Abstract
The role of conserved amino acid residues in the polymerase domain of Escherichia coli primase has been studied by mutagenesis. We demonstrate that each of the conserved amino acids Arg146, Arg221, Tyr230, Gly266, and Asp311 is involved in the process of catalysis. Residues Glu265 and Asp309 are also critical because a substitution of each amino acid irreversibly destroys the catalytic activity. Two K229A and M268A mutant primase proteins synthesize only 2-nucleotide products in de novo synthesis reactions under standard conditions. Y267A mutant primase protein synthesizes both full-size and 2-nucleotide RNA, but with no intermediate-size products. From these data we discuss the significant step of the 2-nucleotide primer RNA synthesis by E. coli primase and the role of amino acids Lys229, Tyr267, and Met268 in primase complex stability.
Primase is one of the essential enzymes of cellular DNA replication mechanism. It initiates leading-strand synthesis once and lagging-strand synthesis multiple times during the course of replication (2, 12) by synthesizing short RNA molecules (pRNA) that are used by DNA polymerase as primers to begin complementary strand synthesis. Most primases are classified into two groups on the basis of amino acid sequence similarities. One group contains primases from bacteria and their phages, and the second group includes eukaryotic primases. All primases share many properties, but the proteins in the two classes differ both in structure and in their relationship with other proteins in the replication complex (5). The bacterial primases and the phage primases are distantly related but share many biochemical properties and contain several consensus signature sequence motifs (1, 9).
Prokaryotic primase proteins are composed of three domains (4, 25, 26). These are the N-terminal zinc binding domain, the oligonucleotide synthesis domain, and a C-terminal domain responsible for interaction with a helicase. Several conserved sequence motifs are clustered in the central region of primase (9). Of the six conserved motifs described by Frick and Richardson (5), motifs 3 to 6 appear to be involved in catalysis.
The X-ray structure of the RNA polymerase domain of Escherichia coli primase has been determined independently by two groups (10, 15). It appears that primase is a hollow cashew-shaped molecule with the N-terminal domain and part of the central domain forming a wedge-shaped cleft. The conserved sequence motifs and amino acids cluster in the cleft (10).
In experiments presented here, we analyzed the role of the evolutionarily conserved amino acids and unconserved amino acids that are located within a conserved sequence EGxxD (19) of E. coli primase. We showed that all residues studied here play an important role in the process of catalysis. Amino acids Glu265 and Asp309 appeared to be vital for primase function. Our data suggest that the 2-nucleotide (nt) RNA synthesis step is essential for the primase synthesis reaction and that amino acids Lys229, Tyr267 and Met268 may be involved in stability of the primase synthesis complex.
MATERIALS AND METHODS
Site-directed mutagenesis.
Mutations were made by PCR mutagenesis (8) using the parent dnaG plasmid pGNG1 (6). 5′-NcoI and 3′-EcoRI ends were added to the reconstructed DNA fragments to clone them into the pET21d vector. Oligonucleotide primers were purchased from Integrated DNA Technologies, Inc., Coralville, IA. All mutant genes were completely sequenced to establish that no additional substitutions had been induced by the mutagenesis procedure.
R221A, E265Q, D309A, and D311A mutations already existed in the P47 N-terminal fragment of primase cloned into the pGEX2T vector (7). The complete P47 fragments were excised from the pGEX2T vectors and recloned into pET21d. The missing C-terminal end of E. coli primase was added to make them full-size P66 primase-coding sequences.
Primase purification.
Wild-type and mutant primase proteins were prepared as His-tagged fusions from E. coli BL21(DE3) cells containing pET21d-P66 plasmids. Transformed cells were grown in LB medium containing 100 μg/ml of ampicillin to a density of 0.5 (A590), and expression of the proteins was induced by incubation with 0.4 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 37°C. The harvested cells were sonicated in phosphate-buffered saline. The His-tagged proteins were affinity purified from cell lysates by incubation with His-binding Talon resin (Clontech, Mountain View, CA) according to the manual.
pRNA synthesis on SSB/R199G4oric template.
The reaction mixture contained 10 pmol of primase, 4 pmol of single-stranded DNA (ssDNA)-binding protein (SSB) (purchased from U.S. Biochemicals), and 0.12 pmol of R199/G4oric ssDNA in 20 μl of pRNA synthesis buffer (20 mM Tris-HCl, pH 7.5, 8 mM dithiothreitol, 8 mM MgCl2, and 4% sucrose; MgCl2 and Tris-HCl were substituted for 2.5 mM MnCl2 and 20 mM HEPES in the case of pRNA synthesis reaction mixture in the presence of Mn2+ ions). Then 100 μM ATP, 20 μM of each GTP, CTP, and UTP, and 10 μCi of [α-32P]GTP (3,000 Ci/mM; Perkin-Elmer, Boston, MA) were added, and the mixture was incubated for 30 min at 30°C. A denaturing buffer (formamide containing 0.05% bromophenol blue) was added to the samples. Aliquots were analyzed by electrophoresis on 30% polyacrylamide-5 M urea gel. The gels were frozen and autoradiographed.
Electrophoretic mobility gel shift assay.
Electrophoretic mobility gel shift assay was done as previously described (20). The standard pRNA synthesis conditions (see above) were used, except that [α-32P]GTP was not added. Approximately 22 pmol of γ-32P-labeled G4oric 278-nt ssDNA fragment was incubated with 0.8 pmol of SSB and 3.8 pmol of primase in pRNA synthesis buffer with four ribonucleoside triphosphates added in a 12.5-μl volume. The samples were incubated at 30°C for 10 min and immediately applied to a prerun nondenaturing 4% polyacrylamide gel. The gels were frozen and autoradiographed.
Oligonucleotide-primed RNA synthesis.
Standard pRNA synthesis conditions were used, except for introduction of an annealing step prior to the synthesis reaction to generate a primed template. R199/G4oric ssDNA template (0.2 pmol) was mixed with 2.8 pmol of RNA oligonucleotide in pRNA synthesis reaction buffer. The mixture was heated at 85°C for 2 min and then cooled down slowly to 35°C. SSB protein (26 pmol), 10 pmol of primase, and appropriate ribonucleoside triphosphates were added to the oligonucleotide-primed template. Synthesis reactions were carried out at 30°C for 30 min. The products were analyzed on 30% polyacrylamide-5 M urea gels.
RESULTS
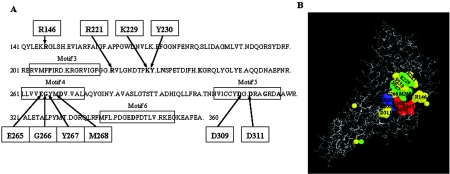
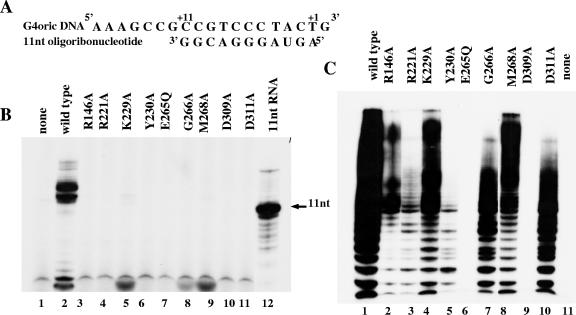
Ten mutant primase proteins were studied (Fig. 1A and B). Arg146, Lys229, Tyr230, Gly266, Tyr267, and Met268 of Escherichia coli primase were changed to Ala, and the altered primases were purified as His-tagged proteins. Previously constructed R221A, E265Q, D309A, and D311A (7) P47 (N-terminal primase fragment) mutants were recloned from pGEX2T vector into pET21d and restored as P66 full-size His-tagged proteins. All these amino acids are conserved among bacterial primase core domains. Tyr267 and Met268 belong to the completely conserved EGxxD motif (24).
FIG. 1.
Conserved amino acids of the catalytic center of E. coli primase. (A) Conserved residues in bacterial primase core domains are in bold type. The data were taken from Keck et al. (10). Arg146, Arg221, Lys229, Tyr230, Gly266, Tyr267, Met268, Asp309, and Asp311, which were changed to Ala by oligonucleotide mutagenesis, are indicated with arrows. Glu265 was changed to Gln, not Ala. (B) Crystal structure of the catalytic core fragment of E. coli primase. The locations of conserved amino acids studied are shown. Asp269, Asp345, and Asp347 amino acids bind Mg2+ ions and create the active site of the enzyme (7). They are shown in red. Glu265 and Asp309 amino acids are shown in blue. Lys229, Tyr267, and Met268 amino acids are shown in green. Arg146, Arg221, Tyr230, Gly266, and Asp311 amino acids are shown in yellow.
pRNA synthesis by mutant primases in the presence of Mg2+ ions.
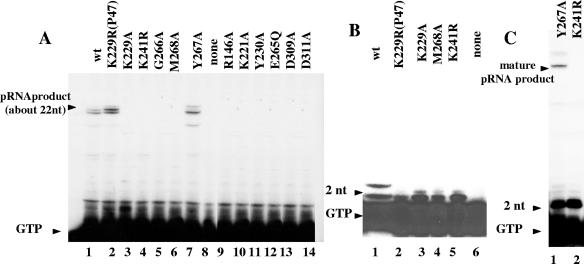
pRNA synthesis activity of the mutant proteins was tested on the R199/G4oric template with Mg2+ ions added to the reaction mixture. Only Y267A mutant protein (Fig. 2A, lane 7) was able to synthesize pRNA of the same size as the wild-type primase; the other mutant proteins did not. (In this study, the size of major pRNA product was about 22 to 25 nucleotides, which is normal for the template we used.) Some mutant proteins (K229A and M268A) synthesized pRNA products of much smaller size (Fig. 2B, lanes 3 and 4). The small products were identified as 2-nucleotide RNA (dimer) by comparison with pRNA product made by K241R mutant protein (Fig. 2B, lane 5, and Fig. 2C, lane 2), which is known to make dimer products only (22). Dimer products are normal for primase and are a result of abortive synthesis (18, 22). Y267A mutant protein synthesized pRNA of both full-size and 2-nucleotide size (Fig. 2C, lane 1).
FIG. 2.
Synthesis of pRNA by wild-type primase and mutant proteins in the presence of Mg2+ ions. (A) pRNA synthesized by wild-type (wt) and mutant primase proteins. In this study, the major products were about 22 to 25 nucleotides long. The position of free [α-32P]GTP not incorporated into the pRNA product is indicated by the arrowhead labeled GTP. Primase protein was absent in the control reaction mixture (lane 8). (B) pRNA synthesized by K229A and M268A mutant proteins compared with the product synthesized by the K241R mutant protein, the previously described primase mutant which was demonstrated to synthesize only 2-nucleotide RNA (22). (C) pRNA synthesized by the Y267A mutant protein compared with the product synthesized by the previously described K241R mutant protein (22).
It is worth noting that a conserved amino acid substitution to a residue other than Ala may have a different effect on the oligonucleotide synthesis activity of primase. For instance, the previously described mutant P47-K229R protein (22) shows basically the same pattern of pRNA synthesis as the wild-type protein (Fig. 2A, lane 2), but P66-K229A mutant protein synthesizes 2-nucleotide-long products only (Fig. 2B, lane 3). The reason could be the nature of the amino acids. Lysine (K) and arginine (R) belong to the same group of amino acids with a positively charged R group, but alanine has a nonpolar R group. It is possible that the change from positive charge to no charge caused the change of the protein function. Another explanation could be that P66-K229A is a full-size primase protein, whereas P47-K229R lacks the P16 C-terminal primase domain. However, this is unlikely, because P66 and P47 primase proteins always display the same activity in G4oric-SSB system (23).
Thus, we could distinguish three types of mutant proteins: (i) Y267A mutant protein, synthesizing normal-size pRNA; (ii) K229A and M268A mutant proteins, synthesizing 2-nucleotide products (dimers) (Y267A synthesizes dimers in addition to normal-size pRNA products.); and (iii) R146A, R221A, Y230A, E265Q, G266A, D309A, and D311A mutant proteins showing no oligonucleotide synthesis activity.
Restoration of oligonucleotide synthesis by mutant primases in the presence of Mn2+ ions.
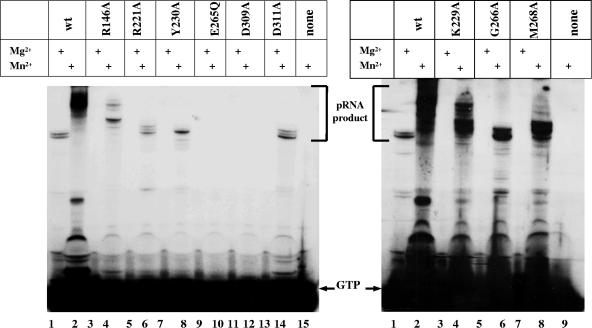
We then used Mn2+ ions instead of Mg2+ in the synthesis reaction mixtures. It is known that the larger Mn2+ ion may be better retained or more efficiently oriented than Mg2+ in the active center of the enzyme (3, 16, 17). Wild-type primase synthesizes considerably more pRNA after the addition of Mn2+ ions than it does after the addition of Mg2+ ions. pRNA molecules are larger in the first case also (Fig. 3, lanes 1 and 2). R146A, R221A, Y230A, G266A, and D311A mutant proteins, which were unable to synthesize oligonucleotides in the presence of Mg2+ ions, did synthesize pRNA products in the reaction mixture with Mn2+ ions added. The products were the same length as the ones made by wild-type primase in the presence of Mg2+ (22 to 25 nt) (Fig. 3). Thus, Mn2+ appears to restore normal oligonucleotide synthesis of these mutant primases. K229A and M268A mutant proteins, which were able to synthesize dimer products in the reaction mixture when Mg2+ ions were added, made pRNA products larger than 22 to 25 nt in the presence of Mn2+ ions (Fig. 3). The pRNA synthesis activity of E265Q and D309A mutant proteins was not changed during the incubation reaction with Mn2+ ions. The amount of pRNA product made by the mutants in the reaction mixture with Mn2+ ions, however, was less (for K229A and M268A mutant proteins) or much less (for other mutants) than the amount synthesized by wild-type primase (Fig. 3).
FIG. 3.
Synthesis of pRNA by wild-type primase and mutant proteins in the presence (+) of Mn2+ ions. The gels show a comparison of pRNA synthesis by wild-type (wt) primase and mutant proteins with Mg2+ or Mn2+ ions added to the reaction mixture. The majority of the products were 22 to 25 nucleotides long. The position of free [α-32P]GTP not incorporated into the pRNA product is indicated by the arrows labeled GTP. No primase protein was added to control reaction mixtures (lane 15 of the left panel and lane 9 of the right panel).
Primed synthesis on an 8-nt oligoribonucleotide by mutant proteins.
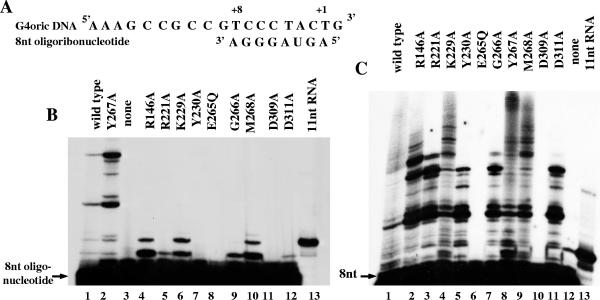
It has been shown that in the G4oric-SSB complex, primase can synthesize RNA using a preexisting oligoribonucleotide as a primer (21). To study the ability of the mutant proteins to elongate already preexisting RNA oligonucleotide on G4oric DNA, we used 8-nt RNA 5′-AGUAGGGA-3′, which corresponded to the first eight nucleotides (+1 to +8) of pRNA product made by primase on the R199/G4oric template (Fig. 4A).
FIG. 4.
Primed RNA synthesis on the 8-nt oligoribonucleotide by wild-type primase and mutant proteins. (A) Sequence of the 8-nt oligoribonucleotide primer and G4oric DNA template. The 8-nt oligoribonucleotide 5′-AGUAGGGA-3′ corresponded to the first 8 nucleotides (+1 to +8) of the pRNA product made by primase on the R199/G4oric template. (B) Primed synthesis on γ-32P-labeled 8-nt oligoribonucleotide (5′-AGUAGGGA-3′) by wild-type and mutant primase proteins in the presence of Mg2+ ions. CTP and GTP were added as a source of nucleotides for the elongation reaction. γ-32P-labeled 11-nt RNA was used as a size marker. The control reaction contained no primase protein (lane 3). (C) Primed synthesis on γ-32P-labeled 8-nt oligoribonucleotide (5′-AGUAGGGA-3′) by wild-type and mutant primase proteins in the presence of Mn2+ ions. CTP and GTP were added as a source of nucleotides for the elongation reaction. γ-32P-labeled 11-nt RNA was used as a size marker. The control reaction mixture contained no primase protein (lane 12).
We used γ-32P-labeled 8-nt oligoribonucleotide and CTP and GTP as a source of nucleotides for the primed synthesis reaction in the reaction buffer with Mg2+ ions added. By this procedure, all the labeled products detected were the elongation products (i.e., not the ones synthesized de novo), because ATP, which is required for de novo synthesis to incorporate into position +1, is eliminated from the reaction mixture (Fig. 4B). Only the Y267A altered protein showed a pattern of primed products similar to that of the wild-type primase (Fig. 4B, lane 2). This was expected because Y267A protein has basically the same activity as wild-type primase in the de novo pRNA synthesis reaction. R146A, K229A, and M268A mutant proteins produced pRNA products with the highest intensity (Fig. 4B, lanes 4, 6, and 10). Other mutant proteins also showed some elongation activity. These were the R221A and G266A mutant proteins, which gave a clear labeled band (Fig. 4B, lanes 5 and 9). There was also a very faint band in the case of the D311A mutant protein (Fig. 4B, lane 12). These latter three mutant proteins as well as the R146A mutant protein did not show any pRNA synthesis activity in de novo synthesis reaction mixtures in the presence of Mg2+ ions (see above). R146A, K229A, and M268A mutant proteins synthesized pRNA products of two different sizes during the reaction (Fig. 4B). R221A, G266A and D311A made only one product, the smaller one (Fig. 4B). The larger primed oligoribonucleotide product represents the 8-nt RNA primer extended by 2 nucleotides. The small pRNA product appears when only 1 nucleotide is added to the 8-nt RNA primer during the primed synthesis reaction. Y230A, E265Q, and D309A mutant proteins were incapable of function in the primed synthesis experiment (Fig. 4B). None of the mutant proteins (except for the Y267A mutant protein) reached the level of pRNA synthesis of wild-type primase.
To confirm the results of the above experiment, we tested the mutant proteins and 8-nt 5′-AGUAGGGA-3′ oligoribonucleotide in a primed synthesis reaction mixture with Mn2+ ions added instead of Mg2+. γ-32P-labeled oligoribonucleotide and CTP and GTP were added to the reaction mixture. As expected, all the mutant proteins, except for E265Q and D309A mutant proteins, had their pRNA synthesis ability restored, and long pRNA products were synthesized in all cases (Fig. 4C).
Primed synthesis on an 11-nt oligoribonucleotide by mutant proteins.
A second oligoribonucleotide was used in the primed synthesis reaction mixtures. This was the 11-nt oligoribonucleotide 5′-AGUAGGGACGG-3′ corresponding to the first 11 (+1 to +11) nucleotides of pRNA product made by primase on the R199/G4oric template (Fig. 5A). The 11-nt oligoribonucleotide had the same 5′ terminus as the 8-nt oligoribonucleotide used before, but with 3 nucleotides added to its 3′ terminus (Fig. 5A).
FIG. 5.
Primed synthesis on the 11-nt oligoribonucleotide by wild-type primase and mutant proteins. (A) Sequence of the 11-nt oligoribonucleotide primer used for the reaction and G4oric DNA template. The 11-nt RNA oligonucleotide 5′-AGUAGGGACGG-3′ corresponded to the first 11 nucleotides (+1 to +11) of the pRNA product made by primase on the R199/G4oric template. (B) Primed synthesis on unlabeled 11-nt oligoribonucleotide, annealed to G4oric ssDNA template, by wild-type and mutant primase proteins in the presence of Mg2+ ions. CTP and [α-32P]GTP were added to the reaction mixture as a source of nucleotides for the elongation. γ-32P-labeled 11-nt RNA was used as a size marker. The control reaction contained no primase protein (lane 1). (C) Primed synthesis on unlabeled 11-nt oligoribonucleotide of the same sequence by wild-type and mutant primase proteins in the presence of Mn2+ ions. GTP and [α-32P]CTP were added as a source of nucleotides for the elongation reaction. The control reaction contained no primase protein (lane 11).
First, we used unlabeled oligoribonucleotide for the primed synthesis reaction with the mutant proteins in reaction buffer containing Mg2+ ions. CTP and [α-32P]GTP were added to the reaction mixture as a source of nucleotides. Surprisingly, none of the mutant proteins studied synthesized any products in this reaction mixture (Fig. 5B). (We did not test Y267A mutant in these experiments, because it was behaving basically the same way as the wild-type primase in the previous experiments.) Even K229A and M268A mutant proteins, which showed elongation of the 8-nt oligoribonucleotide, were unable to extend the 11-nt oligoribonucleotide. Only wild-type primase generated a pRNA product, showing that at least this system worked.
Then, we used γ-32P-labeled 11-nt oligoribonucleotide for the next experiment, because in the previous one with unlabeled 11-nt primer oligonucleotide and [α-32P]GTP, it was impossible to detect the addition of the first nucleotide (i.e., C) to the RNA chain. CTP and GTP were added to the reaction mixture as a source of nucleotides. The reaction was carried out in the presence of Mg2+ ions. The result was the same as in the previous experiment (data not shown). None of the mutant proteins could add even a single nucleotide by primer extension. Only wild-type primase was able to elongate the 11-nt oligoribonucleotide.
To confirm the results of previous experiments, we tested the mutant proteins and 11-nt 5′-AGUAGGGACGG-3′ oligoribonucleotide in the primed synthesis reaction mixture with Mn2+ ions added instead of Mg2+. Unlabeled oligoribonucleotide, GTP, and [α-32P]CTP were used in the reaction mixture. As in the case with 8-nt oligoribonucleotide, all the mutant proteins, except for E265Q and D309A, synthesized primer extension products (Fig. 5C).
As a conclusion of the primed synthesis experiments, it appears that there is a difference in the ability of the mutant proteins to elongate an oligoribonucleotide and that this difference depends on the size of the primer.
Comparison of de novo and primed pRNA synthesis by wild-type and Y267A mutant primase.
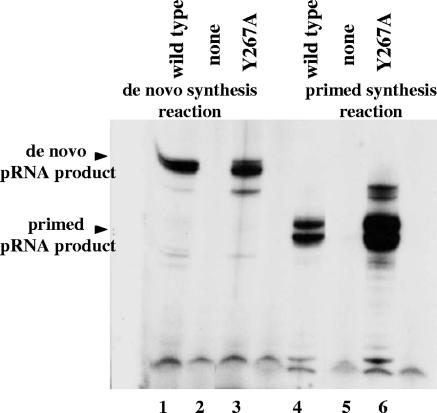
During the primed synthesis experiments, we noticed that Y267A mutant protein was more efficient at elongation of the 8-nt oligoribonucleotide than the wild-type primase protein was (Fig. 4B, lanes 1 and 2).
We decided to compare de novo and primed synthesis by wild-type primase protein and Y267A mutant protein using the same 11-nt oligoribonucleotide and the same synthesis conditions as before. The reactions were carried out in the presence of Mg2+ ions. As shown in Fig. 6, while the amounts of full-size pRNA product made by wild-type primase and Y267A mutant protein are about the same in de novo synthesis reactions (Fig. 6, lanes 1 and 3), the enzymatic activity of the Y267A mutant protein during primed synthesis is increased than that of wild-type primase (Fig. 6, lanes 4 and 6).
FIG. 6.
Comparison of de novo and primed RNA synthesis by wild-type primase and Y267A mutant proteins. Lanes 1 to 3 represent de novo synthesis. Lanes 4 to 6 represent primed synthesis. The 11-nt RNA oligonucleotide 5′-AGUAGGGACGG-3′ (the same as in the previous experiments) was used for the primed synthesis reaction. CTP and [α-32P]GTP were added to the primed synthesis reaction as a source of nucleotides for the elongation. The reactions were carried out in the presence of Mg2+ ions. No primase protein was added into the control reaction mixtures (lane 2 and 5).
Taken together, the results of the primed synthesis experiments with both 8- and 11-nt oligoribonucleotides show that the Y267A mutant protein is more efficient in elongation of preexisting oligonucleotides than the wild-type primase protein is. At the same time, wild-type primase and Y267A mutant proteins make about the same amount of full-size pRNA during de novo synthesis reactions (Fig. 2A, lanes 1 and 7, and Fig. 6, lanes 1 and 3). The explanation could be that during de novo synthesis by Y267A mutant protein, the majority of the pRNA product is 2-nucleotide RNA (Fig. 2C, lane 1). Only a small portion of pRNA reaches the size of mature product. This suggests that the Y267A mutant protein is in general more active in pRNA production than the wild-type primase is.
DNA template binding by mutant primase proteins.
The inability of some mutant proteins to synthesize pRNA could be caused by impairment of their capacity to bind a DNA template. To test this possibility, we used a gel retardation assay. All the mutant proteins were capable of binding to the SSB-G4oric complex to generate a shifted band (data not shown).
We concluded, therefore, that regardless of their ability to synthesize primer RNA, all mutant proteins were capable of binding to the DNA template.
DISCUSSION
In the experiments presented in this paper, we studied the role of the evolutionarily conserved amino acids in the catalytic activity of E. coli primase.
We showed that changing Arg146, Arg221, Tyr230, and Gly266 amino acids to Ala completely abolished pRNA synthesis by these mutant proteins in de novo synthesis reaction mixtures in the presence of Mg2+ ions. We also confirmed the results that have been shown before on P47 protein (the N-terminal domain of E. coli primase [7]) that E265Q, D309A, and D311A mutant proteins, expressed as whole P66 primase proteins, are unable to synthesize primer RNA under the same conditions. We had the activity of R146A, R221A, Y230A, G266A, and D311A mutant proteins restored to some degree (but still not as high as that of wild-type primase) by using Mn2+ ions instead of Mg2+ in the pRNA synthesis reaction mixture. We also showed that the same mutant proteins (R146A, R221A, G266A, and D311A), except for Y230A, had some ability (also not as high as wild-type primase) to elongate an 8-nt oligoribonucleotide annealed primer in the reaction mixture with Mg2+ ions added. This demonstrates that each of these amino acids (Arg146, Arg221, Tyr230, Gly266, and Asp311) is involved in the process of catalysis by E. coli primase.
Two E. coli primase mutant proteins, E265Q and D309A mutant proteins, showed no pRNA synthesis activity in any of our experiments, even in the presence of Mn2+ that can enhance synthesis by wild-type primase and all other mutant primases. From this, we conclude that these two amino acids are extremely important and directly participate in the catalytic function of E. coli primase. Amino acids Glu265 and Asp309 (as well as Asp311) belong to the Torprim domain (1), which is a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases, and RecR proteins. It has been shown previously that the activity of DNA primases encoded by plasmid RP4 and by E. coli satellite phage P4 was completely abolished by the E-to-Q substitution of the Glu265 equivalent (19). A previous study on E. coli primase using the P47 fragment carried out by our laboratory also showed that E265Q and D309A mutant proteins lost their catalytic activity and that both amino acids were not involved in Mg2+ binding (7). It has been proposed that Glu265 most likely functions as a general base, which abstracts the proton from the 3′-OH of the growing chain. The resulting 3′-O− may then attack the incoming 5′-nucleoside triphosphate, resulting in chain elongation (1).
We showed here that two primase mutant proteins, the K229A and M268A mutant proteins, synthesized only 2-nucleotide pRNA (dimer) products in de novo synthesis reactions in the presence of Mg2+ ions. Y267A mutant protein synthesized both dimer and full-size pRNA products under the same conditions. Also, previously, another E. coli primase mutant (K241R) was found (22), which synthesizes only 2-nucleotide pRNA products. Thus, from a total of 13 primase mutant proteins studied here and previously (22), four synthesized 2-nucleotide RNA products. We do not think that this number could be a coincidence. Also, we have not observed any mutant proteins that synthesized 3-, 4-, 5-nucleotide, etc., pRNA products. This suggests that the 2-nucleotide pRNA synthesis step is critical for the reaction.
It would be possible to assume that these mutant proteins (K229A and M268A mutant proteins) are simply defective in elongation, that they are unable to move the catalytic center along the template, move the second nucleotide to the “i” site, and let the third nucleotide enter the “i+1” site. The results of the primed synthesis experiment show, however, that K229A and M268A mutant primase proteins can add 1 or 2 nucleotides to the preexisting 8-nt oligoribonucleotide. This confirms that the mutant proteins are capable of moving the catalytic center along the DNA template. In the case of the Y267A mutant protein, if it would have difficulties to move itself along the template, it would stop at each step during the synthesis reaction and make 3-, 4-, 5-nucleotide, etc., pRNA products. However, once the Y267A mutant protein passes the 2-nucleotide RNA synthesis step, it produces mature oligoribonucleotide products only.
We suggest that these mutant proteins (K229A, Y267A, and M268A) form unstable complexes at the 2-nucleotide pRNA synthesis step, which easily dissociates, releasing the 2-nucleotide product. This idea is supported by the observation that K229A and M268A mutant proteins were capable of 8-nt oligoribonucleotide elongation. In this case the preexisting oligonucleotide stabilizes the complex by creating RNA-DNA hybrids and interacting with primase protein. Changing the Tyr267 amino acid to alanine disrupts the complex stability only to a degree.
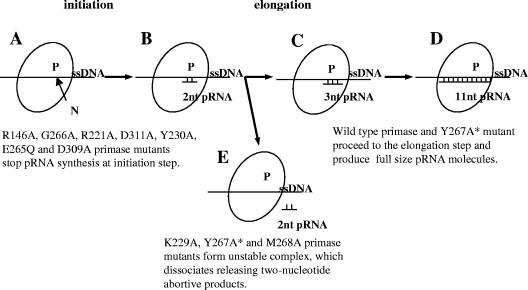
Why do these mutant proteins stop synthesis at the 2-nucleotide pRNA formation step, but not at any other? We assume that primase has the same type of synthesis cycle as RNA polymerase. The first step is initiation where the RNA polymerase complex with promoter DNA is not stable and can dissociate at any time, releasing the abortive products. The next step is elongation where the complex is stable and the enzyme continues the synthesis till termination. pRNA synthesis by the Y267A mutant protein clearly resembles this cycle. From this, we suggest that it is possible, that at the 2-nucleotide pRNA synthesis stage, the primase enzyme is at the initiation step where the complex is unstable and pRNA product can be easily released (Fig. 7B and E). Once the complex passes this stage and proceeds with further synthesis, it becomes stable and primase synthesizes mature pRNA products (Fig. 7C and D). This idea is supported by the observation that wild-type E. coli primase normally synthesizes dimer pRNA products as well as the full-size ones (18, 22).
FIG. 7.
Model of primer oligoribonucleotide synthesis by E. coli primase. Stages A and B show the initiation step of primer RNA synthesis by E. coli primase. At stage A, the first nucleotide binds to the active center of E. coli primase. P, E. coli primase; N, first nucleotide; ssDNA, single-stranded DNA template. Stage B is formation of the bond of the first nucleotide. At stage B, the enzyme complex is unstable. The enzyme complex can go to stage C or E at this step. At stage C, the primase enzyme enters the elongation step of the synthesis reaction. At stage C, the primase complex is stable and it continues to elongate pRNA product till the final stage (stage D), when it reaches the 11-nt size (for in vivo reaction). Stage E shows abortive synthesis. The primase complex dissociates, releasing the 2-nucleotide pRNA product. The Y267A* mutant primase protein synthesizes both full-size and 2-nucleotide pRNA.
We propose that Lys229, Tyr267, and Met268 amino acids are involved in the stability of the enzymatic complex, because proteins with these mutations release 2-nucleotide RNA abortive product. For RNA polymerase, the stability of the transcription elongation complex is determined by the existence of a RNA-DNA hybrid and protein-DNA and protein-RNA interactions (14) as well as locking a complementary protein surface tightly around the RNA-DNA hybrid (13). A similar mechanism may function in primase. Thus, amino acids Lys229, Tyr267, and Met268 could be involved in some of these interactions.
Our experiments suggest that the Y267A mutant protein is more active in pRNA production than wild-type primase. It has been shown previously that changing Y to F in the Tyr267 equivalent caused a two- to threefold increase in the activity of DNA primase encoded by plasmid RP4, but primase activity of E. coli satellite phage P4 was not changed by the substitution (19).
During our experiments we have observed a difference in the primed synthesis procedure by the mutant proteins and wild-type primase. Some of the mutant proteins (R146A, R221A, K229A, G266A, M268A, and D311A) were able to elongate 8-nt oligoribonucleotide, but none except the wild-type primase (and the Y267A mutant protein) was able to elongate the 11-nt oligoribonucleotide (5′-AGUAGGGACGG-3′). The size of the oligonucleotide chosen may be significant here, as 11 nt is the size of the mature pRNA product made by E. coli primase in vivo (11). It has also been predicted that E. coli primase is able to accommodate about 10 base pairs of RNA-DNA duplex in its catalytic domain (15). It is possible that after 11-nt oligoribonucleotide is synthesized, primase starts a new synthesis cycle completely independent of the first one. That is why, probably, mutant primase proteins whose catalytic activity was impaired were unable to elongate 11-nt oligoribonucleotide. The larger (doubled) size of pRNA synthesized by wild-type primase in this system (i.e., 22 to 25 nt) suggests that under certain conditions primase can reinitiate a round of synthesis, which further extends the pRNA chain either by using a new molecule of primase or reprogramming initiation of the same molecule.
Acknowledgments
We thank Arkady Mustaev of the New York Public Health Research Institute and Wuliang Sun and Vitaly Epshtein of the New York University Medical Center for helpful comments and discussion throughout this work. We also thank Wuliang Sun for making a series of reviews of and suggestions for the manuscript.
This work was supported by NIH grant 5RO1GM38292-19 to G.N.G.
REFERENCES
- 1.Aravind, L., D. D. Leipe, and E. V. Koonin. 1998. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26:4205-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouche, J. P., L. Rowen, and A. Kornberg. 1978. The RNA primer synthesized by primase to initiate phage G4 DNA replication. J. Biol. Chem. 253:765-769. [PubMed] [Google Scholar]
- 3.Copeland, W. C., and T. S. Wang. 1993. Mutational analysis of the human DNA polymerase alpha. The most conserved region in alpha-like DNA polymerases is involved in metal-specific catalysis. J. Biol. Chem. 268:11028-11040. [PubMed] [Google Scholar]
- 4.Crouch, E., S. Miller, V. Wilson, and D. Busbee. 1997. A DNA polymerase alpha accessory protein exhibits structural and functional similarities to SV40 large tumor antigen. Mutat. Res. 374:109-123. [DOI] [PubMed] [Google Scholar]
- 5.Frick, D. N., and C. C. Richardson. 2001. DNA primases. Annu. Rev. Biochem. 70:39-80. [DOI] [PubMed] [Google Scholar]
- 6.Godson, G. N. 1991. An over-expression plasmid for Escherichia coli primase. Gene 100:59-64. [DOI] [PubMed] [Google Scholar]
- 7.Godson, G. N., J. Schoenich, W. Sun, and A. A. Mustaev. 2000. Identification of the magnesium ion binding site in the catalytic center of Escherichia coli primase by iron cleavage. Biochemistry 39:332-339. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Snisky, and T. J. White (ed.), PCR protocols. A guide to methods and applications. Academic Press, New York, N.Y.
- 9.Ilyina, T. V., A. E. Gorbalenya, and E. V. Koonin. 1992. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J. Mol. Evol. 34:351-357. [DOI] [PubMed] [Google Scholar]
- 10.Keck, J. L., D. D. Roche, A. S. Lynch, and J. M. Berger. 2000. Structure of the RNA polymerase domain of E. coli primase. Science 287:2482-2486. [DOI] [PubMed] [Google Scholar]
- 11.Kitani, T., K. Yoda, T. Ogawa, and T. Okazaki. 1985. Evidence that discontinuous DNA replication in Escherichia coli is primed by approximately 10 to 12 residues of RNA starting with a purine. J. Mol. Biol. 184:45-52. [DOI] [PubMed] [Google Scholar]
- 12.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. Freeman, New York, N.Y.
- 13.Korzheva, N., A. Mustaev, E. Nudler, V. Nikiforov, and A. Goldfarb. 1998. Mechanistic model of the elongation complex of Escherichia coli RNA polymerase. Cold Spring Harbor Symp. Quant. Biol. 63:337-345. [DOI] [PubMed] [Google Scholar]
- 14.Nudler, E. 1999. Transcription elongation: structural basis and mechanisms. J. Mol. Biol. 288:1-12. [DOI] [PubMed] [Google Scholar]
- 15.Podobnik, M., P. McInerney, M. O'Donnell, and L. Kuriyan. 2000. A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J. Mol. Biol. 300:353-362. [DOI] [PubMed] [Google Scholar]
- 16.Pritchard, A. E., and C. S. McHenry. 1999. Identification of the acidic residues in the active site of DNA polymerase III. J. Mol. Biol. 285:1067-1080. [DOI] [PubMed] [Google Scholar]
- 17.Saturno, J., J. M. Lazaro, L. Blanco, and M. Salas. 1998. Role of the first aspartate residue of the “YxDTDS” motif of phi29 DNA polymerase as a metal ligand during both TP-primed and DNA-primed DNA synthesis. J. Mol. Biol. 283:633-642. [DOI] [PubMed] [Google Scholar]
- 18.Sheaff, R. J., and R. D. Kuchta. 1993. Mechanism of calf thymus DNA primase: slow initiation, rapid polymerization, and intelligent termination. Biochemistry 32:3027-3037. [DOI] [PubMed] [Google Scholar]
- 19.Strack, B., M. Lessl, R. Calendar, and E. Lanka. 1992. A common sequence motif, -E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I- and P-type DNA primases and the alpha protein of the Escherichia coli satellite phage P4. J. Biol. Chem. 267:13062-13072. [PubMed] [Google Scholar]
- 20.Sun, W., and G. N. Godson. 1996. Interaction of Escherichia coli primase with a phage G4oric-E. coli SSB complex. J. Bacteriol. 178:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun, W., and G. N. Godson. 1998. Synthesis of polyribonucleotide chains from the 3′-hydroxyl terminus of oligodeoxynucleotides by Escherichia coli primase. J. Biol. Chem. 273:16358-16365. [DOI] [PubMed] [Google Scholar]
- 22.Sun, W., J. Schoneich, and G. N. Godson. 1999. A mutant Escherichia coli primase defective in elongation of primer RNA chains. J. Bacteriol. 181:3761-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun, W., J. Tormo, T. A. Steitz, and G. N. Godson. 1994. Domains of Escherichia coli primase: functional activity of a 47-kDa N-terminal proteolytic fragment. Proc. Natl. Acad. Sci. USA 91:11462-11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szafranski, P., C. L. Smith, and C. R. Cantor. 1997. Cloning and analysis of the dnaG gene encoding Pseudomonas putida DNA primase. Biochim. Biophys. Acta 1352:243-248. [DOI] [PubMed] [Google Scholar]
- 25.Washington, M. T., A. H. Rosenberg, K. Griffin, F. W. Studier, and S. S. Patel. 1996. Biochemical analysis of mutant T7 primase/helicase proteins defective in DNA binding, nucleotide hydrolysis, and the coupling of hydrolysis with DNA unwinding. J. Biol. Chem. 271:26825-26834. [DOI] [PubMed] [Google Scholar]
- 26.Yong, Y., and L. J. Romano. 1995. Nucleotide and DNA-induced conformational changes in the bacteriophage T7 gene 4 protein. J. Biol. Chem. 270:24509-24517. [DOI] [PubMed] [Google Scholar]