasmids to strain UU1117, which lacks Aer but has all four MCPs. In this strain, the high-abundance MCPs Tar and Tsr exert principal control over steady-state rotational behavior. We found that all (nine of nine) CW-biased mutant Aer proteins mediated aerotactic responses in UU1117 (Fig. 4). Not surprisingly, six of six mutant proteins with leaky null defects also mediated aerotactic responses upon induction. However, most (27/32) proteins with null defects were not functionally rescued in strain UU1117, even at high inducer levels. We conclude from these rescue tests that the CW-biased mutant proteins retain input-output control, whereas most null mutant proteins do not. Of the five rescuable null mutants, three (V260A, Q263R, and R268G) had signaling domain lesions that may lie outside the Aer I/O module (see below).
FIG. 4.
Functional rescue of aerotaxis defects. Mutant plasmids carrying the indicated Aer mutations were tested in strain UU1117, which is deleted for the aer gene but carries functional MCP genes (tap, tar, trg, and tsr). Colonies were transferred to minimal succinate soft agar plates and incubated at 30°C for 20 h. Plates contained no IPTG inducer because the basal Aer expression level in UU1117 produces optimal aerotactic responses (8). The L220P null defect is not rescued by the other chemoreceptors in UU1117, whereas Q263R (null) and N228S (fuzzy) are rescued. Note that all three mutant Aer proteins fail to function in the absence of other chemoreceptors (strain UU1250). WT, wild type.
FAD binding.
The Aer PAS domain is believed to sense aerotactic stimuli through its bound FAD moiety. The CW-biased mutant proteins, which showed input-output control in the functional rescue tests, should be competent for FAD binding. Null mutant proteins might also bind FAD, if their signaling defect lies elsewhere in the transduction pathway. We tested FAD binding by the mutant Aer proteins by expressing them at high induction levels in strain UU1250. Overexpression of wild-type Aer causes a concomitant increase in cellular FAD levels that is detectable by spectrophotometric analysis of membrane extracts (8). Using this assay, we found that all CW-biased mutant proteins bound FAD, as expected (Fig. 5). In addition, all mutant proteins with null lesions in the proximal signaling domain bound FAD, consistent with our designation of the PAS-HAMP segment as the input-output module of Aer. In contrast, all proteins with null defects, except for one leaky mutant (Q218R), failed to bind FAD by this test.
The FAD-binding assay requires high-level expression of the mutant Aer protein. Thus, mutant proteins that are poorly expressed or unstable could seem to be defective in FAD binding, even though that is not their primary defect. To explore this possibility, we determined the expression levels of selected mutant Aer proteins under the same high-induction conditions used in the FAD assays. Four null mutant proteins that scored as FAD binding defective (R235P, D237Y, L249F, and L251P) had steady-state expression levels ranging from 39% to 67% that of the wild type. In contrast, Aer D259G, a binding-positive protein, had a fully-wild-type expression level. Thus, it seems that low steady-state expression levels might contribute to the apparent FAD-binding defects of null mutant proteins.
Steady-state protein levels.
To determine whether altered expression levels might be responsible for any of the Aer mutant phenotypes, steady-state protein levels were measured in plasmid-containing UU1250 strains grown at 50 μM IPTG, the inducer concentration optimal for wild-type Aer function. At this inducer concentration, we found that CW-biased mutant proteins had steady-state Aer levels at least 60% that of the wild type (Fig. 5). Mutant proteins with null lesions in the signaling domain, which were FAD positive in the binding tests, had lower steady-state levels, ranging from 20% to nearly 60% that of the wild type (Fig. 5). In contrast, null mutant proteins with PAS, F1, or HAMP mutations, which did not appear to bind FAD, had, with a few modest exceptions, steady-state levels below 20% that of the wild type; many were below 10% (Fig. 5). These low protein levels confound interpretation of the mutant phenotypes, which could reflect defects mainly in maturation or stability rather than in functional activities of a native molecule.
Protein degradation patterns.
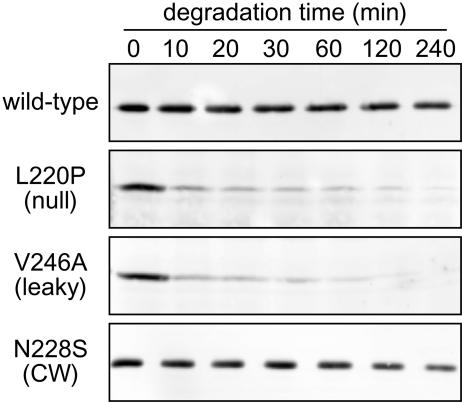
Assuming that the rate of synthesis of null mutant proteins is comparable to that of the wild type, their low steady-state expression levels most likely result from enhanced rates of degradation. We tested this hypothesis by following the disappearance of full-length Aer molecules after blocking new protein synthesis in intact cells (see Materials and Methods). Strain UU1250 carrying a mutant Aer plasmid was induced with 50 μM IPTG, and then protein synthesis was blocked with chloramphenicol and full-length Aer molecules were quantified over a 4-h period. Under these conditions, wild-type Aer molecules proved remarkably and consistently stable, with little apparent degradation over 4 h (Fig. 6). CW-biased mutant proteins were also quite stable, whereas proteins with null or leaky mutations disappeared rapidly, usually within the first 10 min (Fig. 6).
FIG. 6.
Examples of unstable Aer proteins. Mutant Aer proteins were expressed from pMB1 derivatives in strain UU1250 and tested for intracellular stability after blocking new protein synthesis, as detailed in Materials and Methods. Cells were sampled at the times indicated and analyzed by electrophoresis in 10% SDS-containing polyacrylamide gels. Full-length Aer molecules in the lysates were visualized by Western blotting with polyclonal rabbit antiserum directed against Aer residues 1 to 263.
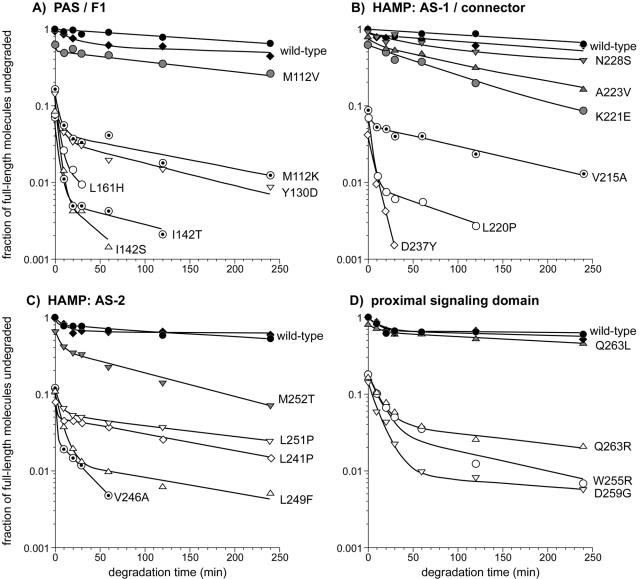
Quantitative analyses of the degradation patterns are presented in Fig. 7. Wild-type Aer and CW-biased mutant proteins generally followed single exponential kinetics, with decay rates ranging from 0.001 to 0.009/minute. By contrast, the null and leaky mutant proteins with PAS, F1, or HAMP lesions typically followed double exponential kinetics, with a high initial decay rate followed by a longer period of slower decay. High decay rates ranged from about 0.15/minute (Y130D and L249F) to more than 1/minute (D237Y and V246A). Low decay rates ranged mainly between 0.004 and 0.007/minute, comparable to the decay rates of CW-biased mutant proteins. Three mutant proteins with null lesions in the proximal signaling domain (W255R, D259G, and Q263R) also exhibited high and low decay rates but with kinetics generally slower than those of the other null mutant proteins. Their high rates were about 0.01/minute; their low rates were 0.003 to 0.01/minute. These lower overall decay rates are consistent with the higher steady-state cellular levels in this group of null mutant proteins (Fig. 5). The ratios of fast- versus slow-decaying molecules varied from one mutant protein to another. As a general rule, mutant proteins with low steady-state expression levels had higher proportions of fast-decaying molecules (e.g., I142S, I142T, L220P, and L249F), whereas null mutant proteins with somewhat higher steady-state levels had a smaller fraction of rapidly decaying molecules (e.g., Y130D and L251P).
FIG. 7.
Degradation profiles of Aer mutant (Aer*) proteins. The degradation data were generated in experiments like those shown in Fig. 6. Black symbols, wild-type Aer; shaded symbols, Aer* proteins with CW-biased outputs; dotted symbols, Aer* proteins with leaky defects; and open symbols, Aer* proteins with null defects. The lines connecting data points represent least-squares best fits to a double exponential decay curve. The degradation curves for the mutant proteins begin at their initial (steady-state) expression levels relative to that of wild-type Aer.
We conclude that the steady-state levels of mutant Aer proteins mainly reflect their susceptibility to cellular proteolysis. The fast-decaying subpopulations of the null mutant proteins suggest that they have maturation defects and seldom reach the native (slow-decay) state. In some experiments we observed that 20 to 30% of wild-type and CW-biased molecules also degraded rapidly (e.g., Fig. 7D), which suggests that variations in synthesis rate might influence the relative proportions of immature and mature Aer molecules in the cell. Indeed, at very high induction levels, the conditions used for FAD-binding assays, the steady-state levels of mutant Aer proteins more closely approximated wild-type values (see above). It seems unlikely that the mutant molecules mature faster under these conditions, so their apparently normal expression levels are probably misleading. We suggest that most of the mutant molecules are still destined for destruction but that at high synthesis rates the degradation machinery is saturated and, therefore, rate limiting.
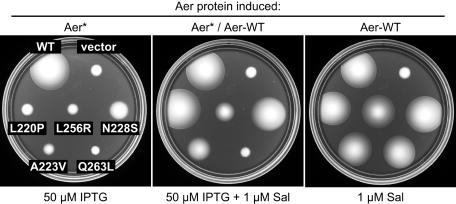
Dominance tests.
To gain further insight into the nature of the functional defects in null and CW-biased Aer lesions, we examined their phenotypic interactions with wild-type Aer supplied from a compatible plasmid. Aer expression from the wild-type plasmid, pKG117, was induced with 1 μM sodium salicylate, the optimum concentration for aerotaxis. The mutant Aer plasmids were tested at optimal and high IPTG concentrations to see if the mutant protein could interfere with wild-type function. The mutants fell into five classes, either recessive (e.g., Fig. 8, L220P) or dominant to different extents (Fig. 8). Some dominant lesions reduced wild-type colony size but did not prevent formation of an aerotactic band of cells at the colony edge (e.g., Fig. 8, L256R), some abolished the sharp ring of aerotactic cells but did not reduce wild-type colony size (e.g., Fig. 8, N228S), some both reduced colony size and abolished the aerotactic ring (e.g., Fig. 8, A223V), and some caused a complete null morphology (e.g., Fig. 8, Q263L).
FIG. 8.
Dominance patterns of Aer mutant (Aer*) defects. The tryptone soft agar colony morphology of UU1250 strains carrying both a pMB1-Aer* plasmid and pKG117, a wild-type Aer plasmid, is shown after 17 h of incubation at 30°C. (Left) Aer* only induced. (Right) Aer wild type (WT) only induced. (Middle) Both Aer* and Aer WT induced. The scoring patterns are given in parentheses for the following mutants: L220P (recessive), L256R (dominant, reduced size), N228S (dominant, impaired ring), A223V (dominant, reduced size and impaired ring), and Q263L (dominant, null phenotype). Sal, sodium salicylate. The vector control colonies contain both vector plasmids (pCJ30, pKG116) and no WT Aer plasmid.
We expected that proteins with null defects would be recessive because of their rapid turnover and low steady-state expression levels. Indeed, the 10 null mutations in the PAS and F1 regions proved recessive, but four of them (S113P, I123N, Y130D, and H143P) exhibited partial dominance at high inducer levels. All but two null mutations in the HAMP region were also recessive. The V215D mutant protein seemed to slow growth (data not shown), which could account for its apparent dominance. The other exceptional mutation (L251P) lies at the C terminus of AS-2 and impaired wild-type ring formation but not colony size. The nearby W255R null mutation in the proximal signaling domain exhibited the same type of dominant behavior (Fig. 5). All other null mutations in the proximal signaling domain exhibited codominant behavior, reducing wild-type colony size but not its aerotactic morphology.
All CW-biased mutations proved dominant, abrogating aerotactic band formation by wild-type Aer (Fig. 5). The conventional explanation for dominant behavior in a multimeric protein such as Aer is subunit spoiling, the inability of wild-type subunits to contribute to function in heterodimers that also contain a mutant subunit. However, because Aer lacks a robust bias correction mechanism (9), we cannot be certain that CW-biased Aer subunits incapacitate Aer heterodimers. Instead, dominant behavior might reflect the inability of wild-type Aer homodimers to modulate the excessive CW signal output from mutant homodimers. This situation is in sharp contrast to the functional rescue of CW-biased Aer defects in cells containing MCP chemoreceptors, which can adjust the rotational bias of the flagellar motors through feedback control of the MCP methylation state.
DISCUSSION
To explore the roles of the F1 and HAMP segments in Aer signaling, we isolated aerotaxis-defective mutations in a host strain lacking all MCP transducers. Under these conditions, Aer alone is responsible for setting the cell's run/tumble swimming pattern and for modulating that behavior in response to oxygen gradients. We found two phenotypic classes of Aer mutants, designated null and CW biased. The majority of mutants exhibited the null phenotype, reflecting a complete loss of Aer function. Null mutant proteins failed to elicit episodes of CW rotation, indicative of an inability to activate the CheA kinase, and failed to raise cellular FAD levels when overexpressed, consistent with a defect in FAD binding. In contrast, CW-biased mutant proteins exhibited wild-type FAD binding but appeared to activate CheA to a much greater extent than wild-type Aer. CW-biased Aer proteins, unlike those with null defects, were able to mediate aerotactic responses in a host strain containing MCPs. Functional rescue of CW-biased defects accounts for the fact that such mutations were not noted in previous Aer mutant hunts done with MCP-containing tester strains (7, 16).
Nature of Aer null defects.
All null mutations in the F1 and HAMP segments, as well as those in the proximal regions of PAS and the signaling domain, severely reduced steady-state levels of the mutant Aer proteins. A survey of representative null mutants indicated that the low expression levels were caused by rapid degradation of Aer subunits. The instability and resultant low intracellular levels of null mutant proteins raise the distinct possibility that Aer null phenotypes arise mainly from the absence of a protein product. As discussed below, such mutations can provide misleading clues about structure-function relationships in native Aer molecules.
Role of F1 and HAMP in FAD binding.
Previous studies of Aer null mutants suggested that the F1 and HAMP segments played a direct role in FAD binding, perhaps by stabilizing or contributing to the FAD-binding pocket of the PAS domain (7, 16). However, this conclusion may be unwarranted because the FAD-binding assay requires high-level expression of Aer molecules, which, through binding, deplete the cellular supply of free FAD, causing a detectable up-regulation of FAD synthesis. Unstable mutant proteins that are actually competent for FAD binding might appear to be binding defective by this assay if proteolytic turnover of the mutant protein returns FAD molecules to the intracellular pool, thereby avoiding up-regulation of the FAD supply. In fact, most null mutant proteins with leaky, MCP-rescuable defects (M112K, I142T, V215A, R235C, and V246A), which presumably bind FAD, appeared to be binding defective with this test. Resolution of this issue requires a more direct method for measuring FAD binding by Aer molecules.
Sources of instability in mutant Aer proteins.
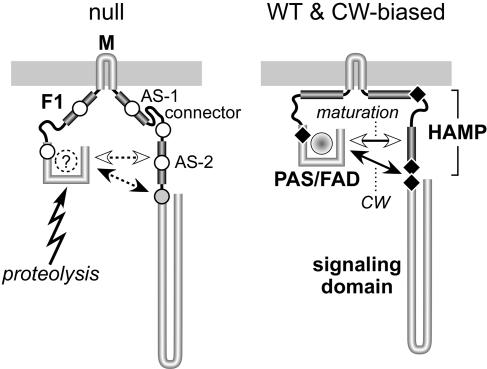
Wild-type Aer undergoes a complex maturation process (13). The nascent PAS domain associates with the GroEL chaperone, presumably to prevent misfolding until all PAS structural determinants have been synthesized (13). Release of the PAS domain from GroEL seems to occur after translation of the AS-1/connector segment; however, FAD binding seems to require translation of the entire HAMP domain (13). These observations suggest that the Aer PAS domain may interact with HAMP, most likely the AS-2 segment, to complete a critical maturation step. The PAS/HAMP interaction could serve to stabilize PAS structure or perhaps to render PAS competent for FAD binding. We suggest that the instability of most null mutant proteins reflects failure of this PAS/HAMP maturation process. Conceivably, a misfolded PAS domain or one without an FAD ligand is quite susceptible to proteolysis. Accordingly, null lesions in PAS and AS-2 might directly impair PAS/HAMP structural interactions needed for maturation. Null mutations in F1, AS-1, and the connector might affect PAS/HAMP interactions less directly, for example, by destabilizing Aer membrane topology, which in turn retards the maturation process (Fig. 9). Although we cannot exclude the possibility of direct interaction between these segments and the PAS domain, their low sequence conservation, particularly in the F1 segment, argues against specific structural interactions with PAS and in favor of a less specific interaction with the cytoplasmic membrane.
FIG. 9.
Possible structural basis for functional defects of null and CW-biased mutant Aer proteins. Although native Aer molecules most likely function as homodimers, only one subunit is shown. Some of the postulated interactions of Aer structural elements might take place between dimer subunits, rather than within one subunit, as illustrated. We propose that in native Aer molecules, the F1 and HAMP AS-1 segments contribute targeting and topology determinants that stabilize the membrane-associated conformation, thereby optimizing CW-inducing interactions between the PAS input domain and the AS-2/SD output region, which are modulated in response to redox changes in the PAS-associated FAD moiety to elicit aerotactic responses. Mutational lesions (indicated by black diamonds) can enhance the CW-inducing PAS interaction in several different ways. CW-biased mutations in the PAS and AS-2/SD segments could conceivably affect the interacting surfaces directly, whereas CW-biased mutations in the AS-1/connector portion of HAMP probably alter the geometry or flexibility of the Aer molecule to favor the CW interaction. Different mutational lesions in many of the same Aer structural elements destabilize the Aer molecule, leading to rapid degradation and apparent null defects. We propose that these mutant proteins are unstable because their PAS domains cannot achieve native structure, perhaps because they fail to bind FAD. The nonnative PAS domain is the primary target for proteolytic cleavage of Aer molecules that fail to mature. Maturation requires a structural interaction between the nascent PAS domain and the AS-2 segment of HAMP. Null mutations in the PAS and AS-2 segments (indicated by unfilled circles) may directly abrogate that interaction. Null mutations in the F1, AS-1, and connector segments (unfilled circles) may influence PAS/AS-2 interactions indirectly, for example, through altered membrane interaction, geometry, or flexibility of the molecule. Null mutations in the proximal signaling domain (indicated by a shaded circle) also destabilize the Aer molecule, but to a less drastic extent. These lesions could distort the structure or dynamics of the adjacent AS-2 segment and thereby affect the efficiency of the PAS maturation process. WT, wild type.
Aerosensing functions in mature null mutant molecules.
The degradation profiles of null mutant proteins revealed a small subpopulation of stable molecules that presumably had completed the maturation process, thereby escaping destruction. Several lines of evidence indicate that these stable mutant molecules are competent for Aer function. First, leaky null mutants had substantial subpopulations of unstable molecules, implying that their residual function emanated from the minority of stable molecules, despite their mutational lesions. Second, most null lesions in the HAMP domain (26; M. Burón-Barral, K. G. Gosink, and J. S. Parkinson, unpublished data) as well as some in the PAS and F1 domains (K. G. Gosink, M. Burón-Barral, and J. S. Parkinson, unpublished data) regain Aer function when combined with a “general suppressor” in the PAS domain (e.g., S28G). The S28G alteration stabilizes mutant Aer molecules to proteolysis, resulting in near-wild-type steady-state levels (Burón-Barral, Gosink, and Parkinson, unpublished). These findings indicate that the suppressible Aer mutations slow maturation but do not affect function of the mature protein. Thus, we conclude that slow maturation and rapid degradation suffice to account for the functional defects of most Aer null mutants characterized in this study and in previous studies (7, 16, 26). The severity of the mutant protein instability problem was not apparent in earlier work because steady-state expression levels were assessed at full induction, where degradation capacity is rate limiting (e.g., reference 7).
Dominant null defects.
The L251P mutation at the C terminus of AS-2 exhibited dominant complementation properties (Fig. 5) and might affect both the maturation and subsequent function of Aer subunits. The expression level of the mutant protein (Fig. 5) and its subpopulation of mature molecules were no larger than for other null mutants, roughly 1 to 5% of normal wild-type levels (Fig. 7). It seems unlikely, therefore, that this null mutant makes enough full-length mature molecules to cause dominance via subunit spoiling in heterodimers. However, it might generate high levels of mutant Aer fragments through proteolysis of the maturation-defective molecules. We would not have detected such fragments in our degradation assays because the Aer antibodies we used in Western analyses were directed against Aer residues 1 to 263. Most epitopes lie in the PAS domain, which is probably the site of initial proteolytic cleavage during degradation (12). At this point, we cannot be certain that dominance involves mutant Aer fragments because the presence of wild-type Aer subunits could conceivably modify or alleviate the maturation defects of some mutant Aer subunits. Nevertheless, functional spoiling of wild-type Aer subunits by mutant fragments seems to be the most parsimonious explanation for the dominant properties of L251P.
Nature of CW-biased Aer defects.
CW-biased lesions were obtained at the C terminus of the PAS domain, in HAMP at the AS-1/connector and AS-2/signaling domain junctions, and in the HAMP-proximal portion of the Aer signaling domain (Fig. 9). All CW-biased mutant proteins exhibited essentially normal expression and FAD-binding levels, although in degradation tests some were noticeably less stable than wild-type Aer (e.g., Fig. 7C, M252T). All CW-biased proteins also retained input-output control, as demonstrated by functional rescue in a host containing MCP transducers. Thus, CW-biased Aer proteins resemble wild-type Aer in most respects but activate the CheA kinase to a much greater extent in the absence of aerotactic stimuli.
Conventional MCP molecules appear to oscillate between two signal states corresponding to kinase-on and kinase-off activity modes. Because Aer has an MCP-like signaling domain, we assume that it also conforms to a two-state model. The CW-biased Aer mutations described in this report lie upstream of the CheA/CheW contact sites, which are located in the hairpin portion of the signaling domain (Fig. 9), so they most likely shift the signaling equilibrium toward the CheA-activating state, either by destabilizing the kinase-off state or by stabilizing the kinase-on state. Although this alteration results in an apparent gain-of-function phenotype (elevated output activity), the underlying structural lesions could act by impairing rather than augmenting a wild-type function or structural interaction.
All CW-biased lesions were dominant over wild-type Aer. The inability of Aer to modify its signaling properties through methylation-demethylation changes, as MCPs can do, is probably ultimately responsible for the dominance of CW-biased defects: the cell's overly tumbly swimming pattern cannot be corrected by sensory adaptation, precluding aerotactic behavior. Dominance might be caused by subunit spoiling in mutant/wild-type heterodimers, for example, through impairment of an intersubunit signaling interaction. Alternatively, mutant homodimers might spoil the function of wild-type homodimers in mixed trimers of dimers. Thus, dominance does not necessarily mean that CW-biased mutations impair signaling interactions between the subunits of an Aer dimer; dominance could arise from mutant homodimers at the trimer-of-dimers level.
Control inputs to the Aer signaling domain.
Regulation of the CCW/CW output equilibrium in MCPs and Aer most likely involves modulation of coiled-coil and four-helix bundle interactions within and between signaling domain (SD) subunits in dimers and, in turn, modulation of dimer-dimer interactions within trimers of dimers (14, 18, 23). CW-biased mutations in the proximal signaling domain of Aer (Q263L, V264A, and V267I) probably directly affect the coiled-coil and intersubunit interactions (SD/SD′) within Aer dimers. The HAMP AS-2 segment in MCPs also appears to have a coiled-coil structure (11), and AS-2/AS-2′ interactions most likely modulate MCP signal domain output (6). Aer signal output might be regulated similarly, through modulation of either AS-2/AS-2′ or SD/SD′ interactions (see below) by a direct, perhaps competing, interaction with the PAS sensory domain (25, 26). We suggest that CW-biased mutations in PAS (M112I and M112V) and in AS-2 (M252T) augment a PAS interaction responsible for promoting CW signal output. Oxygen increases presumably attenuate this CW interaction to elicit smooth-swimming aerotactic responses. According to this model, CW output in Aer should be PAS dependent. Analysis of epistatic interactions between Aer mutations supports this view (Gosink, del Carmen Burón-Barral, and Parkinson, unpublished).
The CW-biased Aer mutations at the AS-1/connector junction in HAMP (K221E, A223V, and N228S) are more difficult to understand. There is no evidence that this portion of HAMP interacts directly with either PAS or AS-2/SD, so we suggest that AS-1/connector lesions modulate PAS interactions with AS-2/SD indirectly by influencing their spatial relationship in Aer molecules. For example, one role of AS-1 might be to stabilize Aer membrane topology through interaction with the cytoplasmic face of the inner membrane (Fig. 9; also see below). The F1 domain flanking the other end of the membrane-embedded M segment might play a similar role (Fig. 9). These membrane interactions could position PAS and AS-2/SD for optimal interaction, either within or between Aer subunits. To account for CW-biased AS-1/connector mutations, we propose a second structural state in which AS-1 interacts with connector residues rather than the membrane (Fig. 9, left). Lesions that disrupt this structure would favor the membrane-associated, CW-signaling conformation (Fig. 9, right). These AS-1/connector structural interactions most likely occur within Aer protomers, but the subsequent PAS/AS-2/SD interaction may occur between subunits of the Aer dimer (25).
Null mutations in the HAMP-proximal signaling domain.
Mutant proteins with null mutations in the proximal signaling domain had steady-state expression levels severalfold higher (Fig. 5) and degradation rates severalfold lower (Fig. 7) than those caused by null lesions in other Aer segments. Unlike other null proteins, the signal domain mutants explicitly bound FAD and were dominant over wild-type Aer (Fig. 5). These properties suggest that the signal domain lesions may still impair the PAS/HAMP maturation interaction, but less severely than other null mutations. Perhaps their structural alterations destabilize the neighboring HAMP region. Moreover, their dominant behavior probably reflects subunit spoiling in either full-subunit or fragment-containing heterodimers, consistent with the idea that SD/SD′ interactions regulate Aer signal output.
These signal domain null lesions fell into two distinct classes with respect to residual I/O control capability: W255R, L256R, D259G, V260D, and S261P were not functionally rescued by MCPs, whereas V260A, Q263R, and R268G were rescued (Fig. 5). Although the sample size is small, the distinct locations of these two mutation types may be significant. The unrescuable lesions, presumably defective in I/O control, lie adjacent to the C terminus of AS-2. Because the AS-2/SD junction appears to be the most likely target for PAS output control interactions (26), the unrescuable lesions may affect determinants or structures important for that PAS interaction.
In summary, the PAS domain of Aer appears to engage in two different structural or functional interactions with AS-2 and the HAMP-proximal signaling domain. One of those interactions is needed for Aer maturation; when it fails, the mutant Aer subunits are rapidly degraded. The other interaction promotes CW output from the Aer signaling domain and is modulated by aerotactic stimuli. Most aerotaxis-defective mutations in these regions affect only maturation, indicating that these two interactions with PAS involve structurally distinct determinants in the HAMP/SD target region.
Acknowledgments
We thank Nicole Pershing for assistance with the HAMP mutant hunt.
This work was supported by research grant GM62940 from the National Institute of General Medical Sciences. The Protein-DNA Core Facility at the University of Utah receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute.
REFERENCES
- 1.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, P., Y. A. Yu, and J. S. Parkinson. 1996. Methylation segments are not required for chemotactic signalling by cytoplasmic fragments of Tsr, the methyl-accepting serine chemoreceptor of Escherichia coli. Mol. Microbiol. 19:737-746. [DOI] [PubMed] [Google Scholar]
- 3.Amin, D. N., B. L. Taylor, and M. S. Johnson. 2006. Topology and boundaries of the aerotaxis receptor Aer in the membrane of Escherichia coli. J. Bacteriol. 188:894-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appleman, J. A., L. L. Chen, and V. Stewart. 2003. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J. Bacteriol. 185:4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 6.Bass, R. B., and J. J. Falke. 1999. The aspartate receptor cytoplasmic domain: in situ chemical analysis of structure, mechanism and dynamics. Structure 7:829-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bibikov, S. I., A. C. Miller, K. K. Gosink, and J. S. Parkinson. 2004. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J. Bacteriol. 186:3730-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biran, D., E. Gur, L. Gollan, and E. Z. Ron. 2000. Control of methionine biosynthesis in Escherichia coli by proteolysis. Mol. Microbiol. 37:1436-1443. [DOI] [PubMed] [Google Scholar]
- 11.Danielson, M., R. Bass, and J. Falke. 1997. Cysteine and disulfide scanning reveals a regulatory α-helix in the cytoplasmic domain of the aspartate receptor. J. Biol. Chem. 272:32878-32888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gosink, K. K., M. D. C. Burón-Barral, and J. S. Parkinson. 2006. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli. J. Bacteriol. 188:3487-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann, S., Q. Ma, M. S. Johnson, A. V. Repik, and B. L. Taylor. 2004. PAS domain of the Aer redox sensor requires C-terminal residues for native-fold formation and flavin adenine dinucleotide binding. J. Bacteriol. 186:6782-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, S. H., W. Wang, and K. K. Kim. 2002. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. USA 99:11611-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Ma, Q., M. S. Johnson, and B. L. Taylor. 2005. Genetic analysis of the HAMP domain of the Aer aerotaxis sensor localizes flavin adenine dinucleotide-binding determinants to the AS-2 helix. J. Bacteriol. 187:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkinson, J. S. 1976. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 126:758-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkinson, J. S., P. Ames, and C. A. Studdert. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8:116-121. [DOI] [PubMed] [Google Scholar]
- 19.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slominska, M., A. Wahl, G. Wegrzyn, and K. Skarstad. 2003. Degradation of mutant initiator protein DnaA204 by proteases ClpP, ClpQ and Lon is prevented when DNA is SeqA-free. Biochem. J. 370:867-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sourjik, V. 2004. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 12:569-576. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watts, K. J., M. S. Johnson, and B. L. Taylor. 2006. Minimal requirements for oxygen sensing by the aerotaxis receptor Aer. Mol. Microbiol. 59:1317-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watts, K. J., Q. Ma, M. S. Johnson, and B. L. Taylor. 2004. Interactions between the PAS and HAMP domains of the Escherichia coli aerotaxis receptor Aer. J. Bacteriol. 186:7440-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams, S. B., and V. Stewart. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 33:1093-1102. [DOI] [PubMed] [Google Scholar]
- 28.Yen, K. M. 1991. Construction of cloning cartridges for development of expression vectors in gram-negative bacteria. J. Bacteriol. 173:5328-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]