Abstract
Among the rhizobia that establish nitrogen-fixing nodules on the roots of host plants, many contain multiple copies of genes encoding the sigma factor RpoH and the chaperone GroEL/GroES. In Sinorhizobium meliloti there are two rpoH genes, four groESL operons, and one groEL gene. rpoH1 mutants are defective for growth at high temperature and form ineffective nodules, rpoH1 rpoH2 double mutants are unable to form nodules, and groESL1 mutants form ineffective nodules. To explore the roles of RpoH1 and RpoH2, we identified mutants that suppress both the growth and nodulation defects. These mutants do not suppress the nitrogen fixation defect. This implies that the functions of RpoH1 during growth and RpoH1/RpoH2 during the initiation of symbiosis are similar but that there is a different function of RpoH1 needed later during symbiosis. We showed that, unlike in Escherichia coli, overexpression of groESL is not sufficient to bypass any of the RpoH defects. Under free-living conditions, we determined that RpoH2 does not control expression of the groE genes, and RpoH1 only controls expression of groESL5. Finally, we completed the series of groE mutants by constructing groESL3 and groEL4 mutants and demonstrated that they do not display symbiotic defects. Therefore, the only groESL operon required by itself for symbiosis is groESL1. Taken together, these results suggest that GroEL/GroES production alone cannot explain the requirements for RpoH1 and RpoH2 in S. meliloti and that there must be other crucial targets.
Sinorhizobium meliloti can be found as a free-living bacterium residing in the soil or as a nitrogen-fixing symbiont residing in nodules on the roots of leguminous host plants, such as alfalfa. The S. meliloti genome contains 14 genes for sigma factors (14), which are subunits of RNA polymerase that direct transcription initiation by recognizing promoters. Two of these genes, rpoH1 and rpoH2, encode members of the RpoH family of secondary sigma factors. RpoH (σ32) was originally identified in Escherichia coli as a sigma factor that responds to heat shock. In response to a sudden increase in temperature or other stresses, the levels of RpoH rise transiently, inducing transcription of a subset of genes encoding heat shock proteins (HSPs). HSPs include chaperones involved in protein folding, such as GroEL/GroES and DnaK/DnaJ/GrpE, and proteases, such as FtsH and Lon (48). Although RpoH and the HSPs were identified as part of the heat shock response, these proteins are present at low temperature and play important roles in cellular processes under nonstress conditions, such that the rpoH gene in E. coli is essential above 20°C (49). The requirement for RpoH in E. coli can largely be explained as a requirement for expression of the groESL operon, because overexpression of groESL is sufficient to suppress the temperature-sensitive growth defect of the rpoH mutant from 20 to 40°C (25).
Although the E. coli genome only contains one rpoH gene and one groESL operon, other bacterial genomes contain multiple copies of these genes. In particular, many Rhizobium species have multiple rpoH and groESL genes, and mutations in some of them result in symbiotic defects. In S. meliloti, in addition to the two rpoH genes (35, 36), there are four groESL operons (14, 33, 34, 40) and one groEL gene (7). rpoH1 and groESL1 mutants are unable to fix nitrogen (Fix−) (33, 35, 36), and rpoH1 rpoH2 double mutants are unable to form nodules (Nod−) (36). In Bradyrhizobium japonicum, there are three rpoH genes (30, 31), five groESL operons (10), and two groEL genes (23). groESL3Bj is regulated with nitrogen fixation genes (10), and a groESL3Bj groESL4Bj double mutant is unable to fix nitrogen (11). Rhizobium sp. strain TAL1145 has at least one rpoH gene, and the rpoH mutant exhibits reduced nodulation, resulting in stunted plant growth (24). Rhizobium leguminosarum has at least three groESL operons (39, 44), and Mesorhizobium loti has two rpoH genes and five groESL operons (22, 24). Interestingly, the genome of the closely related plant pathogen Agrobacterium tumefaciens, also a member of the Rhizobiaceae, only contains single copies of these genes (46). The reason for multiple rpoH and groESL genes in these plant endosymbionts is unclear. Are the genes regulated differentially but encode proteins with similar functions, or do they encode proteins with specialized functions?
The two rpoH genes in S. meliloti were identified as members of the rpoH family by sequence analysis and by the ability to complement an E. coli rpoH mutation (35, 36). Under free-living conditions, rpoH1 mutants exhibit a slight growth defect at the optimum growth temperature (30°C) and a severe defect at higher temperatures (35). During symbiosis, rpoH1 mutant cells invade the nodule and differentiate into bacteroids but undergo early senescence (28), resulting in a Fix− phenotype (35, 36). rpoH2 mutants have no phenotype under free-living or symbiotic conditions (35, 36). However, Ono et al. (36) discovered that an rpoH1 rpoH2 double mutant is unable to form nodules.
Transcriptional reporter fusions to rpoH1 and rpoH2 have shown that rpoH1 is transcribed during stationary phase in Luria-Bertani (LB) rich medium and M9 minimal medium and that rpoH2 is transcribed during stationary phase only in M9 medium. During symbiosis, rpoH1 is strongly expressed throughout the nodule, whereas rpoH2 is not expressed in the nodule, except for low levels at the tip and variable punctate spots at other locations (35). The phenotypes and expression data suggest that rpoH1 and rpoH2 have distinct but overlapping functions.
The presence of a family of four to five groEL genes in S. meliloti was initially discovered by Southern blot analysis (40). Additional work by other groups and subsequent sequencing of the S. meliloti genome has led to a final count of four groESL operons and one groEL gene (7, 14, 33, 34, 40). The names of the groE genes used in this paper are those given in the genome annotation, although groES5 was not annotated (14).
A connection between GroEL/GroES and symbiosis was uncovered when groESL1 was identified in a genetic screen for S. meliloti genes required for full induction of nod genes, which are required for formation of a bacterial signal that initiates nodule formation by host plants (33). The groESL1 mutation affects the activities of several related transcription factors (NodD1, NodD3, and SyrM) that activate gene expression of nod genes, and GroEL copurifies with NodD1 and NodD3 (12, 33). In vitro work has demonstrated that the NodD proteins are substrates for GroEL/GroES, resulting in modulation of the DNA binding activity (47). groESL1, groESL2, and groESL5 mutants have been studied. groESL1 mutants have a slight growth defect, are delayed for nodulation, and form Fix− nodules (33). A groESL2 mutant displays neither a growth nor a symbiotic defect, but the groESL1 groESL2 double mutant is not viable (32). The groESL5 mutant has no symbiotic defect (28). GroES1/GroEL1 and GroES2/GroEL2 are the most similar to each other (97% identical for GroES and 99% identical for GroEL), whereas GroES3/GroEL3 is the most dissimilar from any other S. meliloti homolog (75 to 78% identical for GroES and 72 to 74% identical for GroEL).
Mitsui et al. (28) tested whether RpoH1 or RpoH2 controls expression of the groESL genes in S. meliloti during heat shock. groESL5 was the only groESL operon whose transcription was controlled by RpoH1, and none of the genes were controlled by RpoH2. However, this work did not explore regulation during stationary phase and within the nodule, other conditions where we know that rpoH1 and rpoH2 are expressed (35).
Given that groESL is a crucial target of RpoH in E. coli and that groESL1, rpoH1, and rpoH1 rpoH2 S. meliloti mutants have symbiotic phenotypes, we hypothesized that groESL might also be a key target of RpoH in S. meliloti. However, in this paper we use suppressor mutant analysis and overexpression experiments to demonstrate that the relationships between RpoH and GroEL/GroES are different in the two organisms. Specifically, our results suggest that GroEL/GroES production is not sufficient to bypass the requirements for RpoH1 or RpoH1/RpoH2 during growth and symbiosis and that there must be other crucial targets. In addition, we show that only groESL5 is controlled by RpoH1 during free-living growth and stationary phase at 30°C, which agrees with results obtained by Mitsui et al. during growth and heat shock (28). Finally, we demonstrate that groESL3 and groEL4 mutants are able to nodulate and fix nitrogen like wild-type cells.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. Bacterial cultures were grown in LB medium, LB medium supplemented with MgSO4 and CaCl2 (LB/MC medium) (17), or M9 minimal medium containing 0.2% sucrose, 0.5 μg biotin ml−1, 1 mM MgSO4, and 0.25 mM CaCl2. Antibiotics were added to the media as follows: 100 μg ampicillin ml−1, 25 μg gentamicin ml−1, 5 to 50 μg hygromycin ml−1, 25 μg kanamycin ml−1, 50 or 200 μg neomycin ml−1, 50 or 200 μg spectinomycin ml−1, 500 μg streptomycin ml−1, and 2 or 10 μg tetracycline ml−1. S. meliloti cells were grown at 30°C unless otherwise indicated. Plasmids were introduced into S. meliloti cells by triparental conjugation (17). Chromosomally located constructs were moved between S. meliloti strains by generalized transduction using N3 phage (26). Although rpoH2::aacC1 containing strain BY294 (36) was constructed in the Rm1021 background, we transferred the mutation by transduction into our own Rm1021 strain, creating AB3, to ensure isogenicity.
TABLE 1.
Strains
| Strain | Relevant characteristic(s) | Reference |
|---|---|---|
| AB3 | rpoH2::aacCI | This study |
| AB4 | Wild type/pAB1 (Plac-groESL1) | This study |
| AB9 | rpoH1::aadA rpoH2::aacCI | This study |
| AB16 | Wild type/pAB2 (Plac-groESL3) | This study |
| AB35 | rpoH1::aadA rpoH2::pVO101 NDS-3 | This study |
| AB36 | rpoH1::aadA rpoH2::pVO101 NDS-4 | This study |
| AB37 | rpoH1::aadA rpoH2::pVO101 NDS-5 | This study |
| AB38 | rpoH1::aadA rpoH2::pVO101 NDS-6 | This study |
| AB39 | rpoH1::aadA rpoH2::pVO101 NDS-7 | This study |
| AB40 | rpoH1::aadA rpoH2::pVO101 NDS-8 | This study |
| AB41 | rpoH1::aadA rpoH2::pVO101 NDS-9 | This study |
| AB42 | rpoH1::aadA rpoH2::pVO101 NDS-10 | This study |
| AB43 | rpoH1::aadA rpoH2::pVO101 NDS-11 | This study |
| AB44 | rpoH1::aadA rpoH2::pVO101 NDS-12 | This study |
| AB92 | Wild type/pAB7 (Ptrp-groESL1) | This study |
| AB103 | Wild type/pAB8 (Ptrp-groESL3) | This study |
| AB129 | groEL2::pAB10 (groEL2-gfp-gus transcriptional fusion, groEL2 not disrupted) | This study |
| AB140 | groEL1::pAB11 (groEL1-gfp-gus transcriptional fusion, groEL1 not disrupted) | This study |
| AB145 | groEL3::pAB12 (groEL3-gfp-gus transcriptional fusion, groEL3 not disrupted) | This study |
| AB147 | groEL4::pAB13 (groEL4-gfp-gus transcriptional fusion, groEL4 not disrupted) | This study |
| AB150 | groEL5::pAB14 (groEL5-gfp-gus transcriptional fusion, groEL5 not disrupted) | This study |
| AF14 | groESL3Δ::tet | This study |
| AR12 | rpoH1::aadA rpoH2::pVO194 (rpoH2-gus transcriptional fusion, rpoH2 not disrupted) | This study |
| AR13 | rpoH1::aadA/pBGR86 (rpoH1-gus transcriptional fusion) | This study |
| AR14 | rpoH2::aacCI/pBGR86 (rpoH1-gus transcriptional fusion) | This study |
| AR15 | rpoH1::aadA rpoH2::aacCI/pBGR86 (rpoH1-gus transcriptional fusion) | This study |
| B4T1 | groEL1::Tn5 | 33 |
| BY294 | rpoH2::aacCI | 36 |
| Rm1021 | Wild type | 27 |
| VO2012 | Wild type/pBGR86 (rpoH1-gus transcriptional fusion) | 35 |
| VO2148 | rpoH2::pVO101 (rpoH2 disruption) | 35 |
| VO2257 | rpoH2::pVO194 (rpoH2-gus transcriptional fusion, rpoH2 not disrupted) | 35 |
| VO3128 | rpoH1::aadA | 35 |
| VO3148 | rpoH1::aadA rpoH2::pVO101 | This study |
| VO3149 | rpoH1::aadA rpoH2::pVO101 | This study |
| VO3150 | rpoH1::aadA rpoH2::pVO101 NDS-1 | This study |
| VO3151 | rpoH1::aadA rpoH2::pVO101 NDS-2 | This study |
| VO3165 | rpoH1::aadA GDS-1 | This study |
| VO3166 | rpoH1::aadA rpoH2::pVO101 GDS-2 | This study |
| VO3170 | rpoH1::aadA rpoH2::pVO101 GDS-1 | This study |
| VO3193 | groEL4Δ | This study |
Plant assays.
Alfalfa plants (Medicago sativa GT13R plus) were grown on nitrogen-free buffered nodulation medium and inoculated with S. meliloti cells as previously described (34). Plant height, leaf color, and nodule color were scored at 6 weeks postinoculation to determine the status of nitrogen fixation. Inoculation with Fix+ bacteria results in tall, green plants with pink nodules. Inoculation with Fix− bacteria results in stunted, chlorotic plants with white nodules. Bacteria were isolated from nodules by surface sterilizing nodules in 20% Clorox bleach for 5 min, washing two times with water and one time with LB medium, crushing with forceps, and then streaking on LB medium.
Western blot analysis.
To obtain samples for Western blot analysis, cells were grown overnight at 30°C in LB/MC medium with streptomycin, diluted back to an optical density at 595 nm (OD595) of 0.1, grown to mid-log phase (0.6 ≤ OD595 ≤ 0.8), harvested, and stored at −80°C. Cells were resuspended in 1× phosphate-buffered saline at 0.1 ml per OD595 unit. The cells were disrupted by sonication, and the resulting extracts were combined with 2× Laemmli sample buffer. Equal volumes of extract were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose. Blots were probed with rabbit polyclonal antibodies to the E. coli proteins at the following dilutions: anti-GroEL (Stressgen) at 1:5,000, anti-DnaK (gift from J. Brodsky) at 1:5,000 or anti-DnaK (Upstate Biotechnology) at 1:2,500, and anti-DnaJ (Stressgen) at 1:1,250. Blots were then probed with a 1:15,000 dilution of anti-rabbit horseradish peroxidase-conjugated secondary antibody, developed with enhanced chemiluminescence reagents (Pierce), and imaged using a Fujifilm LAS-3000 imaging system.
To quantify relative protein levels, band density was determined by ImageGauge software (Fuji). Protein concentration of cell lysates was determined by bicinchoninic acid protein assay (Pierce), and band intensities were then normalized to protein concentration.
Statistical analysis.
Significance of differences in bacterial growth levels and protein levels was determined by using both the Student's t test and the Wilcoxon rank sum test, which does not assume a normal distribution. Although the P values varied, differences were significant with both tests unless otherwise noted.
Construction of plasmids for overexpression of groESL1 and groESL3.
To place groESL1 under the control of the E. coli lac promoter, a 2.1-kb DNA fragment that extends from 68 bp upstream of the groES1 start codon to 30 bp downstream of the groEL1 stop codon was amplified using primers that generate ApaI and XbaI restriction sites. The fragment was inserted into ApaI-SpeI-digested pMB403 (3), a broad-host-range vector that contains the lac promoter, creating pAB1 (Plac-groESL1). To place groESL3 under the control of the lac promoter, a 2.2-kb fragment that extends from 78 bp upstream of the groES3 start codon to 85 bp downstream of the groEL3 stop codon was amplified with primers generating ApaI and XbaI restriction sites. The fragment was inserted into ApaI-SpeI-digested pMB403, creating pAB2 (Plac-groESL3).
To place groESL1 and groESL3 under the control of the Salmonella enterica serovar Typhimurium trp promoter, the lac promoter was removed from pAB1 and pAB2 and replaced with a fragment containing 141 bp of S. enterica serovar Typhimurium DNA containing the trp promoter. First, a 220-bp EcoRV-Acc65I fragment containing Ptrp was isolated from pVO131. To delete the lac promoter, pAB1 and pAB2 were digested with NsiI, blunted with T4 DNA polymerase, and digested with Acc65I. The Ptrp fragment was then inserted into pAB1 to create pAB7 (Ptrp-groESL1) and into pAB2 to create pAB8 (Ptrp-groESL3).
The expression plasmids were introduced into wild-type (Rm1021), groEL1::Tn5 (B4T1), rpoH1::aadA (VO3128), and rpoH1::aadA rpoH2::pVO101 (VO3148) strains by triparental conjugation.
Construction of groEL-gus fusions.
The groEL-gus fusions were constructed using recombinational cloning as described in House et al. (20). This method is a modification of Invitrogen's Gateway Technology, such that transfer of DNA from an entry vector to a destination vector by the λ recombination system is performed in vivo via a pentaparental mating. In brief, each groEL open reading frame (ORF) was transferred from an entry vector (pESmc00913, pESma0744, pESma0124, pESmc01758, and pESmb21566) (41) to the destination vector pMK2030 (B. K. Schroeder, B. L. House, M. W. Mortimer, and M. L. Kahn, unpublished data) during a pentaparental mating using the helper plasmid pRK2013 (9) and the λ integrase- and excisionase-expressing plasmid pXINT129 (37). This destination vector is a suicide vector that contains attR recombination sites upstream of promoterless gfp and gus genes to allow the formation of transcriptional fusions. Each groEL-gfp-gus-containing plasmid was moved into Rm1021 by triparental mating and integrated at the respective groEL gene by single reciprocal recombination, resulting in a PgroE-groES-groEL-gfp-gus construct. The resulting strains AB140 (groESL1-gfp-gus), AB129 (groESL2-gfp-gus), AB145 (groESL3-gfp-gus), AB147 (groEL4-gfp-gus), and AB150 (groESL5-gfp-gus) were confirmed by Southern analysis. The fusions were transferred into rpoH1::aadA (VO3128), rpoH2::aacCI (AB3), and rpoH1::aadA rpoH2::aacCI (AB9) mutant backgrounds by transduction.
Assay of β-glucuronidase activity.
Cells were collected for β-glucuronidase (GUS) assays at the indicated times and frozen at −80°C until assayed for activity. The cells were permeabilized with lysozyme (200 μg ml−1, 37°C for 10 min), and β-glucuronidase activity was assayed using p-nitrophenyl-β-d-glucuronide as described previously (21). GUS activity is expressed in nanomoles per minute per OD595 unit × 1,000.
Construction of groESL3 and groEL4 null mutants.
To disrupt the groESL3 operon, a 3.3-kb DNA fragment containing groESL3 and flanking DNA was amplified from chromosomal DNA by PCR and inserted into pCR-Blunt II-TOPO using the Zero Blunt TOPO Cloning kit (Invitrogen). The fragment was removed using XbaI and XhoI restriction sites generated by the primers and cloned into pBluescript II KS(−) (Stratagene), resulting in pAF2. A 1.6-kb fragment containing the tet gene encoding tetracycline resistance was amplified from pBR322 DNA and inserted into pAF2 digested with HindIII and blunted with Klenow, deleting 1,606 bp of groESL3. The resulting plasmid, pAF3, contains 161 bp of the 5′ end of groES3, the tet gene in the same orientation as groESL3, and 233 bp of the 3′ end of groEL3. The groESL3Δ::tet construct was removed from pAF3 as an XbaI-XhoI fragment and inserted into pJQ200SK, creating pAF4. This plasmid contains the sacB gene from Bacillus subtilis, allowing negative selection in gram-negative bacteria when grown on sucrose (15, 38). To construct a strain carrying the groESL3 deletion in the chromosome, pAF4 was introduced into Rm1021 by triparental mating. A double recombination event was selected by plating on medium containing tetracycline and 5% sucrose, followed by screening for gentamicin sensitivity to confirm the absence of pJQ200SK. The resulting strain, AF14 (groESL3Δ::tet), was confirmed by Southern analysis.
To disrupt the groEL4 gene, we used a recombinational method (20). The ORFs flanking groEL4, SMc01757 and SMc01759, were transferred from entry plasmids pESmc01757 and pESmc01759 (41) to destination vectors pMK2016 and pMK2017, respectively, by using lambda recombination in vivo as described above. The resulting plasmids were sequentially introduced into Rm1021 by triparental mating and integrated by single reciprocal recombination, creating KB113 (SMc01757::pKB101 SMc01759::pKB100), as confirmed by Southern and PCR analyses. The plasmids contain FLP recombinase target sequences oriented such that expression of FLP recombinase results in deletion of the region between the two ORFs, leaving a single FLP recombinase target sequence. pBH474, which expresses FLP recombinase, was introduced into KB113 by triparental mating, and the cells were grown without selection for pKB100 and pKB101. Multiple colonies were screened for the loss of the drug markers associated with pKB100 and pKB101, indicating a deletion event. pBH474, which contains the sacB gene which is lethal in the presence of sucrose, was removed by streaking the cells on plates containing 5% sucrose and then screening for loss of the drug resistance marker. The deletion in the resulting strain VO3193 (groEL4Δ) was confirmed by PCR analysis. Since the adjacent ORFs are oriented with the stop codons proximal to groEL4, the deletion removes groEL4 and the adjacent intergenic region but leaves SMc01759 and SMc01757 intact.
RESULTS
Suppression of the rpoH1 and rpoH1 rpoH2 mutant defects.
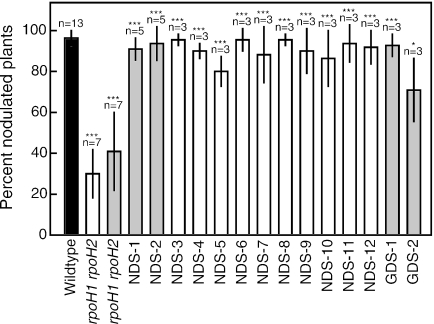
The rpoH1 rpoH2 double mutant RmHM9 was reported to be Nod− (36). Using our rpoH1 and rpoH2 mutant alleles (35), we generated two isolates of an rpoH1 rpoH2 double mutant (VO3148 and VO3149). When we inoculated Medicago sativa GT13R plus alfalfa plants under our growth conditions with these strains, as well as RmHM9, we found that the double mutants varied greatly in the ability to nodulate plants from experiment to experiment (average of 36% nodulated plants with a range from 10 to 78%) (Fig. 1). There were two possible explanations for the variability: either the rpoH1 rpoH2 phenotype is leaky, or the nodules contain suppressor mutants. To distinguish between these possibilities, we isolated bacteria from 12 nodules elicited by the rpoH1 rpoH2 mutants in two independent experiments and confirmed that both mutations were still present (data not shown). We used these strains to inoculate alfalfa and found that they were similar to the wild type in nodulation efficiency (Fig. 1) although still defective in nitrogen fixation (data not shown). Therefore, the nodules are due to suppressor mutants, which we call NDS for “nodulation defect suppressor.”
FIG. 1.
Nodulation by suppressor mutant strains. Alfalfa plants were inoculated with control and suppressor mutant strains, and the percentage of nodulated plants was determined after at least 3 weeks of incubation. The graph depicts the average percentage of nodulation over the indicated number of experiments, and error bars represent the sample standard deviation. At least 50 plants in total were inoculated with each bacterial strain. White bars indicate strains derived from the rpoH1 rpoH2 mutant strain VO3148, and gray bars indicate strains derived from the rpoH1 rpoH2 mutant strain VO3149. Significance was determined using the Student's t test. ***, P < 0.001; *, P < 0.05. The strains from left to right were Rm1021, VO3148-VO3151, AB35-AB44, VO3170, and VO3166.
Cells containing an rpoH1 mutation grow more slowly than wild-type cells in LB medium at 30°C (35). Because we were concerned about the generation of suppressor mutations, we looked for conditions in which the rpoH1 mutant cells would grow as well as the wild-type cells. We switched to LB medium supplemented with MgSO4 and CaCl2 (LB/MC) (17). As shown in Fig. 2, rpoH1 rpoH2 mutant cells grow like wild-type cells at 30°C in LB/MC but display a severe growth defect at 40°C.
FIG. 2.
Comparison of the growth of suppressor mutant cells with wild-type and rpoH1 rpoH2 double mutant cells, as measured using OD595. Cells were grown in LB/MC medium plus streptomycin at 30°C (A) or 40°C (B). The control strains are Rm1021 (wild type; filled circles), VO3148 (rpoH1 rpoH2; filled triangles), and VO3149 (rpoH1 rpoH2; filled diamonds), and the suppressor mutant strains are VO3150 (NDS-1; open diamonds), AB35 (NDS-3; open triangles), VO3170 (GDS-1; crosses), and VO3166 (GDS-2; plus signs). The panels show representative data from one of four experiments.
To determine if the rpoH1 rpoH2 nodulation suppressor mutations also suppressed the growth defect, we compared the growth of two independent suppressors strains (NDS-1 and NDS-3) to the rpoH1 rpoH2 parent strains (VO3149 and VO3148, respectively) at 30°C and 40°C in LB/MC. As shown in Fig. 2, NDS-1 grows slightly more poorly than the wild type at 30°C, whereas NDS-3 is indistinguishable. At 40°C neither NDS-1 nor NDS-3 cells grow as well as wild-type cells, but NDS-1 cells do grow better than the rpoH1 rpoH2 parent strain. To determine if the difference was significant, we compared the amount of growth as measured by OD595 at 48 h and performed the Student's t test. The OD595 of NDS-1 at 48 h was significantly higher than that of the double mutant parent (P < 0.05), whereas NDS-3 was not significantly different.
In E. coli, suppressors of the rpoH growth defect are readily obtained by plating rpoH mutant cells at 30 to 40°C (25). By streaking for single colonies, we found that wild-type S. meliloti cells form colonies on LB/MC plates at 42°C, whereas cells containing an rpoH1 mutation do not (data not shown). Therefore, to select for suppressor mutants we plated rpoH1 and rpoH1 rpoH2 mutant cells at high density at 42°C and obtained colonies. Many of the mutations were not stable, such that the ability to grow at 42°C was lost upon streaking for single colonies at 30°C or 42°C. However, by selecting for growth at 42°C multiple times, we obtained two independent, stable suppressor mutants that we called GDS-1 (rpoH1 background) and GDS-2 (rpoH1 rpoH2 background) for “growth defect suppressor.” To facilitate characterization of GDS-1 for suppression of the growth defect as well as the nodulation defect, we introduced the rpoH2 mutation into the cells by generalized transduction so that all of the suppressor mutants were in the rpoH1 rpoH2 background. To determine if these plate growth defect suppressor mutants also suppressed the growth defect in liquid medium, we grew the strains at 30°C and 40°C in LB/MC. The suppressor mutant cells grew better than the rpoH1 rpoH2 mutant cells at 40°C but not as well as the wild type (Fig. 2). We compared the amount of growth as measured by OD595 at 48 h and performed the Student's t test. The OD595 at 48 h was significantly higher than that of the rpoH1 rpoH2 parent strain for both GDS-1 (P < 0.001) and GDS-2 (P < 0.05).
To determine whether the growth defect suppressor mutations also suppress the nodulation and nitrogen fixation defects, we inoculated alfalfa plants with GDS-1 and GDS-2. The growth defect suppressor mutants nodulated alfalfa plants at levels significantly higher than those of the parent strains (Fig. 1), indicating suppression of the nodulation defect. However, the strains were still unable to fix nitrogen (data not shown).
In E. coli, an rpoH mutant cannot grow above 20°C (49). Suppressor mutants selected at 30°C to 40°C display increased expression of the groESL operon, and suppressor mutants selected at 42°C display increased expression of both groESL and dnaK. The increased transcription and subsequent synthesis of these HSPs in the suppressor mutants is independent of heat shock, such that high levels are observed at 30°C, unlike in wild-type cells (25). To test whether our NDS or GDS suppressor mutants function by a similar mechanism, we grew cells to mid-log phase at 30°C and performed Western blot analysis for GroEL and the DnaK/DnaJ chaperone complex using polyclonal antibodies to the E. coli proteins (Fig. 3). Each antibody recognized a major band of the appropriate molecular weight in S. meliloti cell extracts. In the case of GroEL, we know that the polyclonal antibody recognizes the S. meliloti GroEL1, GroEL2, and GroEL5 proteins (A. N. Bittner and V. Oke, unpublished results). It is likely that the antibody also recognizes GroEL3 and GroEL4, since all of the S. meliloti GroEL proteins are 57 to 62% identical to E. coli GroEL. Although the level of total GroEL appears lower in the rpoH1 rpoH2 mutant than in the wild type and the level of DnaK appears higher in the rpoH1 and rpoH1 rpoH2 mutants than in the wild type, the differences were not significant by the Student's t test and were just significant using the Wilcoxon rank sum test (P = 0.0496).
FIG. 3.
Western analysis of heat shock proteins in suppressor mutant strains. Cells were grown to mid-log phase in LB/MC medium plus streptomycin at 30°C. Equal numbers of cells as measured by OD595 were resuspended in buffer and sonicated. Equal volumes of cell extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting using primary antibodies generated to the following E. coli proteins: GroEL (A), DnaK (B), and DnaJ (C). A representative immunoblot is shown. Graphs depict the mean band intensity normalized to protein concentration and relative to the wild-type signal, with error bars representing the sample standard deviation (n = 3). Gray bars indicate strains with the rpoH1 mutant background, and white bars indicate strains with the rpoH1 rpoH2 double mutant background. The strains from left to right were Rm1021, VO3128, VO3148, VO3150, VO3151, AB35, AB36, VO3165, and VO3166.
Analysis of the suppressor mutants shows that GroEL, DnaK, and DnaJ protein levels were not significantly higher in the mutants compared to those of the rpoH1 and the rpoH1 rpoH2 parent strains by the Student's t test and the Wilcoxon rank sum test. However, it is possible that a small increase of one particular GroEL protein is masked by GroEL1, since groESL1 is expressed at the highest levels (Fig. 4). In the case of DnaJ, there was a significant decrease in protein levels relative to the rpoH1 rpoH2 double mutant in NDS-1 (P < 0.01) and NSD-2 (P < 0.05). Since the S. meliloti suppressor mutants do not exhibit the increased production of GroEL and DnaK seen in the E. coli suppressor mutants, the suppression appears to function by a different mechanism.
FIG. 4.
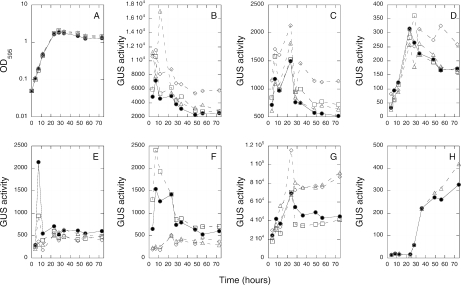
groESL and rpoH expression in rpoH mutant cells grown in M9 sucrose minimal medium. Growth as measured by OD595 and gene expression as monitored by β-glucuronidase (GUS) activity were determined in wild-type (filled circles), rpoH1 (open triangles), rpoH2 (open squares), and rpoH1 rpoH2 (open diamonds) backgrounds. (A) Representative growth curve of cells containing the groEL1-gus fusion. All of the strains in the experiment showed indistinguishable growth patterns. (B to H) GUS activity of cells containing groEL1-gus (B), groEL2-gus (C), groEL3-gus (D), groEL4-gus (E), groEL5-gus (F), rpoH1-gus (G), and rpoH2-gus (H). Each panel shows the data from one representative experiment.
Overexpression of groESL1 or groESL3 does not suppress the mutant phenotypes.
In E. coli, expression of groESL from a multicopy plasmid is sufficient to suppress the temperature-sensitive phenotype of the rpoH mutant (25). In S. meliloti, we know that GroEL/GroES affects NodD activity and that groESL1 mutants form Fix− nodules. We hypothesized that the defects observed for the S. meliloti rpoH1 single mutant and rpoH1 rpoH2 double mutant might be suppressed by overexpression of groESL. Therefore, we created constructs to express groESL1 and groESL3 independently of any possible RpoH control. We chose groESL1 because it is highly expressed (33) (Fig. 4) and the mutant displays a Fix− phenotype (33), and we chose groESL3 because it is the most divergent of the five groESL operons. We chose the E. coli lac promoter and the S. enterica serovar Typhimurium trp promoter because both act constitutively in S. meliloti, they have been successfully used to overexpress other genes in S. meliloti (4, 8, 12, 13), and expression from these promoters on a multicopy plasmid is stronger than expression from the endogenous groESL1 and groESL3 promoters (data not shown). Each construct was introduced separately into wild-type, groEL1, rpoH1, and rpoH1 rpoH2 cells.
To determine whether expression of groESL1 or groESL3 bypasses the symbiotic phenotypes of the rpoH1 and rpoH1 rpoH2 mutants, we inoculated alfalfa seedlings with wild-type and mutant bacteria containing the expression constructs. As shown in Table 2, none of the constructs altered nodulation or nitrogen fixation in the wild-type strain. Both groESL1 constructs were able to complement the Fix− phenotype of the groEL1 mutant. Therefore, these constructs produce active GroEL1 protein. Complementation required the lac promoter (data not shown), confirming that the groESL1 fragment does not contain the endogenous promoter. Neither groESL1 construct was able to suppress the Fix− phenotype of the rpoH1 mutant (Table 2). In terms of the nodulation defect of the rpoH1 rpoH2 double mutant, Plac-groESL1 did not suppress the defect but Ptrp-groESL1 elicited an increase in the number of nodulated plants. However, the nodulation defect was still apparent in the low number of nodules per nodulated plant, which was similar to the rpoH1 rpoH2 mutant. Therefore, overexpression of groESL1 is only able to partially bypass the Nod− phenotype of the rpoH1 rpoH2 mutant and has no effect on the Fix− phenotype of the rpoH1 mutant.
TABLE 2.
Symbiotic phenotypes of strains overexpressing groESL
| Strain/construct | % Plants nodulateda | No. of nodules/nodulated plantb | Fixation status |
|---|---|---|---|
| Wild type | 98 ± 4 | 3.9 ± 1.9 | + |
| Wild type/Plac-groESL1 | 98 ± 3 | 3.8 ± 1.6 | + |
| Wild type/Ptrp-groESL1 | 98 ± 3 | 3.8 ± 2.1 | + |
| Wild type/Plac-groESL3 | 98 ± 3 | 4.0 ± 1.9 | + |
| Wild type/Ptrp-groESL3 | 100 ± 0 | 3.6 ± 2.0 | + |
| groEL1 | 98 ± 3 | 3.7 ± 2.4 | − |
| groEL1/Plac-groESL1 | 98 ± 3 | 3.6 ± 1.8 | + |
| groEL1/Ptrp-groESL1 | 97 ± 6 | 3.6 ± 1.8 | + |
| groEL1/Plac-groESL3 | 90 ± 14 | 3.2 ± 1.8 | − |
| groEL1/Ptrp-groESL3 | 95 ± 6 | 3.4 ± 2.0 | − |
| rpoH1 | 96 ± 4 | 3.7 ± 2.2 | − |
| rpoH1/Plac-groESL1 | 90 ± 4 | 3.7 ± 2.3 | − |
| rpoH1/Ptrp-groESL1 | 100 ± 0 | 3.7 ± 2.0 | − |
| rpoH1/Plac-groESL3 | 97 ± 6 | 3.8 ± 2.2 | − |
| rpoH1/Ptrp-groESL3 | 100 ± 0 | 3.1 ± 1.8 | − |
| rpoH1 rpoH2 | 38 ± 15 | 1.8 ± 1.2 | − |
| rpoH1 rpoH2/Plac-groESL1 | 35 ± 13 | 1.4 ± 0.8 | − |
| rpoH1 rpoH2/Ptrp-groESL1 | 74 ± 23 | 1.8 ± 1.0 | − |
| rpoH1 rpoH2/Plac-groESL3 | 31 ± 30 | 1.3 ± 0.5 | − |
| rpoH1 rpoH2/Ptrp-groESL3 | 47 ± 15 | 1.7 ± 1.1 | − |
Shown are averages and standard deviations (n ≥ 3).
Shown are averages and standard deviations of total nodulated plants from all experiments (n ≥ 3).
Neither the Plac-groESL3 nor the Ptrp-groESL3 construct suppressed the symbiotic phenotypes of the groEL1, rpoH1, and rpoH1 rpoH2 mutants. Since a groESL3 mutant has no phenotype (see below), we could not do a complementation test to prove that our groESL3 constructs were producing active protein. Therefore, to determine whether the groESL3 constructs were functional, we first sequenced the Plac-groESL3 construct and confirmed that no mutations were introduced during amplification of groESL3. We then used site-directed mutagenesis to insert codons generating a hexahistidine tag at the carboxy terminus of GroEL3. Subsequent detection using the SuperSignal West HisProbe kit (Pierce) showed that the Plac-groESL3 construct produced protein (data not shown). Therefore, our groESL3 constructs probably produce active proteins. Thus, the results suggest that groEL3 is not interchangeable with groEL1 and that groESL3 does not bypass the symbiotic phenotypes of the rpoH1 and rpoH1 rpoH2 mutants.
In addition to the symbiotic phenotype, the rpoH1 mutant displays a high-temperature growth defect (Fig. 2). Given that overexpression of groESL bypasses the growth defect of the rpoH mutant in E. coli (25), we tested whether our groESL1 or groESL3 constructs could bypass the growth defect of the rpoH1 mutant. There was no significant increase in growth of cells grown in LB/MC at 30°C or 40°C when the constructs were present (data not shown).
Control of groESL and rpoH gene expression by RpoH1 and RpoH2 under free-living conditions.
To test if RpoH1 or RpoH2 controls expression of the various groESL genes, we constructed a matched set of chromosomal groEL-gus transcriptional fusions by recombinational cloning as described in Materials and Methods. We compared expression of the groEL-gus fusions in wild-type, rpoH1, rpoH2, and rpoH1 rpoH2 cells during growth in M9 sucrose medium (Fig. 4). All five groEL-gus fusions generated GUS activity above background levels, with groEL1 being the most highly expressed. Neither rpoH1 nor rpoH2 was required for expression of groEL1, groEL2, groEL3, or groEL4. However, rpoH1 was required for full expression of groEL5. Similar results were obtained with the rpoH1 mutant grown in LB/MC medium, although we did additionally observe a slight decrease in groEL3 and groEL4 expression (data not shown).
Although rpoH1 is expressed within root nodules, we have been unable to test directly whether RpoH1 controls expression of the groEL genes during symbiosis, because rpoH1 mutant cells undergo early senescence within the nodule (28). Therefore, it is possible that RpoH1 directs transcription of a different subset of these genes within the host plant.
We also tested whether RpoH1 or RpoH2 is autoregulatory or controls expression of the other. rpoH1-gus expression was not dependent on rpoH1 or rpoH2, and rpoH2-gus expression was not dependent on rpoH1 (Fig. 4). In the case of rpoH1-gus, deletion of rpoH1 allows increased expression, perhaps reflecting reduced competition for core RNA polymerase by other sigma factors. We were unable to determine whether RpoH2 was autoregulatory, because our rpoH2-gus fusion, which does not disrupt rpoH2, is located at the rpoH2 locus.
Mitsui et al. (28) defined a consensus sequence for rpoH1-dependent promoters in S. meliloti (cnCTTgAA-n17-CCAnaT) based on the rpoH1-dependent promoters of groES5, lon, and clpB. We used the program dna-pattern (43) to look for the consensus sequence upstream of the five groE loci, rpoH1, and rpoH2. The sequence is only upstream of groESL5, which is consistent with the experimental data.
groESL3 and groEL4 are not required for successful symbiosis.
During symbiosis, groEL1 is required for nitrogen fixation (33), but groEL2 (32) and groESL5 (28) are not required for either nodulation or nitrogen fixation. To determine if the other groESL genes are necessary for symbiosis, we constructed deletions of groESL3 and groEL4 as described in Materials and Methods. Both groESL3 and groEL4 mutant cells were capable of eliciting Fix+ nodules at wild-type levels on alfalfa.
DISCUSSION
A simple hypothesis to explain why RpoH1 is required for growth at high temperature and RpoH1 and RpoH2 are required for nodulation and nitrogen fixation during the S. meliloti-alfalfa symbiosis is that the transcription factors are required for the expression of one or more of the groESL operons and that production of GroEL/GroES is the crucial function. This hypothesis is based on two observations. First, in E. coli groESL is a key target of RpoH. This has been concluded because mutants that suppress the growth defect of rpoH overexpress groESL, and expression of groESL from a multicopy plasmid is sufficient to allow rpoH mutant cells to grow up to 40°C (25). Second, in S. meliloti, groESL1 mutants are delayed in nodulation and form Fix− nodules (33). However, several lines of evidence suggest that this hypothesis is not correct. First, suppressor mutants of the high-temperature growth defect and the nodulation defect do not exhibit increased production of total GroEL protein. Second, overexpression of groESL1 or groESL3 from constitutive promoters does not bypass the defects of the rpoH mutants. Third, at least under free-living conditions (heat shock in Mitsui et al. [28] and rich and minimal media in this study), RpoH2 does not control any of the groESL genes and RpoH1 only controls expression of groESL5. However, groESL5 is not required for nodulation or nitrogen fixation (28). Therefore, groESL5 cannot be a single key target. We conclude that the system is unlike E. coli, which is not surprising given the greater complexity, and that there must be other crucial targets of RpoH1 and RpoH2.
What genes might be under the control of RpoH1 and RpoH2? We envision two scenarios that could be true for either protein. First, the requirements for RpoH could solely be due to the need for properly folded proteins. The requirement during symbiosis may reflect the need to fold specific proteins induced during symbiosis and/or to respond to an increase in unfolded proteins due to stress within the nodule. The regulon would, therefore, be similar to that in E. coli. Second, although RpoH may direct expression of the classic HSPs, the requirement may reflect expression of other genes, perhaps specific to Rhizobium. For example, rpoH2 in Rhizobium sp. strain TAL1145 regulates genes for exopolysaccharide synthesis, which is required for effective nodulation (24). Mitsui et al. (28) determined whether RpoH1 and RpoH2 control expression of nine HSP homologs in S. meliloti (groESL1 through groESL5, dnaK, clpA, clpB, and lon) during heat shock. RpoH1 controlled expression of groESL5 and partially controlled expression of clpB and lon. In contrast, RpoH2 did not control expression of any of these genes. Therefore, the regulon of RpoH1 at least overlaps with the regulon of RpoH in E. coli, but genes under the control of RpoH2 are currently unknown. Microarray experiments to determine the regulons of RpoH1 and RpoH2 in S. meliloti should be illuminating.
Three different phenotypes are associated with rpoH1 and rpoH2 in S. meliloti. The rpoH1 mutant has a growth defect at high temperature and forms ineffective nodules on plants (35, 36), and the rpoH1 rpoH2 double mutant is unable to nodulate (36). Our suppressor mutant analysis suggests that the requirements for RpoH are not the same for all of the phenotypes. We have isolated spontaneous suppressor mutants based on the ability to grow at high temperature (bypassing RpoH1) or to nodulate (bypassing RpoH1 and RpoH2). Interestingly, regardless of how they were initially isolated, most of these mutants are able to suppress both the growth and nodulation defects. In contrast, none of our suppressor mutants are able to suppress the nitrogen fixation defect. This implies that the functions of RpoH1 during growth and RpoH1/RpoH2 during the early stages of symbiosis are similar but that there is a different or additional function of RpoH1 needed later during symbiosis. We do not know what has been altered in these suppressor mutants, although we have shown that the GroEL and DnaK/DnaJ chaperones are not overproduced. Analysis of the differences between the suppressor mutants and the parent strains should provide clues about the roles of RpoH1 and RpoH2 during free-living growth and symbiosis.
Although we cannot explain the requirements for RpoH1 and RpoH2 during symbiosis as a requirement for expression of groESL, the presence of multiple groESL genes and the connections to symbiosis make this gene family particularly interesting in the Rhizobiaceae. All of the nodule-forming rhizobia that have been fully sequenced (S. meliloti, B. japonicum, and M. loti), as well as R. leguminosarum, contain multiple groESL genes. Although many single and double groESL mutants do not have symbiotic defects, some mutants do (11, 28, 32, 39). In S. meliloti, groESL1 mutants form nodules late and the nodules are Fix− (33), and in B. japonicum a groESL3 groESL4 double mutant is unable to fix nitrogen (11). What roles do these genes play in symbiosis? In S. meliloti, genetic and biochemical studies have demonstrated that two key regulatory proteins necessary for early gene expression during symbiosis, NodD1 and NodD3, are substrates of GroEL/GroES (33, 47). In addition, later during symbiosis, GroEL/GroES may help to form active nitrogenase. In B. japonicum, the level of nitrogenase subunits in the groESL3Bj mutant is greatly decreased, although transcription of the genes is unaffected (11). In the free-living bacterium Klebsiella pneumoniae, GroEL regulates nitrogen fixation, possibly as a result of direct interactions with the regulatory protein NifA and nitrogenase subunits (18, 19). Finally, the GroEL/GroES chaperone complex may help to fold other proteins that are newly produced as the cells adapt and differentiate within the plant host.
Currently there is no clear reason why multiple groESL genes are present in these genomes. One possibility is that the genes are simply regulated differentially, providing GroES and GroEL under different conditions. Evidence for differential gene expression has been obtained in S. meliloti (28), B. japonicum (1, 10), and R. leguminosarum (39). Specifically in S. meliloti, only groESL1 and groESL5 are induced by heat shock (28), only groESL5 is controlled by RpoH1 (28 and this study), and only groESL1 and groESL2 are preceded by a CIRCE (controlling inverted repeat of chaperone expression) element that may indicate regulation by the HrcA repressor, which is used to regulate heat-inducible genes in some bacteria (29). An additional possibility is that the encoded chaperones have different ranges of substrates. Although the GroEL/GroES complex can assist in the folding of a wide variety of proteins, it cannot function universally. Directed evolution studies have demonstrated that small numbers of amino acid changes in GroES and GroEL can lead to shifts in the spectrum of substrates (45). Therefore, multiple groESL genes may allow the cell to fold a wider variety of proteins. As an extreme example, bacteriophage T4 encodes a protein of little sequence similarity to GroES that nevertheless substitutes for the host GroES, generating a new chaperone complex that can fold the major capsid protein (2, 42). In R. leguminosarum, the three GroEL proteins have different in vitro properties, including the ability to refold a specific denatured substrate (16). We found that groESL3 is not interchangeable with groESL1, which would be consistent with different substrate specificities, whereas groESL2 is interchangeable with groESL1 (33), suggesting at least overlapping substrate specificities for that pair. As an added complexity, heteromeric complexes as well as homomeric complexes might be made, which would dramatically increase the number of different types of GroEL/GroES chaperones within the cells.
With the construction of the groESL3 and groEL4 mutations described in this paper, we now have mutations in all of the groESL operons in S. meliloti. Although the only single mutant that has a symbiotic defect is groESL1 (28, 32, 33, and this study), we know that all of the genes are expressed within the nodule (5, 6, 32, and A. N. Bittner and V. Oke, unpublished). In addition, previous work has shown that S. meliloti cells need either groESL1 or groESL2 in order to be viable (32). It will be interesting to determine if the two possible quadruple mutants can be constructed and, if so, whether they are proficient in symbiosis.
Acknowledgments
We thank Kristen Butela, Amanda Foltz, and Adam Retchless for help with strain construction and Élan Alford for help with nodulation experiments. We thank Michael Kahn and Brenda Schroeder for providing entry plasmids with S. meliloti ORFs and the gfp-gus destination vector pMK2030 prior to publication, for the strains for pentaparental recombination matings, and for advice on recombinational cloning. We thank Ghideon Ghebregiorgis at The Center for Statistics at the University of Pittsburgh for advice on statistical analysis.
This work was supported by award 2001-35319-10902 from the NRI Competitive Grants Program/USDA to V.O.
REFERENCES
- 1.Babst, M., H. Hennecke, and H.-M. Fischer. 1996. Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol. Microbiol. 19:827-839. [DOI] [PubMed] [Google Scholar]
- 2.Bakkes, P. J., B. W. Faber, H. van Heerikhuizen, and S. M. van der Vies. 2005. The T4-encoded cochaperonin, gp31, has unique properties that explain its requirement for the folding of the T4 major capsid protein. Proc. Natl. Acad. Sci. USA 102:8144-8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, M. J., V. Oke, and S. R. Long. 2000. New genetic tools for use in the Rhizobiaceae and other bacteria. BioTechniques 29:240-245. [DOI] [PubMed] [Google Scholar]
- 4.Barnett, M. J., J. A. Swanson, and S. R. Long. 1998. Multiple genetic controls on Rhizobium meliloti syrA, a regulator of exopolysaccharide abundance. Genetics 148:19-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett, M. J., C. J. Toman, R. F. Fisher, and S. R. Long. 2004. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. USA 101:16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, A., H. Bergès, E. Krol, C. Bruand, S. Rüberg, D. Capela, E. Lauber, E. Meilhoc, F. Ampe, F. J. de Bruijn, J. Fourment, A. Francez-Charlot, D. Kahn, H. Küster, C. Liebe, A. Pühler, S. Weidner, and J. Batut. 2004. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant-Microbe Interact. 17:292-303. [DOI] [PubMed] [Google Scholar]
- 7.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dréano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Pühler, B. Purnelle, U. Ramsperger, C. Renard, P. Thébault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egelhoff, T. T., and S. R. Long. 1985. Rhizobium meliloti nodulation genes: identification of nodDABC gene products, purification of nodA protein, and expression of nodA in Rhizobium meliloti. J. Bacteriol. 164:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer, H.-M., M. Babst, T. Kaspar, G. Acuña, F. Arigoni, and H. Hennecke. 1993. One member of a groESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 12:2901-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, H.-M., K. Schneider, M. Babst, and H. Hennecke. 1999. GroEL chaperonins are required for the formation of a functional nitrogenase in Bradyrhizobium japonicum. Arch. Microbiol. 171:279-289. [Google Scholar]
- 12.Fisher, R. F., T. T. Egelhoff, J. T. Mulligan, and S. R. Long. 1988. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 2:282-293. [DOI] [PubMed] [Google Scholar]
- 13.Gage, D. J., T. Bobo, and S. R. Long. 1996. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 178:7159-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, A. P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dréano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thébault, M. Vandenbol, F.-J. Vorhölter, S. Weidner, D. H. Wells, K. Wong, K.-C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 15.Gay, P., D. Le Coq, M. Steinmetz, T. Berkelman, and C. I. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164:918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George, R., S. M. Kelly, N. C. Price, A. Erbse, M. Fisher, and P. A. Lund. 2004. Three GroEL homologues from Rhizobium leguminosarum have distinct in vitro properties. Biochem. Biophys. Res. Commun. 324:822-828. [DOI] [PubMed] [Google Scholar]
- 17.Glazebrook, J., and G. C. Walker. 1991. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 204:398-418. [DOI] [PubMed] [Google Scholar]
- 18.Govezensky, D., E. S. Bochkareva, A. Zamir, and A. S. Girshovich. 1994. Chaperonins as potential gene regulatory factors: in vitro interaction and solubilization of NifA, the nif transcriptional activator, with GroEL. J. Biol. Chem. 269:14003-14006. [PubMed] [Google Scholar]
- 19.Govezensky, D., T. Greener, G. Segal, and A. Zamir. 1991. Involvement of GroEL in nif gene regulation and nitrogenase assembly. J. Bacteriol. 173:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.House, B. L., M. W. Mortimer, and M. L. Kahn. 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 70:2806-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jefferson, R. A., S. M. Burgess, and D. Hirsh. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. S. M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 24.Kaufusi, P. H., L. S. Forsberg, P. Tittabutr, and D. Borthakur. 2004. Regulation of exopolysaccharide synthesis in Rhizobium sp. strain TAL1145 involves an alternative sigma factor gene, rpoH2. Microbiology 150:3473-3482. [DOI] [PubMed] [Google Scholar]
- 25.Kusukawa, N., and T. Yura. 1988. Heat shock protein GroE of Escherichia coli: key protective roles against thermal stress. Genes Dev. 2:874-882. [DOI] [PubMed] [Google Scholar]
- 26.Martin, M. O., and S. R. Long. 1984. Generalized transduction in Rhizobium meliloti. J. Bacteriol. 159:125-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitsui, H., T. Sato, Y. Sato, N. Ito, and K. Minamisawa. 2004. Sinorhizobium meliloti RpoH1 is required for effective nitrogen-fixing symbiosis with alfalfa. Mol. Genet. Genomics 271:416-425. [DOI] [PubMed] [Google Scholar]
- 29.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1-8. [DOI] [PubMed] [Google Scholar]
- 30.Narberhaus, F., P. Krummenacher, H.-M. Fischer, and H. Hennecke. 1997. Three disparately regulated genes for σ32-like transcription factors in Bradyrhizobium japonicum. Mol. Microbiol. 24:93-104. [DOI] [PubMed] [Google Scholar]
- 31.Narberhaus, F., W. Weiglhofer, H.-M. Fisher, and H. Hennecke. 1996. The Bradyrhizobium japonicum rpoH1 gene encoding a σ32-like protein is part of a unique heat shock gene cluster together with groESL1 and three small heat shock genes. J. Bacteriol. 178:5337-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa, J. 1993. Ph.D. thesis. Rhizobium meliloti nod gene regulation: a role for GroEL in the activation of nod gene expression. Stanford University, Stanford, Calif.
- 33.Ogawa, J., and S. R. Long. 1995. The Rhizobium meliloti groELc locus is required for regulation of early nod genes by the transcription activator NodD. Genes Dev. 9:714-729. [DOI] [PubMed] [Google Scholar]
- 34.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-850. [DOI] [PubMed] [Google Scholar]
- 35.Oke, V., B. G. Rushing, E. J. Fisher, M. Moghadam Tabrizi, and S. R. Long. 2001. Identification of the heat shock sigma factor RpoH and a second RpoH-like protein in Sinorhizobium meliloti. Microbiology 147:2399-2408. [DOI] [PubMed] [Google Scholar]
- 36.Ono, Y., H. Mitsui, T. Sato, and K. Minamisawa. 2001. Two RpoH homologs responsible for the expression of heat shock protein genes in Sinorhizobium meliloti. Mol. Gen. Genet. 264:902-912. [DOI] [PubMed] [Google Scholar]
- 37.Platt, R., C. Drescher, S. K. Park, and G. J. Phillips. 2000. Genetic system for reversible integration of DNA constructs and lacZ gene fusions into the Escherichia coli chromosome. Plasmid 43:12-23. [DOI] [PubMed] [Google Scholar]
- 38.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 39.Rodríguez-Quiñones, F., M. Maguire, E. J. Wallington, P. S. Gould, V. Yerko, J. A. Downie, and P. A. Lund. 2005. Two of the three groEL homologues in Rhizobium leguminosarum are dispensable for normal growth. Arch. Microbiol. 183:253-265. [DOI] [PubMed] [Google Scholar]
- 40.Rusanganwa, E., and R. S. Gupta. 1993. Cloning and characterization of multiple groEL chaperonin-encoding genes in Rhizobium meliloti. Gene 126:67-75. [DOI] [PubMed] [Google Scholar]
- 41.Schroeder, B. K., B. L. House, M. W. Mortimer, S. N. Yurgel, S. C. Maloney, K. L. Ward, and M. L. Kahn. 2005. Development of a functional genomics platform for Sinorhizobium meliloti: construction of an ORFeome. Appl. Environ. Microbiol. 71:5858-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Vies, S. M., A. A. Gatenby, and C. Georgopoulos. 1994. Bacteriophage T4 encodes a co-chaperonin that can substitute for Escherichia coli GroES in protein folding. Nature (London) 368:654-656. [DOI] [PubMed] [Google Scholar]
- 43.van Helden, J., B. André, and J. Collado-Vides. 2000. A web site for the computational analysis of yeast regulatory sequences. Yeast 16:177-187. [DOI] [PubMed] [Google Scholar]
- 44.Wallington, E. J., and P. A. Lund. 1994. Rhizobium leguminosarum contains multiple chaperonin (cpn60) genes. Microbiology 140:113-122. [DOI] [PubMed] [Google Scholar]
- 45.Wang, J. D., C. Herman, K. A. Tipton, C. A. Gross, and J. S. Weissman. 2002. Directed evolution of substrate-optimized GroEL/S chaperonins. Cell 111:1027-1039. [DOI] [PubMed] [Google Scholar]
- 46.Wood, D., J. Setubal, R. Kaul, D. Monks, J. Kitajima, V. Okura, Y. Zhou, L. Chen, G. Wood, N. J. Almeida, L. Woo, Y. Chen, I. Paulsen, J. Eisen, P. Karp, D. S. Bovee, P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Zhao, M. Dolan, F. Chumley, S. Tingey, J. Tomb, M. Gordon, M. Olson, and E. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 47.Yeh, K.-C., M. C. Peck, and S. R. Long. 2002. Luteolin and GroESL modulate in vitro activity of NodD. J. Bacteriol. 184:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yura, T., and K. Nakahigashi. 1999. Regulation of the heat-shock response. Curr. Opin. Microbiol. 2:153-158. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, Y.-N., N. Kusukawa, J. W. Erickson, C. A. Gross, and T. Yura. 1988. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor σ32. J. Bacteriol. 170:3640-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]