Abstract
A DNA microarray was used to identify genes transcribed in Neisseria gonorrhoeae using Ecf, an alternative sigma factor. No differences between the transcriptional profiles of strain FA1090 and a mutant where ecf had been inactivated could be detected when both were grown in vitro. We therefore constructed a gonococcal strain in which Ecf can be overexpressed. Some differentially expressed genes are clustered with ecf on the genome and appear to form a single transcriptional unit. Expression of the gene encoding MsrAB, which possesses methionine sulfoxide reductase activity, was also dependent on Ecf, suggesting that the regulon responds to oxidative damage. Western blotting confirmed that the increased level of MsrAB protein is dependent on the presence of Ecf.
Bacterial sigma factors are essential components of the RNA polymerase holoenzyme and determine promoter selectivity and specificity. Bacteria usually contain at least one essential sigma factor, sigma-70, which is necessary for cell viability, as well as a number of accessory sigma factors that are often involved in responses to environmental stimuli or the phase of growth. The relative amount of RNA polymerase holoenzyme containing each sigma factor determines the amplitude of the expression of a specific collection of genes.
We have searched the genome sequence of Neisseria gonorrhoeae strain FA1090 (GenBank accession number AE004969) for the presence of genes encoding sigma factors. As expected, there is a gene, rpoD, encoding sigma-70 (NGO0999). Only two intact genes encoding alternative sigma factors, rpoH (NGO0288) and ecf (NGO1944), were found. Laskos et al. (11) have shown that there is also an inactive RpoN-like sigma factor, RLS, although this sequence feature (NGO1766) has not been annotated as such in the publicly available annotations of the N. gonorrhoeae strain FA1090 genome sequence (GenBank accession number AE004969; annotations at www.stdgen.lanl.gov,cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi, and www.ncbi.nlm.nih.gov/genomes/lproks.cgi). The ecf gene encodes a member of the extracytoplasmic function (ECF) family of sigma factors. As the name suggests, ECF sigma factors from different bacterial species appear to respond specifically to a variety of extracytoplasmic stimuli. Characteristically, their activity is controlled by anti-sigma factors, and they control relatively small regulons (1). In most examples of this system investigated to date, the ECF sigma factors regulate not only their own expression but also that of the genes encoding the cognate anti-sigma factor, which are located in the same operon.
We have used DNA microarrays to assess the role of the sigma factor Ecf in gonococci and show that it controls a small regulon which contains the msrAB gene, which encodes an unusual methionine sulfoxide reductase.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strain DH5α [F− endA1 thi-1 hsdR17 supE44 relA1 ΔlacU169 (φ80 ΔlacZM15)] was used for genetic manipulation of constructs. E. coli strains were grown in Luria-Bertani (LB) broth (Difco) at 37°C with agitation or on LB agar plates supplemented with 1.5% agar, and where appropriate, with the following antibiotics: 100 μg/ml ampicillin, 25 μg/ml kanamycin, 150 μg/ml erythromycin, or 10 μg/ml tetracycline. Gonococcal strains were grown on GC agar base (Oxoid) or GC broth as described previously (7). Where appropriate, media were supplemented with 7 μg/ml erythromycin, 12.5 μg/ml tetracycline, or 40 μg/ml kanamycin. Transformation and conjugation experiments with N. gonorrhoeae were performed as described previously (12) with the exception that conjugation experiments involved mixing 5 × 108 donor cells and 1.5 × 109 recipient cells.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference |
|---|---|---|
| Plasmids | ||
| Hermes-8 | E. coli/N. gonorrhoeae shuttle vector, erythromycin resistance | 10 |
| pJKD2601 | 2.3-kb PCR product amplified from the N. gonorrhoeae FA1090 genomic DNA with primer pair 5673 and 5566, inserted into HincII-digested pUC18 | This study |
| pJKD2603 | aphA1 cassette ligated into pJKD2601 which was linearized at a HincII site within ecf | This study |
| pJKD2606 | 0.5-kb PCR product containing the ecf gene from N. gonorrhoeae strain FA1090 ligated into BamHI digested Hermes-8 | This study |
| pJKD2623 | 1.7-kb PCR product (containing the msrAB promoter region) amplified with primers 23764 and 23763 and ligated to HincII-digested pUC18 vector | This study |
| pJKD5065 | pJKD2606 linearized and recombined into ptetM25.2 | This study |
| pJKD5133 | Empty Hermes-8 shuttle vector recombined into ptetM25.2 | This study |
| ptetM25.2 | Gonococcal conjugative plasmid | 10 |
| pUC18 | Cloning vector, ampicillin resistance | 31 |
| pUC4K | Cloning vector used as source of aphA1(kanamycin resistance)cassette, ampicillin resistance | 28 |
| N. gonorrhoeae strains | ||
| FA1090 | Wild-type strain | 4 |
| JKD484 | Spontaneous rifampin-resistant mutant of strain N. gonorrhoeae MS11-A, carrying ptetM25.2 | 12 |
| JKD5062 | JKD5069(pJKD5065) | This study |
| JKD5064 | JKD5083(pJKD5133) | This study |
| JKD5065 | JKD484 carrying pJKD5065, resulting from recombination between pJKD2606 and ptetM25.2 | This study |
| JKD5069 | ecf mutant of strain FA1090 | This study |
| JKD5083 | Spectinomycin-resistant mutant of N. gonorrhoeae strain FA1090 | This study |
| JKD5133 | JKD5083 carrying Hermes-8 shuttle vector recombined into the ptetM25.2 (pJKD5133) | This study |
Construction of an ecf mutant of N. gonorrhoeae.
All DNA manipulations were performed by standard methods as described previously (7). Oligonucleotide primers are listed in Table 2. The ecf gene from N. gonorrhoeae strain FA1090 was amplified using the Expand long-template PCR system (Roche Diagnostics) and primer pair 5673 and 5566 (Table 2). The 2.3-kb PCR product was purified using QIAquick PCR purification spin columns (QIAGEN), treated with T4 DNA polymerase, and ligated to HincII-digested pUC18 (31) to create pJKD2601 (Table 1). The aphA1 cassette was excised from pUC4K (28) by HincII/EcoRI digestion, gel purified, and treated with T4 DNA polymerase to remove nucleotide overhangs. This purified cassette was ligated into pJKD2601, which was linearized at the HincII site within ecf and used to transform E. coli DH5α. Ampicillin- and kanamycin-resistant colonies were screened by PCR for the presence of the insert. The resultant plasmid, pJKD2603 (Table 1), was linearized with SphI, and the mutated gene was recombined into the genome of N. gonorrhoeae strain FA1090 by homologous recombination. Transformants were isolated by selection for kanamycin resistance, and the presence of the aphA1 cassette in ecf was confirmed by PCR. The ecf mutant was designated JKD5069.
TABLE 2.
Oligonucleotide primers
| Oligonucleotide | Sequence (5′-3′) | Reference | Usea |
|---|---|---|---|
| 3260 | CACACTGGGACTGAGACATG | 12 | Northern blotting; amplification of 16S rRNA probe |
| 3261 | CGGCAGTCTCATTAGAGTGC | 12 | Northern blotting; amplification of 16S rRNA probe |
| 5566 | GGCCTGTCCTACGAGTAG | This study | Amplification of ecf region |
| 5673 | ACCGACCGTTTGGGACTTGCTC | This study | Amplification of ecf region |
| 16764 | GCTGCCGTCCATTTTCATG | This study | qRT-PCR of recA |
| 16766 | TGGCGCAAATCGAAAAAAGT | This study | qRT-PCR of recA |
| 20398 | CGGGATCCCGTTTACTTCGGGTTTTCTTG | This study | Amplification of ecf gene |
| 20399 | CGGGATCCAGGAGGATACTATGCCGCTACCCGACCTG | This study | Amplification of ecf gene |
| 23108 | CTTGGCAGACGGCATCCG | This study | Northern blotting; amplification of NGO1948 probe |
| 23110 | AAGGCAGCATCAACGAAGCG | This study | Northern blotting; amplification of msrAB probe |
| 23111 | CATTTTATAACCGTTGTCGC | This study | Northern blotting; amplification of NGO1948 probe, 5′ RACE |
| 23112 | TTGCTGCTCGCGTTTGAGGG | This study | Northern blotting; amplification of msrAB probe |
| 23488 | CGCCGTATGATGCACCATT | This study | qRT-PCR of NGO1946 |
| 23489 | AACGACCATCAGCCCCAAT | This study | qRT-PCR of NGO1948, 5′ RACE |
| 23494 | ACGGCGGTCATCTTTACGAT | This study | qRT-PCR of NGO1946 |
| 23495 | CACCCTGACCGAAGAGCAAT | This study | qRT-PCR of msrAB |
| 23496 | TCGTGGCTGAAGGCGTATTC | This study | qRT-PCR of msrAB |
| 23497 | GCTGCCCGTACTGTTGCTTT | This study | qRT-PCR of NGO1948 |
| 23573 | TTCCCAAAACGGGCATTG | This study | qRT-PCR of ecf |
| 23574 | TGATTTTTCCGGAGTGTTCCA | This study | qRT-PCR of ecf |
| 23762 | ACGTGGATAAAGTGTGCG | This study | Primer extension of msrAB tsp |
| 23763 | TCTTCGTAGCTCGGGTTTTCC | This study | Amplification of msrAB promoter region |
| 23764 | CAAGCACGATGTCGATCC | This study | Amplification of msrAB promoter region |
| DT88 | GAAGAGAAGGTGGAAATGGCGTTTTGG | 27 | 5′ RACE |
| DT89 | CCAAAACGCCATTTCCACCTTCTCTTC | 27 | 5′ RACE |
qRT-PCR, quantitative real-time RT-PCR.
Construction of a strain of N. gonorrhoeae that can overexpress the ecf gene.
JKD5062, in which ecf can be expressed from an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Ptrc promoter, was created in the following manner. The ecf gene from N. gonorrhoeae strain FA1090 was amplified using oligonucleotide primers 20399 and 20398 (Table 2). Each primer contained a BamHI site to assist in cloning, and primer 20399 also contained a modified ribosome binding site for improved translation of the mRNA. The PCR product was gel purified, digested with BamHI, and ligated to a BamHI-digested, alkaline phosphatase-treated Hermes-8 shuttle vector (Table 1). Briefly, this shuttle vector has an ermC′ gene capable of being expressed in both E. coli and N. gonorrhoeae, an inducible promoter that is also functional in N. gonorrhoeae and is upstream of a multiple cloning site, and the lacI repressor gene (10). The initial E. coli transformants were isolated on erythromycin plates and screened for the ecf insert by PCR. The resultant plasmid, pJKD2606, was linearized with SacII and used to transform N. gonorrhoeae strain JKD484, which carries the conjugative plasmid ptetM25.2 (10). Transformation of competent N. gonorrhoeae strains involves recombination of Hermes constructs into the permissive region of the conjugative plasmid ptetM25.2 by allelic replacement (10). Erythromycin- and tetracycline-resistant transformants were tested by PCR to confirm that pJKD2606 had recombined into ptetM25.2, and this strain was designated JKD5065 (Table 1). The resulting plasmid, pJKD5065, was transferred into JKD5069 by conjugation. The resulting strain was designated JKD5062 (Table 1). The ecf gene in pJKD5065 was sequenced to verify that no mutations had been introduced. A control strain, JKD5064, was constructed by conjugation of pJKD5133 (empty Hermes-8 shuttle vector recombined into the ptetM25.2 conjugative plasmid) from JKD5133 into JKD5083 (Table 1).
For investigation of the Ecf regulon, gonococcal strain JKD5062 was grown to mid-exponential phase (optical density at 560 nm = 0.6), and the culture was then split between two prewarmed flasks. One of the cultures was induced with 2 mM IPTG for 30 min. Samples from cultures were harvested and stored in RNAlater RNA stabilization solution (Ambion). The RNA from these samples was extracted using the RNeasy midi kit (QIAGEN) with an optional on-column DNase I digestion step according to the manufacturer's instructions. The quality and quantity of RNA were determined by gel electrophoresis and spectrophotometry.
cDNA synthesis and fluorescent labeling.
Total RNA (30 μg) was mixed with 30 μg of random hexamers, heated to 70°C for 10 min, and then rapidly chilled on ice. To this mixture 0.5 μl (20 units) of RNasin (Promega), 6 μl of Superscript II buffer (Life Technologies, Inc.), 3 μl dithiothreitol (DTT), 1 mM dATP, 1 mM dCTP, 1 mM dGTP, 0.4 mM dTTP (Promega), 0.6 mM amino allyl-dUTP (Sigma), and 2 μl (400 units) Superscript II reverse transcriptase (Life Technologies, Inc) were added, and the mixture was incubated for 2.5 h at 42°C. Reactions were terminated by the addition of 10 μl of 1 M NaOH and 10 μl of 0.5 M EDTA, and the mixtures were heated to 65°C for 15 min and neutralized by the addition of 25 μl of 1 M Tris, pH 7.4. Unincorporated amino allyl-dUTP was removed with Microcon 30 columns (Millipore) according to the manufacturer's instructions. The purified cDNA was concentrated to 12 μl, and the quality of labeling and quantity of cDNA were determined using a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies). Cy3 and Cy5 dyes were dissolved in dimethyl sulfoxide (Sigma) to a final concentration of 14 μg/μl, and a 4-μl aliquot was added to each cDNA sample in 0.1 M sodium bicarbonate buffer (pH 9). Reaction mixtures were incubated for 1 h at room temperature in the dark, labeled cDNA was purified using Microcon columns, and the eluted samples were concentrated to 10 μl using a SpeedVac SVC100 (Savant). Cy3 and Cy5 dye incorporation was estimated by spectrophotometry.
Microarray hybridization and data analysis.
The labeled cDNA was hybridized to a pan-Neisseria microarray which contains 2,704 PCR products, spotted in triplicate, corresponding to potential coding sequences from N. gonorrhoeae strain FA1090, N. meningitidis strains MC58 and Z2491, the N. gonorrhoeae strain MS11 gonococcal genetic island (8), and various controls. Details of the construction of this array will be published elsewhere, but details and arrays can be obtained from the corresponding author. Prior to prehybridization, the slide was plunged into a 95°C water bath for 2 min, centrifuged for 5 min at 2,000 × g, and used immediately. Prehybridization was carried out in a volume of 30 μl containing 25% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), 10 mg/ml bovine serum albumin (BSA; fraction V), and 30 μg of herring sperm DNA (Promega) under a coverslip in a humidified Corning CMT hybridization chamber for 45 min at 42°C. The slide was rinsed in water, dried by centrifugation, and used immediately for hybridization. The labeled cDNA was added to the hybridization fluid (prehybridization solution without BSA) in a total volume of 30 μl and denatured at 95°C for 5 min. The hybridization fluid was placed on the surface of the microarray under a coverslip. The slide was enclosed in the hybridization chamber and submerged in a 42°C water bath overnight. After hybridization, slides were washed once in 2× SSC-0.1% SDS for 5 min at 42°C, once in 0.1× SSC-0.1% SDS for 10 min at room temperature, and four times in 0.1× SSC for 1 min at room temperature. The slides were rinsed with water for 10 seconds, and excess fluid on the surface of the slide was removed by centrifugation before scanning. A GMS 418 array scanner (Genetic Microsystem) was used to scan the images. The Cy3 and Cy5 images were combined, and fluorescent and background intensity for each spot was determined using ImaGene version 5 software (BioDiscovery) as outlined previously (2). Data from poor spots that were manually or automatically flagged in ImaGene were removed from further analysis.
The individual ImaGene data files were uploaded to a web site created with BASE (19) and converted to a common BASE format using a series of custom-made applications. The spot intensities were found to be most reliable when no background correction was performed. Analysis was done using Bioconductor (5) and Limma (23). The normalization of the data to remove various biases involved two steps. Firstly, each array was normalized independently using print-tip loess normalization (Y. H. Yang, S. Dudoit, P. Luu, and T. P. Speed, presented at SPIE BiOS 2001, San Jose, Calif.). Secondly, diagnostic plots suggested a variation in scale between arrays, so the log ratios were scaled in such a way that each array had the same median-absolute-deviation. The normalized data were then used to fit a linear model (23) for each gene using a generalized least-squares method which took into account the correlation between replicate spots (24). The coefficient of the fitted model for each gene describes the inferred difference in RNA expression between the two strains. Empirical Bayes method was then used to calculate the moderated t statistics and associated P values. The P values were adjusted for multiple testing using the false-discovery rate. Genes with an absolute ratio greater than 1.5-fold and significant at the 0.001 level were selected as differentially expressed.
Quantitative real-time reverse transcriptase PCR (RT-PCR).
cDNA was generated from 5 μg of the total RNA samples used for the microarray experiments. The RNA preparation was subjected to a second DNase I treatment before cDNA synthesis. The cDNA synthesis was performed as described above with the exception that a 1 mM concentration of each dNTP and 7.8 μg of random hexamers were used. Oligonucleotide primer pairs specific for each gene of interest were designed using ABI PRISM Primer Express software (Applied Biosystems) and are listed in Table 2. To quantitate the cDNA, a gene-specific standard curve method was employed using serial dilutions of strain FA1090 genomic DNA as the template. All assays included 12.5 μl of SYBR Green PCR master mix (Applied Biosystems), 2 μl of each primer (0.5 nM final), 2 μl of template, and diethyl pyrocarbonate-treated water made up to a final volume of 25 μl. Controls lacked reverse transcriptase or template. Reactions were run on an ABI 7700 sequence detection system (Applied Biosystems), and recA was used as the normalizer for all reactions. All reactions amplified a single product, as determined by melting curve analysis (Applied Biosystems).
Northern blots.
Northern blotting was performed on 10 μg of the total RNA preparations used in the microarray experiments. RNA was electrophoresed on formaldehyde-MOPS (morpholinepropanesulfonic acid) 1% agarose gels and transferred to Hybond N nylon membranes (Amersham) as previously described (20). The transferred RNA was fixed to the nylon membrane with the auto-cross-link setting on a UV Stratalinker 1800 (Stratagene). Probe labeling and detection were performed using a digoxigenin nonradioactive DNA labeling and detection kit (Roche Diagnostics) according to the manufacturer's instructions. Probes were amplified by PCR from each gene using oligonucleotides listed in Table 2.
Identification of transcription start points.
Oligonucleotide primer 23762 (Table 2), which is complementary to the msrAB sequence, was 5′-end labeled with 30 μCi of [γ-32P]dATP (Amersham) and T4 polynucleotide kinase. Primer extension reactions were performed as described in the instructions for the use of the primer extension system (Promega) using avian myeloblastosis virus reverse transcriptase. The precipitated primer extension products were subjected to electrophoresis in an 8% polyacrylamide gel containing 8 M urea, next to a sequencing ladder generated from plasmid pJKD2623 (Table 1) with oligonucleotide primer 23762 (Table 2). Plasmid pJKD2623 was constructed by amplifying a PCR product encompassing the promoter region of msrAB with primers 23764 and 23763 (Table 2), T4 filling, and ligation to HincII-cut pUC18 vector. Primer extension gels were dried onto chromatography paper and subjected to autoradiography.
To confirm the transcription start points, 5′ RACE (rapid amplification of cDNA ends) experiments were performed (27) with the same RNA preparations used in the microarray experiments. cDNA synthesis was performed as described previously (27) using a specific oligonucleotide primer, 23111 (Table 2), located 150 to 200 bp downstream from the initiation codon of NGO1948. Briefly, 5′-phosphorylated, 3′-blocked primer DT88 (27) (Geneworks) (Table 2) was anchored to the 5′ end of the cDNA, and a first-round PCR product was amplified using primer DT89 (Table 2), which was complementary to DT88 (27), and oligonucleotide 23111 (Table 2). A second round of PCR used oligonucleotides DT89 and a nested primer, 23489 (Table 2), that was internal to 23111. Controls with and without reverse transcriptase and T4 RNA ligase were included. Purified PCR product was sequenced to determine the 5′ end of the transcript, which represents the transcription start point.
Western blots.
Western blotting was performed as described previously (11). The samples were prepared from whole-cell extracts of induced and noninduced gonococcal strains JKD5062 and JKD5064 (Table 1) grown in liquid medium. Prior to electrophoresis, 1 × 109 cells were harvested by centrifugation at 5,000 × g at 4°C for 5 min, resuspended in 500 μl of phosphate-buffered saline and SDS sample buffer (0.2 M Tris, 20% glycerol, 25% SDS, 10% β-mercaptoethanol, 0.1% bromophenol blue; pH 6.8), and boiled for 5 min. For equivalent loading, total protein concentration was determined by the microplate procedure using the bicinchoninic acid protein assay kit (Pierce). Primary polyclonal anti-MsrAB rabbit antiserum was used at a 1:20,000 dilution (22). Peroxidase-conjugated anti-rabbit immunoglobulin was used as the secondary antibody (Chemicon) at a 1:3,000 dilution. The Western blot was completed using the protocol provided with the ECL detection reagent kit (Amersham).
RESULTS AND DISCUSSION
The ecf gene of N. gonorrhoeae strain FA1090 was amplified by PCR, cloned, and insertionally inactivated, and the resultant construct was used to transform strain FA1090, creating the ecf mutant JKD5069 (Table 1). Inactivation of the ecf gene did not result in any noticeable change in colony morphology or growth rate on solid or in liquid media (data not shown). DNA microarrays were used to assess whether there were any detectable differences in the transcriptome of JKD5069 compared to FA1090, when both strains were in the mid-exponential phase of growth. Despite the fact that a basal level of transcription of the ecf gene could be detected in strain FA1090, when the data from two biological repeat experiments (four hybridizations) were combined and analyzed, no genes were found to be differentially expressed in the mutant compared to the wild type at the 1.5-fold level with a P value of less than 0.001. This result suggests that, under the growth conditions used, Ecf activity might be suppressed by an anti-sigma factor, as is the case in other bacterial species (3, 25). It may also be that the microarray experiment was simply not sensitive enough to detect the expected drop in transcription of Ecf-regulated genes, from a low basal level.
In the absence of any knowledge of the physiological signals to which Ecf responds, we reasoned that activation of the regulon could be obtained by overexpression of the sigma factor. To this end we used an IPTG-inducible Ptrc promoter in the E. coli/N. gonorrhoeae Hermes shuttle vector system (10), in the background of the ecf mutant JKD5069. The strain constructed, JKD5062, allows exclusive and inducible expression of a plasmid-borne copy of the ecf gene (see Materials and Methods). We again used microarrays to measure changes in gene expression when the ecf gene in strain JKD5062 was induced for 30 min compared to an uninduced control.
Four genes, NGO1946, NGO1947, NGO1948, and NGO2059, were up-regulated along with ecf (NGO1944) itself when ecf was overexpressed (Table 3). As might be expected with the overexpression of an alternative sigma factor, no down-regulated genes were detected. Quantitative real-time RT-PCR was used to validate the microarray data for three of the up-regulated genes (Table 3). NGO1946, NGO1947, and NGO1948 are annotated as encoding proteins with no known function. The presence of possible transmembrane domains, as predicted by PSORT (15), suggests that NGO1948 might encode an integral membrane protein. A conserved-domain search (13) with the derived amino acid sequence from NGO1948 also revealed the presence of a motif that is found in the DoxD-like family of proteins. DoxD is a subunit of quinone oxidoreductase, which is involved in the oxidation of sulfur (14, 17). NGO1947 is predicted by PSORT (15) to encode a periplasmic protein. Genes encoding proteins closely related to Ecf and the genes NGO1946, NGO1947, and NGO1948 can be found clustered in other bacterial genomes, where the gene order also seems to have been at least partially conserved (data not shown). The expression ratio for NGO1945, the gene located between ecf and NGO1946, was just below the 1.5-fold cutoff used in these experiments.
TABLE 3.
Genes up-regulated in strain JKD5062 as a result of overexpression of Ecf
| ORF IDa | Gene name | Fold changeb
|
Proposed function | |
|---|---|---|---|---|
| Microarrayc | qRT-PCR | |||
| NGO1944d | ecf | 2.2 | 5.7 | ECF family RNA polymerase sigma factor |
| NGO1946 | 1.8 | 3.7 | Conserved hypothetical protein | |
| NGO1947 | 1.8 | Hypothetical periplasmic protein | ||
| NGO1948 | 1.8 | 4.0 | Conserved hypothetical protein (possible membrane protein) | |
| NGO2059 | msrAB | 3.1 | 5.7 | Methionine sulfoxide reductase |
ORF ID, identification from the annotation of the GenBank entry (AE004969).
Ratio of mRNA transcript levels in induced relative to uninduced JKD5062.
Expression ratio for genes that were up-regulated 1.5-fold with a P value of <0.001.
Overexpression of a plasmid-borne copy of this gene is driven by an IPTG-inducible Ptrc promoter. The ratio is a reflection of the engineered levels of transcription of this gene in induced relative to uninduced JKD5062.
We also observed an up-regulation of the msrAB gene (NGO2059), and this was confirmed by real-time RT-PCR (Table 3), suggesting that transcription of this gene is also dependent on Ecf. The protein now termed MsrAB (previously called PilB) appears to be involved in survival in the presence of oxidative damage (22). In addition, it has been recently reported that the expression of msrAB in gonococci is induced by exposure to hydrogen peroxide (26). The MsrAB protein is unusual in that it contains three domains with separate enzymatic activities that are often found in different proteins in other bacterial species (6). The carboxy-terminal domain possesses MsrB methionine sulfoxide reductase activity specific for the R isomer of methionine sulfoxide, and a central domain has the complementary MsrA activity with specificity for the S isomer (16). The amino-terminal domain contains a signal sequence and possesses a disulfide reductase activity that can recycle methionine sulfoxide reductases (30). This activity allows recycling of MsrA and MsrB activity in the periplasm, a task usually performed by thioredoxin in other species where methionine sulfoxide reductases are cytoplasmic (6).
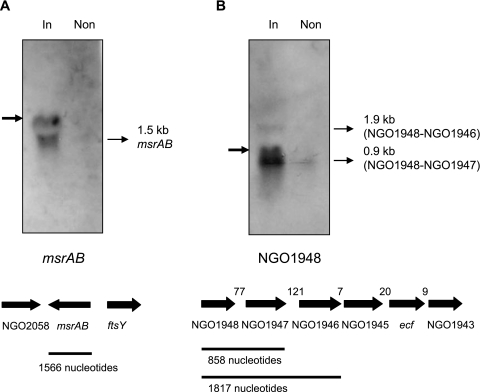
One component of the anti-sigma factors controlling ECF activity is an inner membrane protein with at least one transmembrane domain (18). NGO1948 encodes a protein with a predicted transmembrane domain. However, as is the case with the sigma E proteins from E. coli and Streptomyces coelicolor, multiple genes can have a role in the anti-sigma function (3, 29). Taking into account the observation that Ecf appears to be relatively inactive in strain FA1090 during the exponential phase of growth, the possibility exists that NGO1946 to NGO1948 encode proteins that function to inhibit Ecf activity. In addition, the genomic arrangement of NGO1944 (ecf), NGO1945, NGO1946, NGO1947, and NGO1948 suggests the possibility of cotranscription of at least some of these genes. Genes encoding several ECF sigma factors have been shown to be cotranscribed with a gene(s) encoding a specific anti-sigma factor(s) (3, 9, 21, 25). Northern hybridization was performed with total RNA isolated from strain JKD5062, with or without induction of ecf. A strong signal was observed from RNA prepared after induction of ecf with probes derived from both msrAB and NGO1948, suggesting the presence of Ecf-controlled promoters upstream of these genes (Fig. 1). The probe specific for NGO1948 hybridized to a major 0.9-kb band and a minor 1.9-kb mRNA transcript. The presence of the latter suggests that at least NGO1946, NGO1947, and NGO1948 can be cotranscribed. Larger transcripts are unlikely to have been detected in this experiment. The major 0.9-kb transcript corresponds in nucleotide length to NGO1948 and NGO1947, which suggests the presence of a transcriptional attenuator at the end of NGO1947. An inverted repeat sequence can be found in this intergenic region (data not shown). The minimal spacing between the remaining genes in the cluster (Fig. 1) suggests that they may be cotranscribed. An inverted repeat sequence that may act as a terminator is found downstream of NGO1943. With this strain, we were unable to look directly for cotranscription of the other genes with ecf, as this gene is transcribed from the multicopy vector in strain JKD5062. As expected, given the orientation of the genes neighboring msrAB (Fig. 1), this probe hybridized to a band of the correct size for a single gene transcript.
FIG. 1.
Detection of msrAB and NGO1948 transcripts by Northern hybridization. Total RNA was isolated from N. gonorrhoeae strain JKD5062, both with (In) and without (Non) overexpression of ecf, and separated on a 1.2% denaturing agarose gel. The separated RNA species were probed with an msrAB-specific (A) or an NGO1948-specific (B) probe. The msrAB-specific probe detected a 1.5-kb transcript (A, thin arrow). The NGO1948-specific probe detected two transcripts (B, thin arrows) of 0.9 kb and 1.9 kb. Nonspecific bands (bold arrows) were artifacts comigrating with rRNA. Below the gels are diagrammatic representations of the putative genes in the region, with approximate sizes of transcripts (B), in nucleotides.
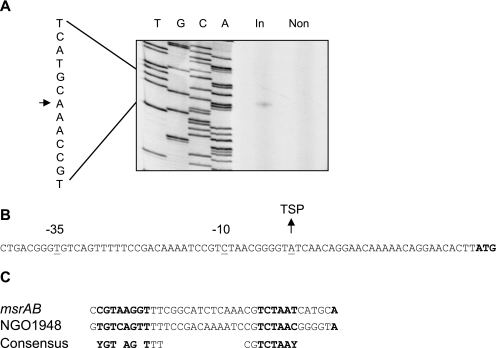
Each ECF sigma factor, even within a bacterial species, recognizes specific promoter sequences. The promoters upstream of msrAB and NGO1948 were mapped using primer extension analysis and 5′ RACE. The transcription start point (TSP) of msrAB was mapped to an A residue located 25 nucleotides upstream of the putative translational start site (Fig. 2A), and this was confirmed using 5′ RACE (data not shown). This TSP was observed only when ecf was overexpressed (Fig. 2A). We were unable to identify a TSP for NGO1948 using primer extensions, but 5′ RACE revealed a TSP at an A residue (Fig. 2B) that again was observed in RNA samples only when ecf was overexpressed (data not shown). Examination of regions upstream of the NGO1948 and msrAB TSP revealed a consensus sequence apparently recognized by Ecf (Fig. 2C). This included a region around the −35 box where five of the eight bases were conserved, but this sequence did not contain the common AAC motif found in this region of most ECF-regulated promoters (9). In addition, a well-conserved AT-rich region was identified 16 bases downstream around the −10 region. Using this consensus sequence, we searched the N. gonorrhoeae strain FA1090 genome sequence using FUZZNUC (http://psychro.bioinformatics.unsw.edu.au) but did not identify any additional genes that might belong to the Ecf regulon by this method.
FIG. 2.
Identification of transcription start points of Ecf-regulated genes. (A) Primer extension mapping of the msrAB TSP. The 5′ terminus of the msrAB transcript was determined using RNA prepared from cultures induced for ecf overexpression (In) and noninduced cultures (Non). The lanes marked T, G, C, and A are the sequencing ladder that was generated with the primer used for primer extension and pJKD2623 (Table 1), which contains the putative promoter. In the sequence on the left, the arrow indicates the transcription start point. (B) 5′ RACE mapping of the NGO1948 TSP. The sequence upstream of NGO1948 is shown; the start codon for the putative gene is in bold. (C) Alignment of the promoter regions of NGO1948 and msrAB with the putative ecf consensus sequence shown below. The −10 and −35 regions are in bold.
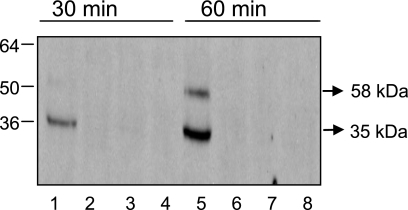
We investigated expression of the gonococcal MsrAB protein to ascertain whether the observed increase in transcription of msrAB translated into increased protein levels. Western blot analysis using an anti-MsrAB polyclonal antibody was performed on whole-cell extracts prepared from strains JKD5062 and JKD5064 (Table 1), which carries an empty vector, grown to mid-exponential phase and harvested with and without induction for 30 min and 60 min. The gonococcal msrAB gene (NGO2059) appears to be responsible for the production of two proteins. In addition to the full-length, three-domain, secreted protein, an internal initiation codon and ribosome binding site allow the translation of a truncated, two-domain cytoplasmic protein with MsrA and MsrB activity (6, 22). Only the secreted form of MsrAB appears to be involved in resistance to oxidative damage (22). JKD5062 induced for 60 min produced a protein of approximately the same size as the full-length MsrAB, suggesting that the transcriptional increase results in elevated levels of MsrAB protein (Fig. 3). However, the majority of the protein detected at both time points appeared to correspond to a truncated form of MsrAB, similar in size to that observed by others (22) using the same antiserum. The antiserum did not detect any protein from JKD5064, reinforcing the finding of dependence on Ecf for expression of msrAB.
FIG. 3.
Western blot of cell extracts from N. gonorrhoeae using a polyclonal antiserum to detect MsrAB. Marker protein sizes are in kDa. Lanes 1, 3, 5, and 7 contain extracts from ecf overexpression strain JKD5062, while lanes 2, 4, 6, and 8 contain extracts from control strain JKD5064, carrying an empty vector. Lanes 1, 2, 5, and 6 contain extracts from cultures that were induced with IPTG, while lanes 3, 4, 7, and 8 contain extracts from noninduced cultures. Lanes 1 to 4 contain extracts that were sampled after 30 min, while lanes 5 to 8 contain extracts that were sampled after 60 min.
In summary, this work has identified a regulon for the only ECF sigma factor of N. gonorrhoeae and a potential consensus sequence for promoters recognized by this sigma factor. This regulon could confer increased protection against oxidative damage. This suggestion is strengthened by the observation that three genes in the ECF regulon (NGO1947, NGO1948 and msrAB) were up-regulated during exposure of gonococci to hydrogen peroxide (26). In addition, this is the first work to describe a regulatory network controlling the expression of msr genes in bacteria.
Acknowledgments
We thank Hank Seifert for the generous gift of the polyclonal antiserum against MsrAB and David R. Powell of the Victorian Bioinformatics Consortium for assistance with analysis of the microarray data. We also thank Dieter Bulach for performing the consensus sequence searches using the N. gonorrhoeae strain FA1090 genome sequence.
This work was supported by an NHMRC Program Grant to the Australian Bacterial Pathogenesis Program. L.A.S.S. was supported by a Wellcome Trust project grant awarded to N.J.S. I.C.G. was the recipient of a Monash University Postgraduate Scholarship. We have made use of data from the Gonococcal Genome Sequencing Project, which was supported by USPHS/NIH grant AI-38399.
REFERENCES
- 1.Bashyam, M. D., and S. E. Hasnain. 2004. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect. Genet. Evol. 4:301-308. [DOI] [PubMed] [Google Scholar]
- 2.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2002. Genomic scale analysis of Pasteurella multocida gene expression during growth within the natural chicken host. Infect. Immun. 70:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Las Penas, A., L. Connolly, and C. A. Gross. 1997. The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol. Microbiol. 24:373-385. [DOI] [PubMed] [Google Scholar]
- 4.Dempsey, J. A., W. Litaker, A. Madhure, T. L. Snodgrass, and J. G. Cannon. 1991. Physical map of the chromosome of Neisseria gonorrhoeae FA1090 with locations of genetic markers, including opa and pil genes. J. Bacteriol. 173:5476-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudoit, S., and Y. H. Yang. 2003. Bioconductor R packages for exploratory analysis and normalization of cDNA microarray data, p. 45-65. In G. Parmigiani, E. S. Garrett, R. A. Irizarry, and S. L. Zeger (ed.), The analysis of gene expression data: methods and software. Springer Verlag, New York, N.Y.
- 6.Ezraty, B., L. Aussel, and F. Barras. 2005. Methionine sulfoxide reductases in prokaryotes. Biochim. Biophys. Acta 1703:221-229. [DOI] [PubMed] [Google Scholar]
- 7.Fyfe, J. A., C. S. Carrick, and J. K. Davies. 1995. The pilE gene of Neisseria gonorrhoeae MS11 is transcribed from a sigma 70 promoter during growth in vitro. J. Bacteriol. 177:3781-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilton, H. L., N. M. Dominguez, K. J. Schwartz, K. T. Hackett, and J. P. Dillard. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704-1721. [DOI] [PubMed] [Google Scholar]
- 9.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 10.Kupsch, E. M., D. Aubel, C. P. Gibbs, A. F. Kahrs, T. Rudel, and T. F. Meyer. 1996. Construction of Hermes shuttle vectors: a versatile system useful for genetic complementation of transformable and non-transformable Neisseria mutants. Mol. Gen. Genet. 250:558-569. [DOI] [PubMed] [Google Scholar]
- 11.Laskos, L., J. P. Dillard, H. S. Seifert, J. A. Fyfe, and J. K. Davies. 1998. The pathogenic neisseriae contain an inactive rpoN gene and do not utilize the pilE sigma54 promoter. Gene 208:95-102. [DOI] [PubMed] [Google Scholar]
- 12.Laskos, L., C. S. Ryan, J. A. Fyfe, and J. K. Davies. 2004. The RpoH-mediated stress response in Neisseria gonorrhoeae is regulated at the level of activity. J. Bacteriol. 186:8443-8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller, F. H., T. M. Bandeiras, T. Urich, M. Teixeira, C. M. Gomes, and A. Kletzin. 2004. Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 53:1147-1160. [DOI] [PubMed] [Google Scholar]
- 15.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-36. [DOI] [PubMed] [Google Scholar]
- 16.Olry, A., S. Boschi-Muller, M. Marraud, S. Sanglier-Cianferani, A. Van Dorsselear, and G. Branlant. 2002. Characterization of the methionine sulfoxide reductase activities of PILB, a probable virulence factor from Neisseria meningitidis. J. Biol. Chem. 277:12016-12022. [DOI] [PubMed] [Google Scholar]
- 17.Purschke, W. G., C. L. Schmidt, A. Petersen, and G. Schafer. 1997. The terminal quinol oxidase of the hyperthermophilic archaeon Acidianus ambivalens exhibits a novel subunit structure and gene organization. J. Bacteriol. 179:1344-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 19.Saal, L. H., C. Troein, J. Vallon-Christersson, S. Gruvberger, A. Borg, and C. Peterson. 2002. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol. 3:SOFTWARE0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning. a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 21.Schurr, M. J., H. Yu, J. M. Martinez-Salazar, J. C. Boucher, and V. Deretic. 1996. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 178:4997-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skaar, E. P., D. M. Tobiason, J. Quick, R. C. Judd, H. Weissbach, F. Etienne, N. Brot, and H. S. Seifert. 2002. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc. Natl. Acad. Sci. USA 99:10108-10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:article 3. [Online.] http://www.bepress.com/sagmb/vol3/iss1/art3/. [DOI] [PubMed]
- 24.Smyth, G. K., J. Michaud, and H. S. Scott. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067-2075. [DOI] [PubMed] [Google Scholar]
- 25.Song, T., S. L. Dove, K. H. Lee, and R. N. Husson. 2003. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol. Microbiol. 50:949-959. [DOI] [PubMed] [Google Scholar]
- 26.Stohl, E. A., A. K. Criss, and H. S. Seifert. 2005. The transcriptome response of Neisseria gonorrhoeae to hydrogen peroxide reveals genes with previously uncharacterized roles in oxidative damage protection. Mol. Microbiol. 58:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tillett, D., B. P. Burns, and B. A. Neilan. 2000. Optimized rapid amplification of cDNA ends (RACE) for mapping bacterial mRNA transcripts. BioTechniques 28:448-456. [DOI] [PubMed] [Google Scholar]
- 28.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 29.Viollier, P. H., G. H. Kelemen, G. E. Dale, K. T. Nguyen, M. J. Buttner, and C. J. Thompson. 2003. Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol. Microbiol. 47:699-714. [DOI] [PubMed] [Google Scholar]
- 30.Wu, J., F. Neiers, S. Boschi-Muller, and G. Branlant. 2005. The N-terminal domain of PILB from Neisseria meningitidis is a disulfide reductase that can recycle methionine sulfoxide reductases. J. Biol. Chem. 280:12344-12350. [DOI] [PubMed] [Google Scholar]
- 31.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]