Abstract
Ralstonia solanacearum, a soilborne plant pathogen of considerable economic importance, invades host plant roots from the soil. Qualitative and quantitative chemotaxis assays revealed that this bacterium is specifically attracted to diverse amino acids and organic acids, and especially to root exudates from the host plant tomato. Exudates from rice, a nonhost plant, were less attractive. Eight different strains from this heterogeneous species complex varied significantly in their attraction to a panel of carbohydrate stimuli, raising the possibility that chemotactic responses may be differentially selected traits that confer adaptation to various hosts or ecological conditions. Previous studies found that an aflagellate mutant lacking swimming motility is significantly reduced in virulence, but the role of directed motility mediated by the chemotaxis system was not known. Two site-directed R. solanacearum mutants lacking either CheA or CheW, which are core chemotaxis signal transduction proteins, were completely nonchemotactic but retained normal swimming motility. In biologically realistic soil soak virulence assays on tomato plants, both nonchemotactic mutants had significantly reduced virulence indistinguishable from that of a nonmotile mutant, demonstrating that directed motility, not simply random motion, is required for full virulence. In contrast, nontactic strains were as virulent as the wild-type strain was when bacteria were introduced directly into the plant stem through a cut petiole, indicating that taxis makes its contribution to virulence in the early stages of host invasion and colonization. When inoculated individually by soaking the soil, both nontactic mutants reached the same population sizes as the wild type did in the stems of tomato plants just beginning to wilt. However, when tomato plants were coinoculated with a 1:1 mixture of a nontactic mutant and its wild-type parent, the wild-type strain outcompeted both nontactic mutants by 100-fold. Together, these results indicate that chemotaxis is an important trait for virulence and pathogenic fitness in this plant pathogen.
Ralstonia solanacearum, a soilborne gram-negative bacterium, causes bacterial wilt disease in many important crop plants, including potato, tobacco, tomato, banana, and peanut plants. The pathogen causes severe losses worldwide due to its wide geographic distribution and unusually broad host range, which spans more than 50 plant families. R. solanacearum is metabolically versatile, surviving and thriving in such diverse habitats as water, soil, and latently infected plants (23). The bacterium normally invades plant roots from the soil through wounds or natural openings, colonizes the intercellular space of the root cortex and vascular parenchyma, and eventually enters the xylem vessel and spreads up into the stem and leaves, where the pathogen cell density commonly surpasses 109 CFU/g of host tissue (33, 55). Affected plants suffer chlorosis, stunting, wilting, and usually die rapidly. Many factors contribute to bacterial wilt virulence, including extracellular polysaccharide, several plant cell wall-degrading enzymes, a set of type III-secreted effector proteins, and a type II secretion system (5, 15, 25, 29, 43).
Many bacteria use a complex behavior called taxis to sense specific chemicals or environmental conditions and move toward attractants and away from repellants (1, 7). Bacterial taxis is directly involved in interactions with both animal and plant hosts (16, 17, 21, 38, 53). Taxis, especially chemotaxis, together with its mechanism of signal transduction and response regulation, has been well studied in Escherichia coli (32, 48). Briefly, cell membrane-associated receptors detect environmental stimuli and respond by changing their conformation. This change triggers autophosphorylation of the cytoplasmic histidine autokinase CheA, which forms a complex with the receptor through a coupling protein called CheW. CheA then transfers its phosphate group to CheY, a diffusible cytoplasmic response regulator. Phosphoryl-CheY interacts with the flagellar motor to switch its direction of rotation, thus altering the bacterial tumbling rate; the net effect of these rate changes is movement toward favorable conditions and away from unfavorable ones. Mutation of CheA, CheY, or CheW results in a completely nontactic phenotype in E. coli (8, 12, 39).
Most soilborne plant-associated bacteria, including R. solanacearum, have swimming motility (54). Swimming motility makes an important quantitative contribution to bacterial wilt virulence in the early stages of host invasion and colonization (51). Nonmotile flagellin (fliC) mutants of R. solanacearum were significantly reduced in virulence on soil-inoculated tomato plants (51). However, because nonmotile bacteria are also perforce nontactic, this experiment could not distinguish between the contribution of random motility to virulence and that of directed movement, or taxis. Furthermore, taxis behavior is likely to play a role in the fitness of R. solanacearum during those parts of its life cycle when it is living in soil or water rather than inside a plant host. Very little is known about any aspect of this bacterium's life outside plants.
In this study, we describe the chemotaxis behavior of R. solanacearum strain K60, which was attracted to various chemicals, to plant root exudates, and to plant roots themselves. Specific tactic responses varied among a set of R. solanacearum strains from different hosts and geographic regions. Motile but nontactic mutants lacking either CheA or CheW had invasive virulence as low as a completely nonmotile fliC mutant. In addition, nontactic mutants competed poorly with the wild-type parent in a soil coinoculation tomato colonization assay. These results suggest that R. solanacearum depends on taxis to locate and infect plant hosts in its natural niches.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Strain K60GFP was constructed by using natural transformation to move the constitutively expressed Tn5::gfp insertion from strain AW1-gfp38 into the K60 chromosome (28). Spontaneous rifampin-resistant mutant K60rif was selected on CPG medium (24) supplemented with rifampin. This strain was then tested to verify its abilities to grow normally in rich and minimal media and in tobacco leaf tissue and to cause disease on tomato, potato, and geranium plants as well as its wild-type parent.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F− endA1 relA φ80 lacZΔM15 hsdR17 supE44 thi-1 recA1 gyrA96 | Invitrogen |
| Ralstonia solanacearum | ||
| K60 | Wild-type race 1, biovar 1, isolated from tomato | 27 |
| K60rif | K60 spontaneous Rifr mutant, motile and tactic, fully virulent | This study |
| K60GFP | K60 constitutively expressing GFP, motile and tactic, fully virulent | This study |
| K701 | K60 fliC::aacC1 Gmr, aflagellate and nonmotile | 51 |
| K750 | K60 cheA::aacC1 Gmr, motile but nontactic | This study |
| K760 | K60 cheW::aphA-3 Kmr, motile but nontactic | This study |
| K760GFP | K760 cheW::aphA-3 Kmr, constitutively expressing GFP, motile but nontactic, virulent like K760 | This study |
| GMI1000, UW141, UW151, UW316, UW373, UW386, UW519, and UW551 | Diverse R. solanacearum strains (see details in Table 3) | 9, 13, 49; Allen lab collections |
| Plasmids | ||
| pSTBlue-1 | Cloning vector, Apr Kmr | EMD Biosciences |
| pBluescript II SK(−) | Cloning vector, Apr | Stratagene |
| pLAFR3 | Broad-host-range cosmid vector, Tcr | 47 |
| pBSaphA | Kmr Apr | 34 |
| pUCGM | Gmr Apr | 45 |
| pSJG | pSTBlue-1Δ1147-2186::aacC1, Gmr | This study |
| 2-89 | pLAFR3 cosmid containing K60 motA-motB-cheY1-cheA-cheW, Tcr | 52 |
| pSJYcheA1.0 | PCR-amplified 1.0-kb internal fragment of K60 cheA in pSTBlue-1, Apr Kmr | This study |
| pSJYcheA1.0::Gm | 0.8-kb Gmr cassette in BglI site of 1.0-kb cheA fragment in pSJYcheA1.0, Apr Kmr Gmr | This study |
| pSJYcheYAW12.0 | 12-kb EcoRI fragment containing K60 motA, motB, cheY1, cheA, and cheW insert from 2-89 in pSTBlue-1, Kmr Apr | This study |
| pBJYcheYAW12.0 | 12-kb BamHI-HindIII fragment from pSJYcheYAW12.0 in pBluescript II SK(−) | This study |
| pBJYcheW::Km | 0.85-kb Kmr cassette in EcoRV site of cheW in pBJYcheYAW12.0, Apr Kmr | This study |
| pSJGcheW::Km | 13-kb EcoRI fragment from pBJYcheW::Km in pSJG, Gmr Kmr | This study |
Apr, ampicillin resistant; Gmr, gentamicin resistant; Kmr, kanamycin resistant; Rifr, rifampin resistant; Tcr, tetracycline resistant.
Escherichia coli was grown in Luria-Bertani medium (36) at 37°C. All R. solanacearum strains were grown either in CPG rich broth or on TZC solid medium (27) at 28°C. To test growth on sole carbon sources, R. solanacearum was grown in quarter-strength M63 medium, also known as Boucher's minimal medium (BMM) (9), buffered with 20 mM morpholineethanesulfonic acid (MES), pH 6.5, and supplemented with one of the carbon sources listed in Table 2. If necessary, antibiotics were used at the following concentrations: ampicillin, 100 mg/liter; kanamycin, 25 mg/liter; tetracycline, 15 mg/liter; gentamicin, 12.5 mg/liter; and rifampin, 25 mg/liter. Growth rates of wild-type and mutant strains were compared in CPG medium, in BMM with 0.2% glucose, and in tobacco leaf tissue (Nicotiana tabacum cv. Bottom Special) as previously described (51). Unless otherwise noted, medium components were purchased from Difco Laboratories (Detroit, MI). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
TABLE 2.
Chemotactic and metabolic response of R. solanacearum strain K60 to various chemicals
| Chemical | Utilization as the sole carbon sourcea | Halo by the semisolid motility plate chemotaxis assayb | Agarose plug assay
|
|
|---|---|---|---|---|
| Relative chemotactic responsec | Chemotactic response ratingd | |||
| d-Sugars | ||||
| Fructose | + | − | 0.92 | − |
| Galactose | + | + | 0.91 | − |
| Glucose | + | − | 0.71 | − |
| Maltose | − | NA | 0.91 | − |
| Sucrose | + | − | 0.93 | − |
| Xylose | + | − | 0.96 | − |
| Organic acids | ||||
| Citric acid | + | + | 1.75 | + |
| Malic acid | + | + | 1.68 | + |
| Succinic acid | + | + | 1.01 | − |
| Glucuronic acid | + | − | 0.55 | − |
| Galacturonic acid | + | + | 0.69 | − |
| l-Amino acids | ||||
| Alanine | ++ | + | 2.21 | + |
| Arginine | + | − | 1.18 | − |
| Aspartic acid | ++ | + | 3.83 | ++ |
| Asparagine | ++ | + | 2.62 | ++ |
| Cysteine | − | NA | 1.01 | − |
| Glutamic acid | ++ | + | 2.76 | ++ |
| Glutamine | ++ | + | 3.01 | ++ |
| Glycine | − | NA | 1.84 | + |
| Histidine | ++ | − | 1.27 | − |
| Isoleucine | − | NA | 2.78 | ++ |
| Leucine | − | NA | 2.76 | + |
| Lysine | − | NA | 1.75 | + |
| Methionine | − | NA | 2.02 | + |
| Phenylalanine | − | NA | 2.06 | + |
| Proline | ++ | + | 3.22 | ++ |
| Serine | + | − | 2.00 | + |
| Threonine | + | − | 1.23 | − |
| Tryptophan | + | − | 0.75 | − |
| Tyrosine | ++ | + | 1.62 | + |
| Valine | + | − | 1.96 | + |
| Other | ||||
| Mannitol | − | NA | 0.70 | − |
| Glycerol | + | − | 0.37 | − |
| Casamino Acids | + | + | 4.58 | ++ |
| Peptone | + | + | 5.41 | ++ |
| Yeast extract | + | + | 1.96 | ++ |
Utilization of the chemicals as the sole carbon source was determined by culturing strain K60 starting at an OD600 of 0.01 in defined BMM broth with the chemicals at the following concentrations: 10 mM for d-sugars, 10 mM for organic acids, 4 mM for l-amino acids, and 0.2% (wt/vol) for the other chemicals. After 24 h, the turbidity of culture was measured by OD600, which indicated the multiplication of bacteria defined as follows: −, no growth (OD600, <0.05); +, moderate growth (OD600 between 0.05 and 0.3); ++, vigorous growth (OD600, >0.3).
Determined by the size of the swimming halo on minimal medium semisolid motility plate plus the tested compounds at the following concentrations: 10 mM for d-sugars, 10 mM for organic acids, 10 mM for l-amino acids, and 0.1% (wt/vol) for the other chemicals. Symbols: −, halo size is equal to that of nonmotile mutant K701; +, halo size at least twice as big as that of nonmotile mutant K701. NA, not assayed, because the chemical was not utilized by K60.
RCR is calculated as the number of bacterial cells per 0.01 mm2 adjacent to the agarose plug containing the tested compound divided by the bacterial number per 0.01 mm2 adjacent to the buffer-only agarose plug (negative control). The compounds were tested at the following concentrations: 1 mM for d-sugars, organic acids, l-amino acids, and 0.1% (wt/vol) for the other chemicals.
The chemotactic response was rated and shown as follows: −, RCR of <1.60; +, RCR between 1.60 and 2.60; ++, RCR of >2.60.
Chemotaxis assays.
Two methods were used to assay the chemotaxis of R. solanacearum. For chemicals that wild-type R. solanacearum can metabolize as the sole carbon source, chemotaxis was measured qualitatively in a semisolid medium. Briefly, overnight cultures grown in CPG medium were collected by centrifugation at 6,000 rpm for 10 min, washed twice in sterile deionized water, and adjusted to 1.0 × 109 CFU/ml (equal to an optical density at 600 nm [OD600] of 1.0); 2 μl of bacterial suspension was placed onto the center of a semisolid plate solidified with 0.3% Noble agar and containing either BMM plus one of the compounds listed in Table 2 or 1% tryptone. The plates were incubated at 28°C for 3 to 5 days. Chemotactic response was considered positive if the size of the halo produced by motile cells around the inoculation site was at least twice as large as that of nonmotile mutant strain K701. All assays were repeated at least three times.
A slightly modified agarose plug method (56) was also used to quantify R. solanacearum chemotaxis. R. solanacearum cells were grown in half-strength CPG medium overnight to an OD600 of 0.3 to 0.7, the cell density corresponding to the maximum proportion of motile cells; the cells were collected by centrifugation, washed twice in sterile chemotaxis buffer (10 mM potassium phosphate, pH 7.0, 0.1 mM EDTA, 1 mM MgSO4), and resuspended to an OD600 of 0.1 for future testing. An 11-μl drop of agarose solution, containing 2% NuSieve GTG agarose (FMC Bioproducts, Rockland, ME) in chemotaxis buffer and one of the compounds listed in Table 2, was placed on the center of an acetone-cleaned microscope slide, framed with two plastic strips, and covered with a coverslip to create a chemotaxis chamber. After the agarose plug solidified, 120 μl of bacterial suspension was added to the chamber surrounding the agarose plug. Following 30 min of incubation at room temperature, the distribution of cells immediately adjacent to the agarose plug was observed under an Olympus BX60 phase-contrast microscope (Olympus America Inc., Melville, NY) and recorded by a charge-coupled device camera. The resulting images were analyzed and quantified with Image-Pro Plus software (Media Cybernetics, Silver Spring, MD) to determine the number of bacteria surrounding the agarose plug; this number was normalized to the number of bacterial cells per area of 0.01 mm2 (BCPA). For each trial, at least two agarose plugs were used per compound; an agarose plug that contained only chemotaxis buffer served as a negative control, and 0.1% yeast extract in chemotaxis buffer was a positive control. The relative chemotactic response (RCR) was calculated as the BCPA of the plug with the tested compound divided by the BCPA of the negative control. An RCR of >1.60 indicated a positive chemotactic response. All agarose plug assays were repeated at least three times.
Collection of plant root exudates.
Tomato (Lycopersicon esculentum Mill. cv. Brandywine) and rice (Oryza sativa L. cv. Drew) seeds were surface sterilized by soaking the seeds in 20% commercial bleach for 5 min, followed by washing them in sterile water for 5 min with gentle shaking; this was repeated. The seeds were then transferred onto 1% water agar plates, which were incubated at 4°C overnight to synchronize germination, and then held in a 28°C dark incubator for 2 or 3 days. For each extraction, 30 germinated seedlings with 2-cm-long roots were transferred into a sterile 50-ml conical tube containing 5 ml of sterile chemotaxis buffer and incubated at 28°C for 24 h. The resulting root exudates were collected, sterilized by passage through a 0.2-μm filter, and stored at −80°C. To normalize the root exudate concentrations, the total protein concentrations were determined by bicinchoninic acid (BCA) assay (Sigma-Aldrich, St. Louis, MO) following the manufacturer's protocol. To study the effects of different treatments on the ability of tomato root exudates (TRE) to attract R. solanacearum, TRE were either boiled for 15 min or digested at 37°C for 1 h with 10 μg/ml proteinase K. The agarose plug chemotaxis assay was used to quantify the R. solanacearum chemotactic response to root exudates.
Microscopic visualization of R. solanacearum strains on tomato roots.
Seeds of wilt-susceptible tomato cv. Bonny Best were surface sterilized as described above and germinated on 1% water agar plates at 28°C in the dark. After the secondary roots were well developed, whole seedlings were placed on a microscopic slide. A suspension of R. solanacearum K60GFP in chemotaxis buffer (OD600 of 0.01, ∼1 × 107 CFU/ml) was used to flood seedling roots. After 30 min of incubation at room temperature, the slide was examined under an Olympus BX60 epifluorescence microscope (Olympus America Inc., Melville, NY) using a U-MNB mirror unit (excitation filter, 470 to 490 nm; emission filter, 515 nm); the images were captured by a charge-coupled device camera and analyzed with Image-Pro Plus software (Media Cybernetics, Silver Spring, MD). All strains were imaged on roots at least five times; images shown are typical results.
DNA manipulations.
DNA extraction, cloning, Southern blotting, and PCR were performed using standard protocols (6). E. coli and R. solanacearum strains were transformed by electroporation as previously described (3). DNA sequencing was performed at the University of Wisconsin Biotechnology Center (Madison, WI) using automated fluorescence sequencing. The R. solanacearum strain GMI1000 genome database (http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/ralsto/index.html) and the Jellyfish 3.0 software package (LabVelocity, Burlingame, CA) were used to analyze DNA sequence data. Molecular biology reagents came from either Promega (Madison, WI) or New England Biolabs (Beverly, MA). Oligonucleotides were from Sigma-Genosys (The Woodlands, TX).
Construction of R. solanacearum cheA and cheW insertion mutants.
A 1,017-bp internal DNA fragment of the R. solanacearum strain K60 cheA gene was amplified by PCR using primers cheA-L (5′-GTCGATCAGCTCATCAAC) and cheA-R (5′-GTTCTTGACCACCACCTG) and cloned into the pSTBlue-1 vector (EMD Biosciences, San Diego, CA) to create pSJYcheA1.0. The gentamicin resistance cassette (aacC1) from pUCGM was inserted into a blunt-ended unique BglI site in the cheA fragment to create the pSJYcheA::Gm mutagenesis construct. To mutate cheW, the 850-bp kanamycin resistance gene cassette (aphA-3) from pBSaphA was inserted into a unique EcoRV site in cheW in plasmid pBJYcheYAW12.0 to create pBJYcheW::Km; the insert was transferred to pSJG to create pSJGcheW::Km. Both the cheA::Gm and cheW::Km constructs were introduced into the chromosome of wild-type R. solanacearum strain K60 by homologous double recombination as previously described (3); this generated cheA mutant strain K750 and cheW mutant strain K760. Correct disruption of cheA in K750 and cheW in K760 were confirmed by Southern blot analysis (data not shown). The swimming motility of mutant strains was checked microscopically.
Complementation of cheA and cheW mutants.
For complementation studies, a 12-kb EcoRI fragment containing the intact cheA and cheW genes was subcloned into pSTBlue-1 from R. solanacearum strain K60 genomic library cosmid 2-89, which contains a large cluster of motility and taxis genes (50, 52), to create pSJYcheYAW12.0. The 12-kb insert of pSJYcheYAW12.0 was transferred into pBluescript II SK(−) with BamHI-HindIII to create pBJYcheYAW12.0. To identify specific DNA fragments that could restore taxis to R. solanacearum chemotaxis mutants, a series of subclones were constructed in the broad-host-range vector pLAFR3 (Table 1 and Fig. 1). All these constructs were electroporated into cheA mutant strain K750, except for pLJYcheW2.0a, which was electroporated into cheW mutant strain K760. Transformants were selected with appropriate antibiotic resistance on TZC plates, and their chemotaxis ability was tested on 1% tryptone semisolid plates containing 6 mg/liter tetracycline to maintain the plasmid.
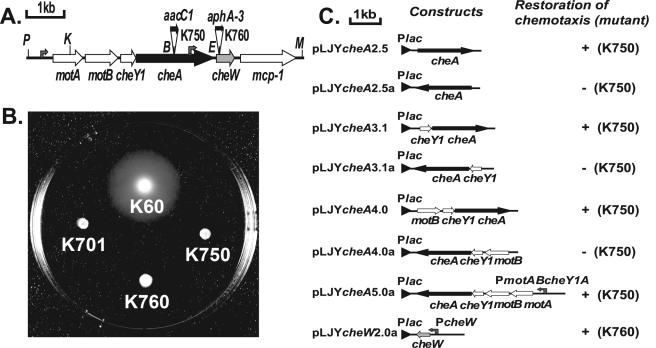
FIG. 1.
Mutagenesis of cheA and cheW in R. solanacearum strain K60. (A) A chemotaxis gene cluster region in R. solanacearum strain K60. Two nontactic mutants were created by inserting a gentamicin resistance cassette (aacC1) into cheA (K750) or a kanamycin resistance cassette (aphA-3) into cheW (K760). P, PmlI; K, KpnI; B, BglI; E, EcoRV; M, MluI. (B) Taxis behavior of R. solanacearum strains on semisolid motility plates. Shown are typical bacterial taxis halos formed after 2 days of incubation at 28°C in a 1.0% tryptone semisolid agar plate inoculated with 2 × 106 bacteria. The halos from four strains, K60 (wild type), K750 (cheA mutant), K760 (cheW mutant), and K701 (nonmotile fliC mutant), are shown. When observed under the microscope, strains K750 and K760 were motile but nonchemotactic. (C) Diagram of complementation plasmid constructs. Shown are various K60 DNA fragments inserted in pLAFR3. cheA2.5, cheA3.1, and cheW2.0 fragments were PCR amplified from cosmid 2-89 using pfx DNA polymerase and primer sets cheA-OL (5′-CGATCCTGATGCTGACCAC)/cheA-OR (5′-ATATCGATGCCGTATTCCTC), cheYA-OL(5′-GCAGAACGGCTACAGCAACT)/cheA-OR, and cheA-L (5′-GTCGATCAGCTCATCAAC)/cheW-OR (5′-CCATCATCAGCGCCAGAC), respectively. cheA4.0 and cheA5.0 fragments were subcloned from pSJYcheYAW12.0 as KpnI-EcoRV and PmlI-EcoRV fragments, respectively. Plac is the lac promoter from pLAFR3, PcheW is the likely cheW promoter, and PmotABcheYA is the likely promoter for the motAB-cheY1A operon. Complementation was measured as the restored ability to form wild-type halos on 1% tryptone semisolid plates (+).
Virulence assays.
The virulence of R. solanacearum chemotaxis mutants on tomatoes was measured using both a direct petiole inoculation and a naturalistic soil soak inoculation method as previously described (50). Briefly, for the soil soak assay, unwounded plants of the susceptible tomato cultivar Bonny Best were inoculated 16 days after planting by pouring a bacterial suspension onto the soil to a final density of around 3 × 107 CFU/g of soil. The plants were maintained at 28°C and rated daily for symptoms on a disease index scale from 0 to 4 by an individual who did not know which plants had been treated by the two methods. For the petiole inoculation, about 2,000 bacterial cells were applied to the cut petiole of the first true leaf of 22-day-old tomato plants, and plants were rated for symptoms as for the soil soak assay. All virulence assays included at least 16 plants per treatment, and each experiment was repeated at least three times.
Population studies of R. solanacearum strains in tomato stems.
To study competitive fitness, R. solanacearum chemotaxis mutants were inoculated by the cut petiole or the soil soak method either alone (single inoculation test) or in a 1:1 mixture with the wild-type strain or the nonmotile mutant strain K701 (competition assay). After tomato plants showed wilt symptoms, a 1-cm stem segment from 1 cm above the soil line (soil soak inoculation) or spanning the inoculation site (petiole inoculation) was collected, weighed, and ground in 1 ml of sterile deionized water. The resulting homogenate was dilution plated on TZC plates supplemented with 100 mg/liter cycloheximide and appropriate antibiotics using Autoplate model 3000 (Spiral Biotech, Norwood, MA). Colonies were counted after 2 days of incubation at 28°C. R. solanacearum population densities were normalized to CFU/g of plant tissue. Our theoretical detection limit was 200 CFU/g tissue. Each experiment contained at least 16 plants per treatment; all experiments were repeated three times.
Data analysis.
Virulence assay data were analyzed by repeated-measure analysis of variance (50). The log10(CFU/g tissue + 1) transformation was used for further analysis of tomato stem population size data. The Bonferroni multiple-comparison method was used to analyze population data from single-strain inoculations, and the paired t test was used to analyze colonization data from competition assays. All statistical calculations were done using MINITAB14 (Minitab, State College, PA).
RESULTS
R. solanacearum strain K60 chemotaxes toward diverse compounds.
To identify specific chemicals that trigger the chemotaxis response in R. solanacearum wild-type strain K60, we used both a qualitative semisolid plate method (Fig. 1B) and a quantitative agarose plug method (Fig. 2) to measure this strain's responses to various chemicals, including most amino acids and sugars known to be present in root exudates from K60's host plant, tomato (31, 46). On plates, strain K60 showed a strong chemotactic response to all organic acids tested, except glucuronic acid, but was not attracted to 0.2% glycerol or to any d-sugars tested, apart from galactose. K60 was strongly attracted to the common complex culture medium components Casamino Acids, peptone, and yeast extract. Of the l-amino acids that K60 can use as the sole carbon source, the bacterium was attracted to alanine, aspartic acid, asparagine, glutamic acid, glutamine, proline, and tyrosine but not to arginine, histidine, serine, threonine, tryptophan, or valine (Table 2).
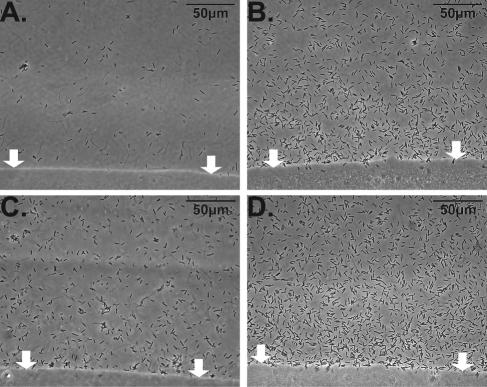
FIG. 2.
An agarose plug method was used to measure chemotaxis of R. solanacearum strain K60. Bacterial cell density around an agarose plug that contained the substance of interest was observed with a 40× phase-contrast objective (Olympus BX60) 30 min after introduction of a bacterial suspension. One plane of focus adjacent to the plug was photographed and analyzed by Image-Pro Plus software (Media Cybernetics, Inc.). R. solanacearum wild-type strain K60 responded differently to (A) chemotaxis buffer (10 mM potassium phosphate buffer, pH 7.0, 0.1 mM EDTA and 1 mM MgSO4) as a negative control; (B) 0.1% yeast extract, a positive control; (C) rice root exudates; and (D) tomato root exudates. The white arrows point to the edge of the agarose plug.
The agarose plug and plate assays gave similar results for most compounds, except that R. solanacearum K60 responded to 1 mM of serine and valine in the plugs, but not on plates, and was attracted to galactose, succinic acid, and galacturonic acid on plates, but not in agarose plugs (Table 2). Curiously, R. solanacearum was attracted to the amino acids glycine, isoleucine, leucine, lysine, methionine, and phenylalanine in agarose plugs, even though it cannot use them as the sole carbon source; however, all of these amino acids except methionine are present in tomato root exudates (46). Conversely, the bacterium was not attracted to maltose, cysteine, or mannitol, which are also not useful carbon sources, although maltose is present in tomato root exudates (31) (Table 2).
Diverse strains in the R. solanacearum species complex have different chemotactic response profiles.
Because R. solanacearum includes thousands of distinct strains with high genetic and phenotypic heterogeneity, it is considered a species complex rather than a single species (18). We tested eight R. solanacearum strains isolated from diverse hosts and geographic origins and coming from all four phylotypes in the species complex to see whether they differed in their chemotactic responses to eight different carbohydrates, each of which could serve as the sole carbon source for these strains. We found considerable diversity in chemotactic response profiles among the eight strains (Table 3). Two strains, UW551 and UW373, were attracted to all the carbohydrates tested. However, the other strains showed positive chemotactic responses to only some of these carbohydrates. Two compounds, citrate and malate, attracted all the strains; however, fructose and sucrose attracted only strains UW551 and UW373 (Table 3).
TABLE 3.
Diverse chemotactic response profiles of eight strains in the R. solanacearum species complex
| Strain | Origin | Host | Phylotype | Biovar | Race | Chemotaxis responsea to:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fructose | Galactose | Glucose | Glycerol | Sucrose | Citrate | Malate | Succinate | ||||||
| K60 | United States | Tomato | II | 1 | 1 | − | + | − | − | − | + | + | + |
| UW551 | Kenya | Geranium | II | 2 | 3 | +/− | + | + | + | + | + | + | + |
| GMI1000 | French Guiana | Tomato | I | 3 | 1 | − | + | + | +/− | − | + | +/− | − |
| UW151 | Australia | Ginger | I | 4 | 1 | − | +/− | + | + | − | + | + | + |
| UW316 | Kenya | Pepper | I | 3 | 1 | − | + | + | + | − | + | + | +/− |
| UW373 | China | Mulberry | I | 5 | 1 | + | + | + | + | + | + | + | + |
| UW386 | Nigeria | Tomato | III | 1 | 1 | − | + | ND | ND | ND | + | + | +/− |
| UW519 | Indonesia | Clove | IV | 1 | 1 | − | + | + | − | − | +/− | + | + |
Chemotactic response was measured as the formation of haloes on minimal medium soft agar plate plus 10 mM tested compound. Symbols: +, strong chemotactic response; +/−, weak chemotactic response; −, no detectable chemotactic response as determined by halo sizes. ND, not determined, because UW386 formed irregular star-like halo shapes on these soft agar plates.
R. solanacearum is more strongly attracted to root exudates from a host plant than to those from a nonhost plant.
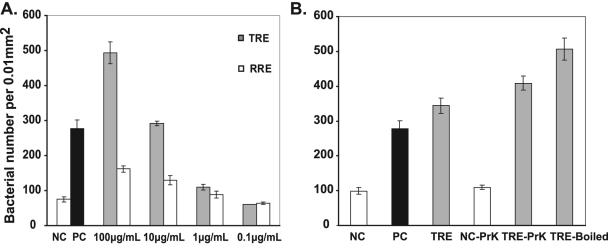
Because R. solanacearum invades host plant roots from the soil, we used the agarose plug assay to determine whether R. solanacearum K60 chemotaxes toward plant root exudates. Strain K60 was attracted to exudates from the roots of both tomato (a host plant) and rice (a nonhost plant) even when the exudates were quite dilute (1-μg/ml protein concentration of tomato root exudates) (Fig. 3A). However, tomato root exudates (TRE) were significantly more attractive than rice root exudates (RRE). The attraction to TRE was three times stronger than the attraction to RRE at exudate protein concentrations of 100 μg/ml, and the attraction was twofold stronger at exudate protein concentrations of 10 μg/ml. This difference disappeared at a concentration of 1 μg/ml (Fig. 3A). The chemoattractant(s) in TRE was heat stable after boiling for 15 min and was not affected by 1 hour of digestion with 10 μg/ml proteinase K at 37°C (Fig. 3B). In fact, these treatments slightly increased the attractiveness of the exudates.
FIG. 3.
R. solanacearum strain K60 was attracted to plant root exudates. Bacterial number per 0.01 mm2 was determined by the agarose plug assay (see the legend to Fig. 2). (A) K60 responded to tomato and rice root exudates. NC, negative control (chemotaxis buffer); PC, positive control (0.1% yeast extract); TRE, tomato root exudates; RRE, rice root exudates. Root exudate concentrations are given as total protein concentrations. (B) Effects of different treatments on the ability of TRE to attract R. solanacearum K60 cells. NC-PrK and TRE-PrK, NC and TRE treated with 10 μg/ml proteinase K at 37°C for 1 h, respectively; TRE-Boiled, TRE boiled for 15 min. Numbers represent the means of three independent assays with two plugs per assay. Error bars represent standard errors of the means.
R. solanacearum strain K60GFP is strongly attracted to tomato seedling roots.
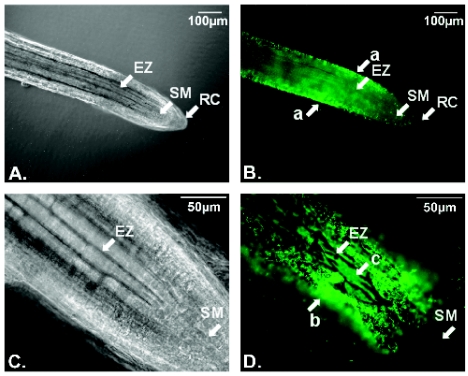
To directly monitor the chemotactic behavior of R. solanacearum cells in the presence of host plant roots, we created strain K60GFP, which constitutively expresses green fluorescent protein (GFP), by moving a Tn5-GFP insertion from R. solanacearum strain AW1-gfp38 (28) into the K60 chromosome. A GFP-expressing nontactic strain was created using the same methods with cheW mutant strain K760. Strains K60GFP and K760GFP had growth rates and virulence traits indistinguishable from those of their parent strains (data not shown). Microscopic observation of seedling roots incubated for 30 min under a coverslip with a bacterial suspension revealed that K60GFP cells were strongly attracted to tomato seedling roots (Fig. 4B and D). Bacterial cells were not evenly distributed on the root surface, avoiding the root tip but forming dense aggregates or thin layers in the elongation zone near the subapical meristem of primary roots (Fig. 4) and at sites of secondary root emergence (data not shown). The motile but nontactic strain K760 (cheW mutant) did not aggregate on seedling roots and was no more densely distributed near the seedlings than anywhere else under the coverslip (data not shown).
FIG. 4.
R. solanacearum strain K60 had strong tactic responses toward tomato seedling roots. Tomato seeds were surface sterilized and germinated on 1.0% water agar plate. After the secondary roots were well developed, the whole seedlings were placed on microscopic slides. Log-phase R. solanacearum K60GFP cells were washed twice with chemotaxis buffer and resuspended in chemotaxis buffer to 1 × 107 CFU/ml. One hundred to 200 μl of bacterial suspension was used to flood the root-containing slide. After 30 min of incubation at room temperature, the roots were examined with epifluorescence microscopy (Olympus BX60). (A) A plant root under regular light (magnification, ×100). Arrows point to the root cap (RC), subapical meristem (SM), and elongation zone (EZ). Magnification, ×100. (B) The same root under UV light using a U-MNB mirror unit (excitation filter, 470 to 490 nm; emission filter, 515 nm). R. solanacearum K60GFP cells were attracted to the tomato root surface in the root elongation zone (a). (C and D) Closer look at the root subapical meristem and elongation zone at a magnification of ×400 under regular and UV light, respectively. R. solanacearum cells formed an aggregate (b) or thin layer (c) on the surface of tomato root, especially in the elongation zone.
R. solanacearum strains with mutations in cheA (K750) and cheW (K760) are no longer chemotactic.
To further explore the role of taxis in the R. solanacearum life cycle, we constructed two site-directed nontactic mutants. R. solanacearum cheA mutant strain K750 and cheW mutant strain K760 were generated by the insertion of the aacC1 (45) and aphA-3 (34) cassettes into cheA and cheW, respectively (Fig. 1A). Neither mutant had a detectable growth defect in rich medium (CPG), minimal medium (BMM plus 0.2% glucose), or in planta in tobacco leaves. This remained true when strains were tested individually or in a 1:1 mixture with the wild-type parent strain (data not shown).
R. solanacearum cheA mutant K750 and cheW mutant K760 strains were completely nonchemotactic. On semisolid 1% tryptone motility medium, strains K750 and K760 formed no detectable halo, indistinguishable from a colony of nonmotile fliC mutant strain K701, with a colony about one-tenth the diameter of the halo formed by wild-type strain K60 (Fig. 1B). Moreover, K750 and K760 no longer chemotaxed toward yeast extract, Casamino Acids, or tomato root exudates in the agarose plug assay (data not shown).
Despite their chemotaxis defect, microscopic study revealed that strains K750 and K760 remained fully motile, swimming in a much smoother format with less frequent changes in swimming speed and direction compared to the wild-type parent K60. At certain cell densities, K750 had a lower proportion of swimming to nonswimming cells than wild-type strain K60 did, but there was no difference in the proportions of swimming cells for strains K760 and K60.
Chemotactic ability was restored to R. solanacearum K750 and K760 by introducing the wild-type cheA or cheW gene in trans.
To identify the complementation units and, by inference, the likely operon structure of the R. solanacearum chemotaxis gene cluster, we used either PCR with a high-fidelity DNA polymerase or subcloning to construct a series of clones containing different parts of the motAB-cheYAW gene cluster in the broad-host-range vector pLAFR3 (Fig. 1C). When under control of the vector lac promoter (Plac), all constructs containing the intact cheA open reading frame (ORF) restored chemotactic ability to cheA mutant K750, which could then form a halo on 1% tryptone semisolid plates that was indistinguishable from that formed by the wild-type strain carrying the empty pLAFR3 vector (data not shown). However, when these fragments were cloned into pLAFR3 in the opposite orientation from Plac, none of them could complement the chemotactic ability of K750 except for pLJYcheA5.0a, which contained the intact upstream region for motA (Fig. 1C). This result suggests that the motAB-cheYA cluster forms a single transcriptional unit. With respect to cheW, pLJYcheW2.0a, which contains only the cheW ORF plus 1,314 bp of upstream sequence, fully restored chemotactic ability to cheW mutant K760 regardless of its orientation in pLAFR3, suggesting that cheW is transcribed separately under its own promoter (Fig. 1C).
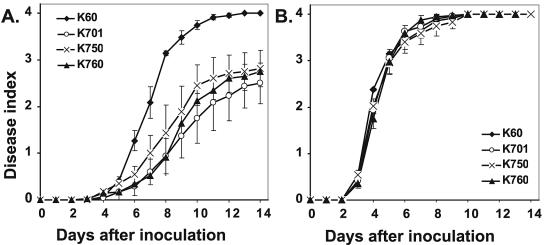
R. solanacearum cheA mutant K750 and cheW mutant K760 had significantly reduced virulence on tomato.
We previously found that flagellum-driven swimming motility makes an important contribution to R. solanacearum virulence early in disease development (51). However, since the nonmotile strain is also necessarily nontactic, that study could not determine whether virulence was reduced because the strain lacked directed movement (taxis) or because it could not move at all. To distinguish between these two possibilities and to determine the specific role of taxis in bacterial wilt disease development, we used two different assays to measure the virulence of R. solanacearum K750 (cheA mutant) and K760 (cheW mutant) on the wilt-susceptible tomato cv. Bonny Best. In the naturalistic soil soak assay, which requires bacteria to locate and invade host roots from the soil, wild-type strain K60 caused a mean disease index of 4.0 (100% wilted or dead) by 14 days postinoculation, whereas chemotaxis mutants K750 (cheA) and K760 (cheW) caused a disease index of only 2.8, significantly lower than that of their wild-type parent (P < 0.01). There was no significant difference in virulence between the two nonchemotactic mutants or between the nonchemotactic mutants and the aflagellate nonmotile mutant K701 (fliC) (Fig. 5A). This result demonstrates that R. solanacearum needs chemotaxis, not just random motion, to effectively locate and colonize host roots. In the petiole inoculation assay, where the normal infection process is bypassed by directly inoculating a few hundred bacteria into the host vascular system, there were no differences among these four strains, all of which killed all plants by 10 days postinoculation (Fig. 5B). This result is in agreement with our previous finding that nonmotile strains are as virulent as the wild type when introduced directly into plant stems (51).
FIG. 5.
Virulence of nonchemotactic R. solanacearum strains on tomato. Wild-type strain K60, K701 (fliC mutant), K750 (cheA mutant), and K760 (cheW mutant) were tested. (A) Soil soak virulence assay. Sixteen-day-old unwounded tomato plants (cv. Bonny Best) were inoculated by pouring bacteria onto the soil to a final concentration about 3 × 107 CFU/g soil. (B) Petiole inoculation virulence assay. Twenty-one-day-old tomato plants were inoculated by introducing about 2,000 cells directly onto the cut petiole of the first true leaf. Plants were rated daily on a disease index scale from 0 to 4. Each point represents the mean disease index for three independent experiments containing 48 plants in total for each treatment. Error bars indicate standard errors of the means.
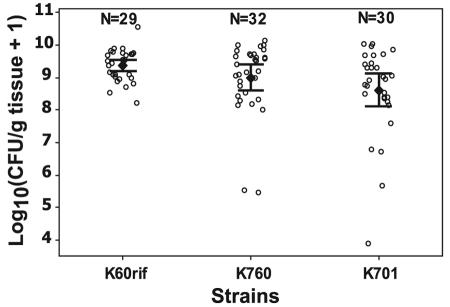
R. solanacearum cheW mutant K760 reached the same population size in tomato stems as wild-type strain K60 and nonmotile fliC mutant K701 did.
To measure pathogen progress through the plant from the soil up into the stem, we quantified the population size of R. solanacearum strains in tomato stems 1 cm above the crown. R. solanacearum K60rif (a spontaneous rifampin-resistant variant of the wild type), cheW mutant K760, and aflagellate fliC mutant K701 were individually inoculated into tomato cv. Bonny Best by soaking the soil as described above. As soon as any wilt disease symptoms appeared (disease index of >0), the population size of R. solanacearum strains in each plant was measured by grinding the stem tissue and dilution plating on the appropriate selective medium. Under these conditions, there was no difference in lower stem populations between strain K760 (1.03 × 109 CFU/g) and strain K60rif (2.09 × 109 CFU/g) (P = 0.52). There was also no significant difference in stem population size between K760 (cheW) and the nonmotile fliC strain K701 (4.27 × 108 CFU/g) (P = 0.47) (Fig. 6). However, the population size of K701 was significantly lower than that of wild-type strain K60 (P = 0.02) (Fig. 6).
FIG. 6.
Population sizes of R. solanacearum strains in tomato stems after wilt symptoms developed. Sixteen-day-old unwounded tomato plants (cv. Bonny Best) were inoculated by pouring a bacterial suspension onto the soil to a final concentration about 3.0 × 107 CFU/g soil. After wilt symptoms developed, 1-cm stem segments from 1 cm above the plant crown were ground in water and dilution plated on TZC plates plus appropriate antibiotics. Colony counts were normalized to CFU/g tissue. The log10(CFU/g tissue + 1) transformation was used to analyze data. Each open circle represents the bacterial population size from one plant. Closed diamonds represent the mean population size of all tested plants in three independent tests. The bars indicate the 95% confidence intervals around the mean. In the stems of plants inoculated with a single strain, there was no significant difference in population size between strains K60rif and K760 (P = 0.52) or between strains K760 and K701 (P = 0.47); however, the difference between K60 and K701 was significant (P = 0.02) when using the Bonferroni multiple-comparison test at the 95% level.
Bacterial taxis contributes to competitive fitness during invasion and colonization of tomato stems.
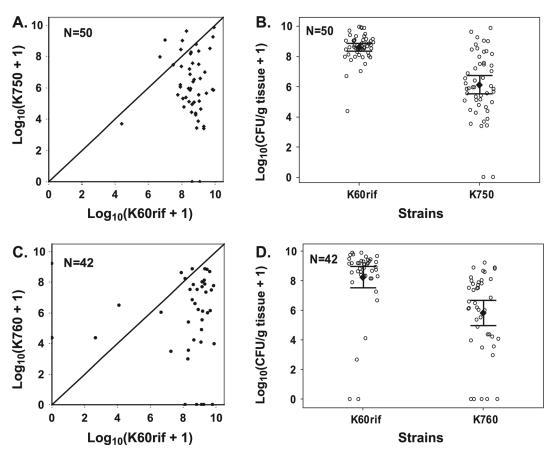
To study the relative fitness of the nonchemotactic mutants, we used competition assays in which tomato plants were inoculated with a 1:1 mixture of various strain combinations. Tomato plants were inoculated by soaking the soil, and lower stem populations of the two strains were quantified after disease symptoms appeared as described above. Wild-type strain K60rif easily outcompeted nontactic cheA strain K750 in the stems of coinoculated tomatoes; in 35 of 50 tested plants, the K60rif population size was at least 10-fold larger than that of K750 (P < 0.001 by the χ2 test; Fig. 7A). Moreover, the average population size of K60rif was 3.89 × 108 CFU/g, more than 200 times larger than that of K750, which was 1.45 × 106 CFU/g (P < 0.001 by the paired t test; Fig. 7B). Similarly, in 29 of 42 tested plants, the K60rif population size was at least 10-fold larger than that of cheW mutant K760 (P < 0.001 by the χ2 test; Fig. 7C). The average population size of the wild-type strain was 1.66 × 108 CFU/g, 500 times larger than that of cheW strain K760, which was 6.31 × 105 CFU/g (P < 0.001 by the paired t test; Fig. 7D). The same soil soak competition assay was also performed using a 1:1 mixture of the motile but nontactic cheW strain K760 and the nonmotile fliC mutant K701 to test the effects of random versus directed motility on competitive fitness. In the 36 plants tested, there was no significant difference between K760 and K701 either in the distribution of the two strains or in their average population sizes (data not shown).
FIG. 7.
Population sizes of R. solanacearum wild-type strain K60rif and cheA mutant K750 or cheW mutant K760 in tomato stem competition assays. Sixteen-day-old unwounded tomato plants (cv. Bonny Best) were inoculated by pouring a 1:1 mixture of K60rif and K750 or K760 onto the soil at about 1.5 × 107 CFU/g soil/strain. After each plant showed wilt symptoms, bacteria inside the stem were quantified by grinding stem sections in sterile water and plating onto selective medium as described in Materials and Methods. (A and C) Each point represents the bacterial population sizes of the two competing strains from a single plant. (B and D) Each open circle represents the population size of one strain from a plant. The closed diamonds represent the mean population size of a strain in the 50 (B) or 42 (D) plants tested in three independent trials. The bars indicate the 95% confidence intervals. (B) The mean population size of K60 in the stems of these coinoculated tomato plants was 3.89 × 108CFU/g, while nontactic cheA mutant K750 reached a mean population size of 1.45 × 106 CFU/g. (D) The mean population size of wild-type strain K60 in coinoculated tomato stems was 1.66 × 108 CFU/g, while cheW mutant K750 reached a mean population size of 6.31 × 105 CFU/g. The mean population sizes of both K750 (B) and K760 (D) were significantly lower than that of K60rif (P < 0.001) at the 95% level using the paired t test.
We also tested the effect of tactic ability on bacterial competitive fitness during colonization following inoculation of a cut tomato petiole. A 1:1 mixture of wild-type strain K60rif and cheW mutant K760 were inoculated directly into the vascular tissue of a 21-day-old tomato plant through the petiole, with an inoculation density of about 100 cells of each strain per plant. Two parts of the tomato stem were sampled, the centimeter spanning the inoculation site and the lower stem 1 cm above the crown. In 28 plants tested, there was no significant difference in population size or strain distribution for the two strains at either the inoculation point or the stem several centimeters below (data not shown).
DISCUSSION
We found that the bacterial wilt pathogen R. solanacearum has a complex and effective chemotaxis system, which it apparently uses to move toward more favorable conditions and increase its ecological fitness. Chemotaxis plays an important role in interactions between plants and bacteria (10, 54). Plants exude diverse and abundant sugars, amino acids, organic acids, aromatics, and secondary metabolites, all of which may be chemoattractants for plant-associated bacteria (10, 54). However, there is no standard set of chemoattractants for plant-associated bacteria. For example, sugars attract Rhizobium leguminosarum (20), Azospirillum brasilense (37), and Agrobacterium tumefaciens (30), but not Erwinia amylovora (41), Pseudomonas syringae pv. tomato (14), or Pseudomonas fluorescens (16). It has been suggested that responses to specific chemoattractants may vary not only by species but within strains of a single species (57). To test this hypothesis, we compared taxis responses among eight R. solanacearum strains selected to cover a wide range of different biovars, phylotypes, hosts, and geographic origins (Table 3). We found a striking diversity of responses to a panel of eight carbohydrates across these strains, a result consistent with the possibility that strain adaptation to different hosts or climate conditions may involve finely tuned changes in the repertoire of chemotactic responses. These preliminary data indicate that future more detailed studies are warranted. These might compare, for example, tactic response profiles of phylogenetically different strains that attack the same host or those of phylogenetically similar strains that attack different hosts.
Our reference wild-type strain K60 was not attracted to glucose, even though it grows, albeit slowly, on glucose as the sole carbon source (Table 2). However, all other isolates tested did chemotax toward glucose (Table 3). Interestingly, although we observed active taxis toward glucose on semisolid 1% tryptone plates, the swimming motility of some R. solanacearum isolates (K60, GMI1000, and UW551) was completely inhibited in minimal medium broth containing glucose as the sole carbon source (data not shown). The presence of some potential attractants, such as Casamino Acids and aspartate, reversed this inhibition, but others, such as alanine, did not (data not shown). The mechanism of this apparent glucose-mediated repression of motility is unknown.
Strain K60 was attracted to galactose and succinate on plates, but not in the 30-min agarose plug assay (Table 2). Chemotactic responses to certain compounds by Pseudomonas putida and Rhodobacter sphaeroides can be induced by preculturing in medium containing the substrate (22, 26), so we speculate that in K60 taxis toward succinate and galactose, chemotaxis is inducible. However, we cannot exclude the possibility that the 1 mM compound concentration in the agarose plug was below a minimal detection level.
R. solanacearum strain K60 is attracted to many compounds that it cannot use as carbon sources (Table 2), and most of these attractants can be found in plant root exudates. Plant root exudates are strongly attractive to many soilborne plant-associated bacteria (10, 54). Some components of plant root exudates are highly specific and active at very low concentrations, such as the flavonoids that attract Rhizobium spp. to their legume hosts at concentrations from 10−8 to 10−10 M (11, 17). We found that plant root exudates strongly attracted R. solanacearum strain K60 at low concentrations (Fig. 3A). Interestingly, root exudates from tomato, the host from which strain K60 was isolated, were more attractive at higher concentrations than exudates from rice, a nonhost plant, although rice exudates were also moderately attractive (Fig. 3A). These results suggest that R. solanacearum uses its chemotaxis system to find and move toward the roots of potential host plants, possibly with some degree of host selectivity. It should be noted that root exudates are complex mixtures and until specific attractants are identified, normalizing their concentrations is necessarily approximate, making direct comparisons between TRE and RRE concentrations difficult. The major elements in tomato root exudates have been characterized (31, 46), and most of these are attractive to strain K60. However, a solution containing the known tomato root exudate constituents in their natural concentrations was not as attractive to K60 as real tomato root exudate (data not shown). This suggests that R. solanacearum's chemotaxis receptors detect and respond to specific trace compound(s) in tomato root exudates. We found that the attractant(s) in tomato root exudates is neither heat labile nor protease sensitive (Fig. 3B). Indeed, heat or protease treatment actually increased attractiveness of the root exudates, suggesting that protein denaturation or degradation generates attractive amino acids or releases an active attractant that is otherwise sequestered by a protein (Fig. 3B).
Genomic sequences of two R. solanacearum strains, GMI1000 (phylotype I, race 1, biovar 3) and UW551 (phylotype II, race 3, biovar 2) are now available (19, 42). Analysis indicates that both genomes encode the four central elements required for bacterial taxis: (i) a set of likely chemotaxis receptors (methyl-accepting chemotaxis proteins [MCPs]) plus two aerotaxis/energy taxis receptors (aer); (ii) the central signal transduction apparatus, composed of histidine autokinase CheA and coupling protein CheW; (iii) three response regulator CheY proteins that interact with the motility apparatus; and (iv) a complete set of flagellar proteins that effect swimming motility. Interestingly, R. solanacearum appears to have one of the most sophisticated taxis sensory systems described to date. Its three distinct CheY homologs may act as a phosphate sink and response regulators, as in Sinorhizobium meliloti (44), but it retains CheZ as a dephosphorylase. It also has both a CheD homolog, presumably to assist in maturation of receptors, and a CheB homolog for receptor adaptation. R. solanacearum has an additional aerotaxis receptor and many more MCPs than the 5 found in E. coli (19 in GMI1000 and 17 in UW551); this implies an ability to perceive multiple and diverse signals that is characteristic of bacteria living in complex, heterogeneous environments (2). Eleven MCPs appear to be identical between the two strains (amino acid identity of >91%), while eight are unique to GMI1000 and six are present only in UW551. These differences in kind and number of MCPs between the two sequenced strains probably explain their diverse responses to different compounds and suggest a mechanism for adaptation to different hosts and/or ecological conditions.
The genes encoding CheA and CheW, the core signaling proteins required for chemotaxis, are clustered with genes encoding several flagellar structural and regulatory proteins in strain K60, with cheA immediately downstream of the motAB-cheY1 cluster and apparently cotranscribed with it (Fig. 1A and C). Although only 81 bp separates the cheA and cheW ORFs, our complementation analysis indicates that cheW is transcribed from a separate promoter (Fig. 1 A and C): this differs from the taxis gene organization in enterobacteria (32).
We created two motile but nontactic mutants by disrupting cheA (K750) and cheW (K760). In a biologically naturalistic soil soak inoculation that required bacteria to find and invade tomato plant roots from the soil, both K750 and K760 strains caused significantly less disease than their wild-type parent. However, their virulence was indistinguishable from that of aflagellate nonmotile fliC mutant K701, demonstrating conclusively that tactic, rather than random, motility is necessary for full virulence in R. solanacearum. The hypothesis that the flagellum and/or flagellin protein contributes separately to virulence, apart from their role in motility, was also disproved by this experiment, since there was no difference in virulence between flagellate but nontactic strains and the aflagellate mutant. This is consistent with our previous finding that R. solanacearum flagellin is not an elicitor of plant defense responses (40).
Interestingly, when nontactic strains were inoculated directly into the vasculature through a cut leaf petiole, they caused the same level of disease as the wild-type strains did. This finding suggests that once bacteria invade the plant vascular system, tactic motility is no longer an important factor for disease development. Again, this is consistent with our findings that the nonmotile mutant was fully virulent in the petiole assay and that R. solanacearum cells are very rarely motile in xylem fluid (51). Together, these results suggest that this pathogen uses taxis and motility to locate and invade plant roots out of the soil and possibly to colonize preferred niches in the developing root protoxylem; after this has occurred, the bacteria apparently no longer need these functions and become nonmotile. Once established in the roots, the pathogen is evidently drawn up into the stem and moved efficiently throughout the plant by the vascular flow. In addition to passive movement in the host transpirational stream, R. solanacearum can also use twitching motility to move within the host (28). Twitching motility contributes to migration against vascular flow in another plant xylem-inhabiting bacterium, Xylella fastidiosa (35); the interaction between the chemotaxis system and twitching motility in R. solanacearum has not yet been explored.
The poor virulence of nontactic mutants clearly is not due to reduced growth rate, since strains K750 and K760 grew as well as the wild-type strain in all circumstances we tested. Once they successfully invaded a tomato plant, nontactic strains reached the same population size in the stem as the wild type did, indicating that the differences in disease indices seen in Fig. 5A resulted from differences in the frequencies of successful colonization, not from differences in the final population size in colonized plants. However, when they were coinoculated with the wild type in the soil soak assay, both nontactic mutants were easily outcompeted by the wild-type strain, suggesting that taxis helps R. solanacearum cells locate, invade, and/or colonize plant hosts more quickly or efficiently. The hundredfold differences in competitive fitness that we measured in this artificial coinoculation assay are likely to have very large biological effects in the field under natural dynamic conditions. There was no difference in competitive fitness between the nontactic cheW mutant and the nonmotile fliC mutant, demonstrating that random motility could not compensate for the fitness reduction associated with loss of taxis. Studies with the animal bacterial pathogen Helicobacter pylori gave similar results; taxis-impaired strains were always outcompeted in the host by wild-type strains (4, 53).
The possible role of bacterial taxis in plant root colonization has been intensively discussed (10, 54), and defined taxis mutants of the nonpathogens P. fluorescens and A. brasilense were found to be significantly reduced in root colonization ability (16, 21). To better understand the early stages of host root colonization by R. solanacearum, we used GFP-expressing R. solanacearum strains to visualize the distribution of bacteria on tomato roots. When sterile tomato roots were incubated with bacteria in vitro, within 30 min the wild-type strain located the plant and clustered densely on the root surface, showing a strong preference for the elongation zone just behind the root tip. In contrast, the nontactic cheW mutant, apparently unable to detect the host root, remained evenly distributed in the observation chamber. It seems likely that the reduction in virulence and competitive fitness that we observed in nontactic mutants results from their inabilities to efficiently sense and move toward host plant roots and to identify and colonize optimal sites in those roots.
Nonetheless, many questions remain about the role of taxis in R. solanacearum-host interactions. What specific compounds in host root exudates are detected by the pathogen's sensory array? Does the R. solanacearum taxis system respond to oxygen levels and cellular energy levels, as is the case for many other bacteria (2)? What causes the differences in chemotactic responses among R. solanacearum strains? How does R. solanacearum regulate taxis-related gene expression in its soil and plant habitats? Does this bacterium use taxis in other parts of its life cycle besides host plant colonization? Further studies are under way to more fully understand the role of this complex behavior in the R. solanacearum life cycle.
Acknowledgments
This research was supported by the National Science Foundation (IBN-0090692), USDA-NRI (35319-13851), by a USDA Floral and Nursery Industry Task Force Specific Cooperative Agreement (58-1230-3-174), and by the University of Wisconsin—Madison College of Agricultural and Life Sciences.
We thank Tim Denny (University of Georgia) for the gift of AW1-gfp38 DNA, Julie Tans-Kersten for selection and analysis of strain K60rif, and Don Waller for statistical advice.
REFERENCES
- 1.Adler, J. 1966. Chemotaxis in bacteria. Science 153:708-716. [DOI] [PubMed] [Google Scholar]
- 2.Alexandre, G., S. Greer-Phillips, and I. B. Zhulin. 2004. Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 28:113-126. [DOI] [PubMed] [Google Scholar]
- 3.Allen, C., Y. Huang, and L. Sequeira. 1991. Cloning of genes affecting polygalacturonase production in Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 4:147-154. [Google Scholar]
- 4.Andermann, T. M., Y.-T. Chen, and K. M. Ottemann. 2002. Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect. Immun. 70:5877-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arlat, M., C. L. Gough, C. Zischek, P. A. Barberis, A. Trigalet, and C. A. Boucher. 1992. Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol. Microbiol. 6:3065-3076. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1995. Short protocols in molecular biology, 3rd ed. John Wiley and Sons, New York, N.Y.
- 7.Blair, D. F. 1995. How bacteria sense and swim. Annu. Rev. Microbiol. 49:489-520. [DOI] [PubMed] [Google Scholar]
- 8.Borkovich, K. A., and M. I. Simon. 1990. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell 63:1339-1348. [DOI] [PubMed] [Google Scholar]
- 9.Boucher, C., P. Barberis, A. Trigalet, and D. Demery. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J. Gen. Microbiol. 131:2449-2457. [Google Scholar]
- 10.Brencic, A., and S. C. Winans. 2005. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 69:155-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caetano-Anollés, G., D. K. Crist-Estes, and W. D. Bauer. 1988. Chemotaxis of Rhizobium meliloti to the plant flavone luteolin requires functional nodulation genes. J. Bacteriol. 170:3164-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conley, M. P., A. J. Wofle, D. F. Blair, and H. C. Berg. 1989. Both CheA and CheW are required for reconstitution of chemotactic signaling in Escherichia coli. J. Bacteriol. 171:5190-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook, D., E. Barlow, and L. Sequeira. 1989. Genetic diversity of Pseudomonas solanacearum: detection of restriction fragment length polymorphisms with DNA probes that specify virulence and the hypersensitive response. Mol. Plant-Microbe Interact. 2:113-121. [Google Scholar]
- 14.Cuppels, D. A. 1988. Chemotaxis by Pseudomonas syringae pathovar tomato. Appl. Environ. Microbiol. 54:629-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denny, T. P., and S. R. Baek. 1991. Genetic evidence that extracellular polysaccharide is a virulence factor of Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 4:198-206. [Google Scholar]
- 16.de Weert, S., H. Vermeiren, I. Mulders, I. Kuiper, N. Hendrickx, G. Bloemberg, J. Vanderleyden, R. De Mot, and B. J. Lugtenberg. 2002. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 15:1173-1180. [DOI] [PubMed] [Google Scholar]
- 17.Dharmatilake, A. J., and W. D. Bauer. 1992. Chemotaxis of Rhizobium meliloti toward nodulation gene-inducing compounds from alfalfa roots. Appl. Environ. Microbiol. 58:1153-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fegan, M., and P. Prior. 2005. How complex is the “Ralstonia solanacearum” species complex?, p. 449-461. In C. Allen, P. Prior, and A. C. Hayward (ed.), Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press, St. Paul, Minn.
- 19.Gabriel, D. W., C. Allen, M. Schell, T. P. Denny, J. T. Greenberg, Y. P. Duan, Z. Flores-Cruz, Q. Huang, J. M. Clifford, G. Presting, E. T. González, J. Reddy, J. Elphinstone, J. Swanson, J. Yao, V. Mulholland, L. Liu, W. Farmerie, M. Patnaikuni, B. Balogh, D. Norman, A. Alvarez, J. A. Castillo, J. Jones, G. Saddler, T. Walunas, A. Zhukov, and N. Mikhailova. 2006. Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol. Plant-Microbe Interact. 19:69-79. [DOI] [PubMed] [Google Scholar]
- 20.Gaworzewska, E. T., and M. J. Carlile. 1982. Positive chemotaxis of Rhizobium leguminosarum and other bacteria towards root exudates from legumes and other plants. J. Gen. Microbiol. 128:1179-1188. [Google Scholar]
- 21.Greer-Phillips, S. E., B. B. Stephens, and G. Alexandre. 2004. An energy taxis transducer promotes root colonization by Azospirillum brasilense. J. Bacteriol. 186:6595-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harwood, C. S., M. Rivelli, and L. N. Ornston. 1984. Aromatic acids are chemoattractant for Pseudomonas putida. J. Bacteriol. 160:622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65-87. [DOI] [PubMed] [Google Scholar]
- 24.Hendrick, C., and L. Sequeira. 1984. Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl. Environ. Microbiol. 48:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, Q., and C. Allen. 2000. Polygalacturonases are required for rapid colonization and full virulence of Ralstonia solanacearum on tomato plants. Physiol. Mol. Plant Pathol. 57:77-83. [Google Scholar]
- 26.Jeziore-Sassoon, Y., P. A. Hamblin, C. A. Bootle-Wilbraham, P. S. Poole, and J. P. Armitage. 1998. Metabolism is required for chemotaxis to sugars in Rhodobacter sphaeroides. Microbiology 144:229-239. [DOI] [PubMed] [Google Scholar]
- 27.Kelman, A. 1954. The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in a tetrazolium medium. Phytopathology 44:693-695. [Google Scholar]
- 28.Liu, H., Y. Kang, S. Genin, M. A. Schell, and T. P. Denny. 2001. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147:3215-3229. [DOI] [PubMed] [Google Scholar]
- 29.Liu, H., S. Zhang, M. A. Schell, and T. P. Denny. 2005. Pyramiding unmarked deletions in Ralstonia solanacearum shows that secreted proteins in addition to plant cell-wall-degrading enzymes contribute to virulence. Mol. Plant-Microbe Interact. 18:1296-1305. [DOI] [PubMed] [Google Scholar]
- 30.Loake, G. J., A. M. Ashby, and C. M. Shaw. 1988. Attraction of Agrobacterium tumefaciens C58C1 towards sugars involves a highly sensitive chemotaxis system. J. Gen. Microbiol. 134:1427-1432. [Google Scholar]
- 31.Lugtenberg, B. J., L. V. Kravchenko, and M. Simons. 1999. Tomato seed and root exudate sugars: composition, utilization by Pseudomonas bicontrol strains and role in rhizosphere colonization. Environ. Microbiol. 1:439-446. [DOI] [PubMed] [Google Scholar]
- 32.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. I. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 33.McGarvey, J. A., T. P. Denny, and M. A. Schell. 1999. Spatial-temporal and quantitative analysis of growth and EPSI production by Ralstonia solanacearum in resistant and susceptible tomato cultivars. Phytopathology 89:1233-1239. [DOI] [PubMed] [Google Scholar]
- 34.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng, Y., Y. Li, C. D. Galvani, G. Hao, J. N. Turner, T. J. Burr, and H. C. Hoch. 2005. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J. Bacteriol. 187:5560-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Okon, Y., L. Cakmakci, I. Nur, and I. Chet. 1980. Aerotaxis and chemotaxis in Azospririllum brasilense: a note. Microb. Ecol. 6:277-280. [DOI] [PubMed] [Google Scholar]
- 38.O'Toole, R., D. L. Milton, and H. Wolf-Watz. 1996. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol. Microbiol. 19:625-637. [DOI] [PubMed] [Google Scholar]
- 39.Parkinson, J. S. 1976. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 126:758-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfund, C., J. K. Tans-Kersten, M. Dunning, C. Allen, and A. F. Bent. 2004. Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol. Plant-Microbe Interact. 17:696-706. [DOI] [PubMed] [Google Scholar]
- 41.Raymundo, A. K., and S. M. Ries. 1980. Chemotaxis of Erwinia amylovora. Phytopathology 70:1066-1069. [Google Scholar]
- 42.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J.-C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 43.Schell, M., D. P. Roberts, and T. P. Denny. 1988. Analysis of the Pseudomonas solanacearum polygalacturonase encoded by pglA and its involvement in phytopathogenicity. J. Bacteriol. 170:4501-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt, R. 2002. Sinorhizobial chemotaxis: a departure from the enterobacterial paradigm. Microbiology 148:627-631. [DOI] [PubMed] [Google Scholar]
- 45.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-833. [PubMed] [Google Scholar]
- 46.Simons, M., H. P. Permentier, L. A. de Weger, C. A. Wijffelman, and B. J. Lugtenberg. 1997. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol. Plant-Microbe Interact. 10:102-106. [Google Scholar]
- 47.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1986. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhardt, R. I. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 49.Swanson, J. K., J. Yao, J. K. Tans-Kersten, and C. Allen. 2005. Behavior of Ralstonia solanacearum race 3 biovar 2 during latent and active infection of geranium. Phytopathology 95:136-143. [DOI] [PubMed] [Google Scholar]
- 50.Tans-Kersten, J., Y. Guan, and C. Allen. 1998. Ralstonia solanacearum pectin methylesterase is required for growth on methylated pectin but not for bacterial wilt virulence. Appl. Environ. Microbiol. 64:4918-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tans-Kersten, J., H. Huang, and C. Allen. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183:3597-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tans-Kersten, J. K., D. Brown, and C. Allen. 2004. Swimming motility, a virulence factor of Ralstonia solanacearum, is regulated by FlhDC and by the plant host environment. Mol. Plant-Microbe Interact. 17:686-695. [DOI] [PubMed] [Google Scholar]
- 53.Terry, K., S. M. Williams, L. Connolly, and K. M. Ottemann. 2005. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect. Immun. 73:803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vande Broek, A., and J. Vanderleyden. 1995. The role of bacterial motility, chemotaxis, and attachment in bacteria-plant interactions. Mol. Plant-Microbe Interact. 8:800-810. [Google Scholar]
- 55.Vasse, J., P. Frey, and A. Trigalet. 1995. Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 8:241-251. [Google Scholar]
- 56.Yu, H. S., and M. Alam. 1997. An agarose-in-plug bridge method to study chemotaxis in the archaeon Halobacterium salinarum. FEMS Microbiol. Lett. 156:265-269. [DOI] [PubMed] [Google Scholar]
- 57.Zhulin, I. B., and B. L. Taylor. 1995. Chemotaxis in plant-associated bacteria: the search for the ecological niche, p. 451-459. In I. Fendrik, M. Del Gallo, J. Vanderleyden, and M. de Zamaroczy (ed.), Azospririllum VI and related microorganisms: genetics, physiology, and ecology, vol. G37. Springer-Verlag, Berlin, Germany. [Google Scholar]