Abstract
Rhizobium sp. strain NGR234 produces a flavonoid-inducible rhamnose-rich lipopolysaccharide (LPS) that is important for the nodulation of legumes. Many of the genes encoding the rhamnan part of the molecule lie between 87° and 110° of pNGR234a, the symbiotic plasmid of NGR234. Computational methods suggest that 5 of the 12 open reading frames (ORFs) within this arc are involved in synthesis (and subsequent polymerization) of l-rhamnose. Two others probably play roles in the transport of carbohydrates. To evaluate the function of these ORFs, we mutated a number of them and tested the ability of the mutants to nodulate a variety of legumes. At the same time, changes in the production of surface polysaccharides (particularly the rhamnan O antigen) were examined. Deletion of rmlB to wbgA and mutation in fixF abolished rhamnan synthesis. Mutation of y4gM (a member of the ATP-binding cassette transporter family) did not abolish production of the rhamnose-rich LPS but, unexpectedly, the mutant displayed a symbiotic phenotype very similar to that of strains unable to produce the rhamnan O antigen (NGRΔrmlB-wbgA and NGRΩfixF). At least two flavonoid-inducible regulatory pathways are involved in synthesis of the rhamnan O antigen. Mutation of either pathway reduces rhamnan production. Coordination of rhamnan synthesis with rhizobial release from infection threads is thus part of the symbiotic interaction.
Symbioses between rhizobia and legumes result in the development of a highly specialized organ, the nodule. Within nodules, soil bacteria convert to an endo-symbiotic form, the bacteroids, in which atmospheric dinitrogen is reduced to ammonia. As ammonia is toxic, it is rapidly converted to amides or ureides that nourish the host plant. Initiation of nodule formation is controlled at several levels: plant roots excrete various flavonoids, some of which activate bacterial regulators of nodulation gene expression, particularly the NodD proteins (3, 42). NodD-flavonoid complexes interact with conserved promoter regions (nod boxes) that are upstream of most nod genes. Transcription follows, and the enzymatic products of the nod genes direct the synthesis and secretion of Nod factors (lipo-chito-oligosaccharides).
Although it is clear that Nod factors allow rhizobia to penetrate the legume root (6, 46, 47), other carbohydrates as well as proteins are needed for infection thread development and subsequent steps in nodule formation. Among these, cell surface polysaccharides of rhizobia are believed to be involved in infection thread initiation, nodule invasion, and host specificity. Unfortunately, only limited structural data on the cell surface components and on how cell surface changes are regulated during infection are available. Cultured cells of a number of Rhizobium and related species produce two forms of LPS: rough LPS (R-LPS), which consists of a lipid A membrane anchor attached to a core oligosaccharide, and smooth LPS (S-LPS), which includes an O antigen (50). In contrast, K antigens lack a lipid anchor and are structurally distinct from the LPS (49). Both types of cell surface polysaccharide can be separated (based on the presence of the hydrophobic lipid A moiety on the LPS), identified by polyacrylamide gel electrophoresis (PAGE) and differential staining (29). The expression of cell surface antigens is modulated symbiotically, and several polysaccharides are at least partially modified during the transition from free-living cells to bacteroids (18, 26, 28, 51). Noel and collaborators have shown that extracts from both seeds and roots of Phaseolus vulgaris induce structural modifications in the LPSs of Rhizobium etli (8, 9, 38, 39). Changes induced by Proteus vulgaris extracts include loss of antigenicity, decreased abundance of the O antigen polysaccharides, and increased 2-O-methylation of the LPS.
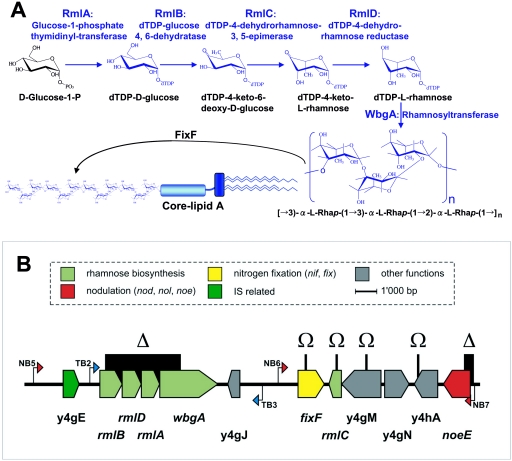
The 87°-110° locus of pNGR234a contains a number of genes involved in flavonoid-inducible modification of cell surface polysaccharides. The proteins encoded by the rmlB-wbgA and fixF genes are necessary for the synthesis of a new, rhamnose-rich, O antigen (35, 48). The short intergenic spacing between the rmlB, rmlD, rmlA, and wbgA open reading frames (ORFs) suggests that they form a small operon (Fig. 1), and the proteins encoded by rmlB, rmlD, and rmlA are predicted to be involved in the synthesis of dTDP-l-rhamnose from d-glucose-1-phosphate (Table 1). Most probably, this rhamnose forms the precursor molecules for the synthesis of the new O antigen. A gene encoding a key enzyme (dTDP-4-dehydrorhamnose 3,5-epimerase) in this synthetic pathway is not found in the rmlB-wbgA cluster; however, an ORF encoding a potential homologue is present approximately 6 kb downstream of wbgA. This ORF, y4gL, has been renamed rmlC in accordance with the nomenclature proposed for enzymes involved in bacterial polysaccharide synthesis (45). How the dTDP-l-rhamnose is then polymerized and exported to form the rhamnan O antigen is less clear. It seems likely, however, that wbgA (encoding a glycosyl transferase) and fixF are involved; a schematic representation of the production of the rhamnan O antigen is shown in Fig. 1A.
FIG. 1.
A. Proposed synthetic pathway of rhamnose and its possible adjunction to the LPS core by enzymes encoded within the 87°-110° locus. The putative roles of the RmlA to -D enzymes in the synthesis of dTDP-l-rhamnose from d-glucose-1-phosphate are shown. A predicted glycosyl transferase, WbgA, could be responsible for the polymerization of the newly synthesized rhamnose residues. FixF is thought to function in the export or attachment of the rhamnose-rich O antigen across the bacterial membrane or onto lipid A core molecules. B. Genetic map of the 87°-110° locus of Rhizobium sp. strain NGR234. Genes are drawn as arrows matching the sense of transcription and are colored according to their proposed function. nod boxes and tts boxes are represented by red and blue arrows, respectively. The positions of the various mutations are shown above the genes as either omega cassette insertions (Ω) or deletions followed by omega cassette insertions (Δ).
TABLE 1.
Properties of proteins encoded by pNGR234a ORFs that lie between NB5 and NB7
| ORF | Function (EC no.) | Transcriptsa | Location and size (kDa)b | Closest homologuec |
|---|---|---|---|---|
| y4gE | Transposase [fragment?] | 0 | Cytosol, 27.7 | M. loti mll9026 (46%, 58%, 1e−27) |
| y4gF (rmlB) | dTDP-glucose 4,6-dehydratase (4.2.1.46) | 1↑(24) | Cytosol, 39.7 | A. tumefaciens RfbB (81%, 90%, 3e−158) |
| y4gG (rmlD) | dTDP-4-dehydrorhamnose reductase (1.1.1.133) | 1↑(24) | Cytosol, 32.2 | A. tumefaciens RfbD (62%, 78%, e−100) |
| y4gH (rmlA) | Glucose-1-phosphate thymidylyltransferase (2.7.7.24) | 1↑(24) | Cytosol, 31.2 | A. tumefaciens RfbA (81%, 90%, e−131) |
| y4gI (wbgA) | Glycosyl transferase [O antigen synthesis protein] | 1-2↑(1-24) | Cytosol, 102.8 | Pseudomonas syringae pv. tomato DC3000 PSPTO 1074 (51%, 69%, 2e−147) |
| y4gJ | No significant homology | 0 | Cytosol, 21.1 | |
| fixF | Involved in O antigen synthesis | 3↑(24) | Cytosol, 45.0 | E. coli KpsS (25%, 42%, 2e−08) |
| y4gL (rmlC) | dTDP-4-dehydrorhamnose 3,5-epimerase (5.1.3.13) | 0 | Cytosol, 21.8 | A. tumefaciens RfbC (72%, 80%, 1e−76) |
| y4gM | ABC transporter ATP-binding protein | 0 | Membrane, 64.3 | A. tumefaciens Atu4600 (60%, 78%, 0) |
| y4gN | Glycosyl transferase [fragment?]d | 0 | Cytosol, 45.0 | Synechococcus sp. strain WH 8102 SYNWO641 (30%, 52%, 8e−15) |
| y4hA | Ionic transporter [Ca2+/H+ antiporter] | 2↑(1) | Membrane, 38.1 | Ralstonia metallidurans COG0387 (45%, 62%, 2e−60) |
| noeE | Sulfyryl transferase (2.8.2.-) | 3↑(1) | Cytosol, 46.6 | Bradyrhizobium elkanii (disrupted) NoeE (54%, 68%, 3e−79) |
Transcript data were taken from Perret et al. (41). Numbers represent the relative transcription intensity: 0, not expressed; 5, high (but not maximum level). The arrow indicates that a transcript is inducible by flavonoids, with the commencement of induction (in hours) shown in parentheses.
The predicted location and size of each ORF were taken from the PEDANT analysis website (http://pedant.gsf.de/).
Homology searches were made using the Blastp program (2) at the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/). The closest homologue is listed, along with the percentages of identity and similarity and then the E value of the homology.
y4gN has homology to the RpgF family of glycosyl transferases from various streptococci. This family is involved in the synthesis of a rhamnose-containing LPS molecule. The conserved domain is drastically truncated, however.
In addition to the appearance of the rhamnan O antigen following flavonoid addition, a transcriptional study showed that rmlB, rmlD, rmlA, wbgA, and fixF are also inducible. Although up-regulation occurred later than genes involved in Nod factor synthesis (e.g., noeE), induction of rmlB, rmlD, rmlA, wbgA, and fixF was not as late as found with those genes that function only within nodules (41). Subsequent work has shown that nod box 6 (NB6) in the promoter upstream of fixF (Fig. 1B) is probably involved in the signal transduction cascade (30). Similarly, a new type of rhizobial cis-acting element called a tts box (TB) has been found upstream of rmlB-wbgA (Fig. 1B). TBs are regulated by TtsI, a two-component regulatory response protein which is also required for type III-dependent protein secretion in NGR234 (35, 56). Thus, there are at least two regulatory pathways governing the production of the rhamnan O antigen.
In this study we have examined other genes in the 87°-110° arc of pNGR234a for their roles in the symbiotic interaction and effects on rhamnan/surface polysaccharide synthesis, as well as extending the functional studies of rhamnan itself.
MATERIALS AND METHODS
Bacterial growth and manipulation.
Escherichia coli and Rhizobium sp. strains along with their relevant characteristics are listed in Table 2. The microbiological techniques used have been described by Lewin et al. (34). Analysis of DNA and cloning and sequencing procedures have been published (19, 46). Various mutants of Rhizobium sp. strain NGR234 were constructed either by inserting an antibiotic resistance omega cassette into specific genes (giving NGRΩtarget gene strains) or by deleting a restriction fragment internal to the target gene and replacing it with an omega antibiotic resistance cassette (resulting in NGRΔtarget gene strains) using standard techniques (12). Promoter constructs cloned into the broad-host-range reporter vector pMP220 (52) were mobilized into NGR234 and its derivatives by triparental matings using pRK2013 as the helper plasmid (16). Flavonoid induction was performed as follows: rhizobial cultures grown to an optical density at 600 nm (OD600) of 0.5 to 0.6 were diluted to an OD600 of 0.1 in RMS medium and induced with 2 × 10−7 M daidzein. β-Galactosidase activity was assayed according to the methods of Miller (37). The results reported represent the means of at least three independent experiments.
TABLE 2.
Strains, plasmids, and vectors used in this study, along with the mutants produced
| Strain, vector, or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| E. coli DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 recA1 | 24 |
| Rhizobium sp. strains | ||
| NGR234 | Rifr derivative of Rhizobium sp. strain NGR234, isolated from Lablab purpureus | 53 |
| NGRΩfixF | fixF mutant obtained by inserting an omega interposon into the unique EcoRI site of fixF (Rifr Spr) | 48 |
| NGRΩgM | y4gM mutant obtained by inserting an omega interposon into the unique EcoRI site of y4gM (Rifr Spr) | This work |
| NGRΩhA | y4hA mutant obtained by inserting an omega interposon into the unique BglII site of y4hA (Rifr Spr) | This work |
| NGRΩnodD2 | nodD2 mutant obtained by inserting an omega interposon into the unique BamHI site of nodD2 (Rifr Kmr) | 13 |
| NGRΔnoeE | Deletion mutant of noeE of NGR234 (Rifr Spr) | 25 |
| NGRΔrmlB-wbgA | rmlB-wbgA mutant obtained by deleting a 3,732-bp EcoRV fragment of pXB285 and replacing it with an omega interposon (Rifr Kmr) | This work |
| NGRΩrmlC | rmlC mutant obtained by inserting an omega interposon into the unique HindIII site of rmlC (Rifr Spr) | This work |
| NGRsyrM2::uidA | syrM2 mutant obtained by inserting a uidA-aph cassette into the unique SacI site of syrM2 (Rifr Kmr) | 30 |
| NGRΩttsI | ttsI mutant obtained by inserting an omega interposon into the unique ApaI site of ttsI (Rifr Spr) | 56 |
| Vectors and plasmids | ||
| Lorist 2 | 5.6-kb cosmid vector (Kmr) | 21 |
| pBBR1-MCS5 | Broad-host-range cloning vector (Gmr) | 31 |
| pBluescript KS(+) | Phage f1, lacαZ+ (Apr) | Stratagene, La Jolla, CA |
| pBR-MZHBgM | 2.5-kb fragment containing y4gM cloned into the HindIII-BamHI sites of pBBR1-MCS5, Gmr | This work |
| pBSgFGHI | pKS+ derivative carrying a 5.6-kb BamHI-ApaI fragment covering a region from rmlB (formerly y4gF) to wbgA (formerly y4gI) (Apr) | This work |
| pBSgFGHIΩKm | pBSgFGHI derivative in which the 3.7-kb EcoRV internal fragment containing rmB-wbgA was replaced by an ΩKmr interposon (Apr Kmr) | This work |
| pJQΩgFGHI | pJQ200SK derivative containing a 4.1-kb BamHI-ApaI fragment carrying the 5′ end of y4gF (rmlB), an ΩKmr interposon, and the 3′ end of y4gI (wbgA) (Gmr Kmr) | This work |
| pJQ200 SK | pACYC184-derived (p15A) suicide vector (Gmr) | 44 |
| pJQMZ1 | pJQ200 SK derivative carrying a 3.9-kb PstI fragment from pXB285 and an ΩSpr interposon inserted into the BglII site of y4hA (Gmr Spr) | This work |
| pJQy4gLΩSp | pJQ200 SK derivative carrying a 2.5-kb EcoRV-BamHI fragment and an ΩSpr interposon inserted into the HindIII site of rmlC (formerly y4gL) (Gmr Spr) | This work |
| pJQy4gMΩSp | pJQ200 SK derivative carrying a 4.5-kb cassette containing y4gM in which an ΩSpr interposon was inserted, Gmr Spr | This work |
| pMIGB | 1.4-kb fragment (amplified using the primer pair 5′-GCAGTGGTCTGCCGCATTTC-3′ and 5′-TCCAGTCCGGTCTTGGAGAG-3′) containing the region upstream of y4gM, cloned into pMP220 (Tcr) | This work |
| pMP220 | IncP expression vector containing a promoterless lacZ gene (Tcr) | 52 |
| pMP-NB5 | NB5 cloned into pMP220 as a 1.4-kb XbaI-PstI fragment | 30 |
| pMP-NB6 | NB6 cloned into pMP220 as a 1.1-kb XbaI-PstI fragment | 30 |
| pMP-NB7 | Also called pNBnoeE; NB7 cloned as a 1.0-kb XbaI-BglII fragment in pMP220 | 25 |
| pRK2013 | Helper plasmid containing the ColE1 replicon with RK2 tra genes (Nmr Kmr) | 16 |
| pRK7813 | Broad-host-range Inc P1 cosmid (Tcr) | 27 |
| pRK7813E/B | pRK7813 with EcoRI to BamHI deleted in the polylinker (Tcr) | This work |
| pXB285 | Lorist 2 clone from pNGR234a (Kmr) | 40 |
Ampr, ampicillin resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance; Nmr, neomycin resistance; Rifr, rifampin resistance; Spr spectinomycin resistance; Tcr, tetracycline resistance.
Plant assays.
Seeds were purchased from the suppliers listed by Pueppke and Broughton (43). Nodulation ability was assayed in modified Magenta jars (33). At harvest (6 to 8 weeks after inoculation), the nodules were sterilized, rolled on TY agar (to check for surface contamination), and cut in half. A small portion of the inner bacteroidal tissue was removed from one half and streaked out on TY (to verify the strain), while the other half was used for light and electron microscopy (22, 57).
Polysaccharide preparation and PAGE.
To extract cell-associated polysaccharides, wild-type and mutant strains were grown in RMS (4) to a final OD600 of 0.8. Induced cultures were grown in the presence of 10−6 M apigenin. Pelleted cells were resuspended in water and mixed with an equal volume of hot phenol (65°C). Water and phenol phases were separated by centrifugation and subsequently dialyzed against water. Polysaccharides were separated by PAGE (18% polyacrylamide) using deoxycholic acid as the detergent. Gels were prerun for 10 min (15 mA/gel) prior to loading samples, then run for 40 to 60 min (15 mA/gel), until the buffer front reached the bottom of the gel, and silver stained for LPS (55).
RESULTS
Putative functions of the ORFs in the 87°-110° arc of pNGR234a.
The ORFs within the 87°-110° region are shown in Fig. 1B. As y4gE represents only a part of a putative transposase gene, it was excluded from all further consideration. At the other edge of the locus, immediately downstream of NB7, is noeE, which encodes a sulfyryl transferase necessary for the sulfation of NGR234 Nod factors (25). Earlier DNA sequence comparisons (19) as well as recent (September 2005) BLAST (2) searches suggest functions for many of the 12 ORFs that lie between nod boxes NB5 and NB7 (Table 1). Six of the ORFs probably take part in rhamnose synthesis and production of the flavonoid-inducible rhamnan O antigen (35, 48). Further analysis of rhamnan O antigen production by these ORFs was undertaken, and the role of this new LPS structure was tested in a variety of NGR234 legume hosts. As no evidence was found for the production of transcripts from y4gJ and y4gN (41) and because they had no significant homology to other ORFs, they were not considered further. Flavonoid-inducible transcripts are made from the y4hA locus, which encodes a Ca2+/H+ antiporter, and this locus was also included in the analysis. Finally, although many homologues of ABC transporter genes exist on pNGR234a (as well as in the genomes of other rhizobia) and despite the absence of detectable transcripts, y4gM, which encodes a member of the MsbA subfamily of ABC transporters, was mutated. This subfamily of ABC transporters contains members known to be involved in the export of lipid A from the inner to outer membrane in Escherichia coli (7).
Role of genes lying in the 87°-110° arc in LPS synthesis.
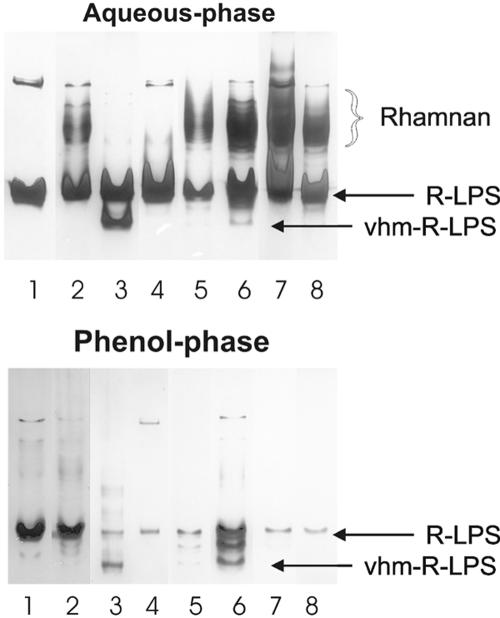
A number of the genes have functions predicted to be involved in polysaccharide synthesis or transport and have been shown to be flavonoid inducible. Demonstration of the parts played in O antigen synthesis by some of the genes was furnished by knocking out eight of the ORFs (rmlB-wbgA, rmlC, y4gM, and y4hA) that lie between NB5 and NB7. All mutants constructed using the omega antibiotic resistance cassettes are collectively called knockout mutants. Two other previously described mutants (NGRΩfixF and NGRΔnoeE) were also included in the analysis (25, 48). To test the role of each gene, rhizobia were extracted with hot phenol-water, and the cell-associated polysaccharides were separated by PAGE. LPSs in the gels were visualized using a highly specific silver stain (Fig. 2). Analysis of the water phase of flavonoid-treated wild-type NGR234 cultures revealed the presence of the rhamnan-containing S-LPS as a dark-staining region that was absent in the control (Fig. 2, aqueous phase, cf. lanes 1 and 2) (48). Noninduced NGR234, in contrast, produces only minor quantities of S-LPS (Fig. 2, aqueous phase, lane 1), in agreement with earlier studies on the structure of NGR234 LPS produced under noninducing conditions (23). The rhamnan was still produced by the knockouts in rmlC, y4gM, y4hA, and noeE, whereas deletion of rmlB to wbgA and mutation in fixF abolished rhamnan synthesis, as reported earlier (35, 48). The presence of rhamnan in the rmlC knockout extractions (Fig. 2, aqueous phase, lane 5) was unexpected as, based upon its homology, it was thought to be essential for rhamnose synthesis (Fig. 1), and this implies that NGR234 must possess another epimerase. Some mutants produced very-high-mobility (vhm) R-LPS, as shown in gels containing extracts of both the H2O and phenol phases (Fig. 2, both phases, lanes 3 and 6). Strain NGRΔrmlB-wbgA produced significant amounts of a single form of vhm-R-LPS, most of which was extracted into the water phase (Fig. 2. aqueous phase, lane 3), while NGRΩgM produced at least three distinct forms of vhm-R-LPS that mostly partitioned into the phenol phase (Fig. 2, phenol phase, lane 6). Minor amounts of vhm-R-LPS were also present in extracts of both phases taken from the mutant NGRΩrmlC. Significantly, the vhm-R-LPSs were not produced by any of the mutants under noninduced conditions (data not shown), suggesting that flavonoids are required for the synthesis of these polysaccharides. Mutation of NGRΔnoeE and NGRΩhA had little obvious effect on the distribution of LPSs as seen in PAGE gels.
FIG. 2.
PAGE analyses of rhamnan-containing LPSs present in the aqueous or phenol phase of hot phenol-water extracts of Rhizobium sp. strain NGR234 (and various mutants thereof). The gels were silver stained for LPS. Lanes contain extracts from the following sources: 1, noninduced wild-type NGR234 (grown in the absence of apigenin); 2, apigenin-induced (IND) NGR234; 3, IND NGRΔrmlB-wbgA; 4, IND NGRΩfixF; 5, IND NGRΩrmlC; 6, IND NGRΩgM; 7, IND NGRΩhA; 8, IND NGRΩnoeE. The position of the rhamnan O antigen produced by induced NGR234 and certain derivatives is indicated. The position of the R-LPS produced by all the strains and present in both phases is also marked. Finally the vhm-R-LPS produced by certain mutants is indicated.
Complex regulation of rhamnan synthesis.
The presence of both NB6 and TB2 upstream of genes involved in the synthesis of the rhamnan O antigen suggest that there are multiple regulatory controls on its production. NB6 is found upstream of fixF and puts FixF production under the control of the NodD1-SyrM2-NodD2 regulatory cascade (30). TB2, which is found upstream of rmlB, has been shown to be required for flavonoid-induced transcription of rmlB and, thus, rhamnose synthesis. TB2 is activated by another regulatory protein, TtsI, which is itself controlled by NodD1 via NB18 (30, 35). We thus examined production of rhamnan by mutants of these regulatory proteins. Although a fixF mutation blocks rhamnan production, mutants of syrM2 and nodD2 still produce some rhamnan (Fig. 3, lanes 2 and 3). Rhamnan was not observed in LPS extracts from the TtsI mutant.
FIG. 3.
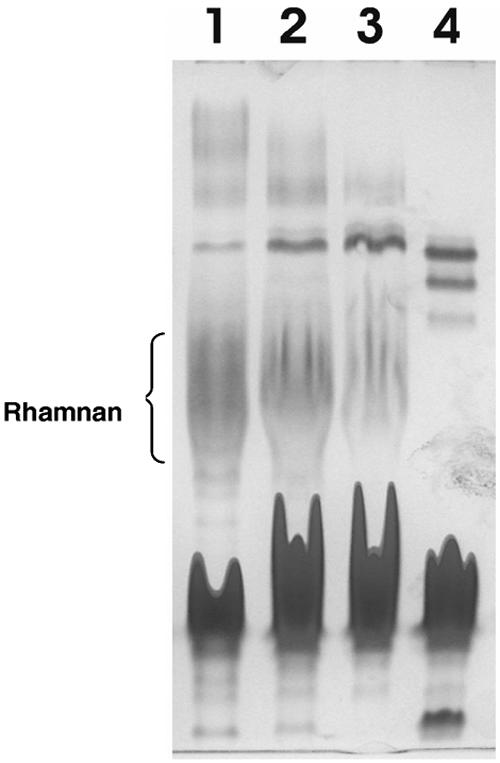
PAGE analyses of rhamnan-containing LPSs present in the aqueous phases of hot phenol-water extracts of Rhizobium sp. strain NGR234 and regulatory mutants involved in transcriptional control of rhamnan production. All cultures were induced with apigenin. Lanes contain extracts from the following sources: 1, wild-type NGR234; 2, NGRsyrM2::uidA; 3, NGRΩnodD2; 4, NGRΩttsI. The position of the rhamnan-containg LPS is indicated in lanes 1 to 3. The gels were silver stained for LPS.
Effects of mutations in the 87°-110° arc on symbiotic phenotypes.
The phenotypes of each mutant were assayed by inoculating them onto five different hosts of NGR234 (Table 3) selected to possess indeterminate nodules (Leucaena leucocephala and Tephrosia vogelii) or determinate nodules (Macroptilium atropurpureum, Pachyrhizus tuberosus, and Vigna unguiculata), as well as to represent different subfamilies of the Leguminosae (all in Papilionoideae except L. leucocephala in Mimosoideae). L. leucocephala and P. tuberosus seemed to be indifferent to the six distinct mutants, although they responded very differently. L. leucocephala formed nitrogen-fixing nodules in all tests, perhaps suggesting that changes in the structures of the LPSs and sulfation of Nod factors are unimportant in the development of mimosoid nodules. P. tuberosus, however, rarely formed nodules with any of the mutants. This plant is a special case, however, as efficient nodulation (between 20 to 40 fixing nodules per plant) is possible only when the type three protein secretion system (T3SS) of NGR234 is inactivated (56). Interestingly, a double mutant of the T3SS and the rhamnose synthetic genes (rmlB-wbgA) does not invoke nodules, suggesting that the rhamnan O antigen is also essential, even if the T3SS block is removed (35). Disruption of the putative ionic transporter y4hA and noeE had no appreciable effects on nodulation relative to the wild-type control (Table 3).
TABLE 3.
Effects of disruption of open reading frames between NB5 and NB7 on nodulation capacity of Rhizobium sp. strain NGR234
| Plantd | Phenotype after inoculation witha:
|
||||||
|---|---|---|---|---|---|---|---|
| NGR234 | NGRΔ rmlB-wbgAb | NGRΩfixF | NGRΩrmlC | NGRΩgM | NGRΩhA | NGRΔnoeE | |
| L. leucocephala (I) | Fix+ | Fix+ | Fix+ | Fix+ | Fix+ | Fix+ | Fix+ |
| M. atropurpureum (D) | Fix+ | Fix+/− | Fix− | Fix+ | Fix− | Fix+ | Fix+ |
| P. tuberosusc (D) | Nod− | Nod− | Nod− | Nod− | Nod− | Nod− | Nod− |
| T. vogelii (I) | Fix+ | Fix− | Fix− | Fix+ | Fix− | Fix+ | Fix+ |
| V. unguiculatan (D) | Fix+ | Fix+/− | Fix− | Fix+ | Fix− | Fix+ | Fix+ |
Results are the means of at least three independent experiments (four Magenta jars per treatment, four plants per jar). Plants were harvested 6 to 8 weeks after inoculation and assessed qualitatively. Leaf color, shoot dry weight, and the presence (or absence) of fixing (pink) nodules per plant were determined at harvest. Inoculated plants with yellow leaves and dry weights similar to those of uninoculated plants were classified as Fix−. Inoculated plants with greenish-yellow leaves and intermediate dry weights were scored as Fix+/−. Fix+ signifies dark green leaves and dry weights equivalent to those of plants inoculated with wild-type NGR234. Nod− means that nodules were not formed.
See text for further description of this inoculant.
P. tuberosus occasionally produced a nodule after inoculation, regardless of the strain, but generally roots appeared uninfected (see text).
dI or D, indeterminate or determinate nodules.
The absence of the rhamnan O antigen (in mutants NGRΔrmlB-wbgA and NGRΩfixF) had profound symbiotic effects on M. atropurpureum, T. vogelii, and V. unguiculata. As reported earlier (35), abolition of rhamnose synthesis led to only nonfixing nodules on T. vogelii and, as shown in this work, NGRΩfixF was similarly unable to efficiently nodulate this legume. NGRΩfixF also only produced nonfixing nodules on M. atropurpureum and V. unguiculata, although the phenotype of NGRΔrmlB-wbgA on these two plants was less severe (Table 3), with the formation of some pink nodules. Thus, despite the common absence of the rhamnan O antigen, there must be other (symbiotically important) differences between these two mutants. The rmlC mutation had no discernible effects on the nodulation, implying that it is not necessary for symbiotically important modifications to LPS structure (despite its predicted role in rhamnose synthesis).
Closer examination of nodules from plants inoculated with NGRΩfixF showed that superficially they were similar to those produced by wild-type strain NGR234, yet light and electron microscopic examination revealed slight differences depending on the plant. Nodulation of M. atropurpureum inoculated with NGRΩfixF was reduced in comparison to the wild type, and the nodules possessed fewer bacteroid-containing cells, which failed to fix nitrogen (data not shown). In contrast, the number of bacteroid-containing cells was appreciably reduced and the cytoplasm of the mutant was extensively degraded (see reference 48).
Mutation of y4gM (NGRΩgM) resulted in nodulation phenotypes reminiscent of NGRΩfixF, i.e., Fix− on M. atropurpureum, T. vogelii, and V. unguiculata. It should be noted that NGRΩgM is still capable of producing the rhamnan O antigen, however (Fig. 2, lane 6). The symbiotic effects of the y4gM mutation are surprising, as this ORF is not obviously under flavonoid control, possesses no known upstream cis-acting promoter elements, and no inducible transcripts were detected in earlier, global transcriptional analyses (41). To shed light on this conundrum, transcription of y4gM was investigated by cloning the intergenic region upstream of y4gM into pMP220 (yielding pMIGB) (see Materials and Methods) (Table 2). β-Galactosidase activities of NGR(pMIGB) transconjugants were unaffected by the addition of inducers but were about 10 times (650 to 700 Miller units) higher than those found in transconjugants harboring the empty vector [NGR(pMP220)]. These data suggest that y4gM is expressed constitutively at low levels. Demonstration that the effects on nodulation were due to disruption of y4gM was shown by complementing NGRΩgM with pBR-MZHBgM, a broad-host-range plasmid carrying a DNA fragment that contains y4gM (as well as a fragment of rmlC). The resulting transconjugant, NGRΩgM(pBR-MZHBgM), was able to induce Fix+ nodules on V. unguiculata roots (data not shown).
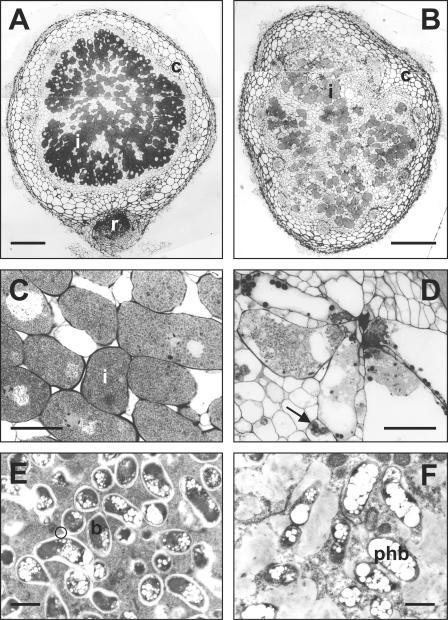
In comparison with the wild type, inoculation of V. unguiculata with NGRΩgM reduced the number of nodules 8- to 10-fold, and those that formed were white inside. Only a few of the central, cortical cells were infected by the mutant (cf. Fig. 4A and B). Those cells that were invaded became enlarged (cf. Fig. 4C and D), and degradation of the cells was evident. Often the bacteroids lacked the peribacteroid membrane (Fig. 4F) and were embedded in material that resembled the matrix of infection threads. In contrast to bacteroids containing NGR234 (Fig. 4E), β-polyhydroxybutyrate accumulated strongly in bacteroids produced by the mutant. Furthermore, the uninfected cells in nodules produced by the mutant nodules were smaller with amyloplasts containing starch grains (Fig. 4D).
FIG. 4.
Structures of Vigna unguiculata nodules formed by Rhizobium sp. strain NGR234 (A, C, and E) and the y4gM mutant NGRΩgM (B, D, and F). (A and B) Low-magnification light micrographs of whole nodules. Magnification, ×35. Bar, 200 μm. (C and D) High-magnification light micrographs of nodule sections. Magnification, ×312. Bar, 32 μm. (E and F) Electron micrographs of bacteroidal tissue. Magnification, ×8,650. Bar, 2 μm. Abbreviations: b, bacteroids; c, nodule cortex; i, infected cells; phb, polyhydroxybutyrate; r, roots. The black arrow points to an amyloplast containing starch granules, and the black circle indicates peribacteroid membranes.
DISCUSSION
As part of our ongoing studies into rhamnan synthesis and function in free-living bacteria, we have examined in more detail the region delimited by NB5 to NB7 of pNGR234a. Of the 12 ORFs identified in this locus, 6 are predicted (or have been shown) to be involved in the synthesis of the flavonoid-inducible rhamnose-rich LPS (rhamnan) (35, 48). Three others have homology to a transposase (y4gE) or do not match other database entries (y4gJ and y4gN) and were excluded from this study. noeE encodes a sulfyryltransferase that modifies NGR234 Nod factors (25). There were no apparent changes to somatic antigens of free-living cells resulting from mutation of noeE, and none of the legumes tested demonstrated any requirement for sulfated Nod factors.
The protein predicted to be encoded by y4hA has homology to calcium proton antiporters from various bacteria, and the ORF itself is inducible by flavonoids (Table 1). Given the important role that calcium ions play during symbiosis (11, 14, 20) and that other rhizobial proteins involved in calcium transport are factors of host specificity (10), y4hA was mutated in this study. The mutant, however, did not cause changes in nodulation of the legumes tested, nor did it appear to affect the somatic antigens. A putative LPS-associated cation exporter has been identified in R. leguminosarum bv. viciae (1) which is thought to transport calcium ions out of the cell to the LPS core. y4hA, being a member of the same transporter family, has homology to CpaA (30% identity and 57% similarity over the total protein), yet here too a mutation in cpaA did not affect the somatic antigens. Unlike y4hA, however, cpaA was shown to be constitutively expressed. The role of calcium ions in the stabilization of the LPS core is still unclear. One hypothesis to explain the function (and induction) of y4hA is that as the somatic antigen undergoes flavonoid-induced changes, y4hA adjusts the quantity of calcium ions associated with the core to stabilize the new molecule.
Half of the genes in the NB5 to NB7 locus are involved in the production of the rhamnan O antigen. The absence of this molecule severely affects nodulation of several legume species, but the exact function of the rhamnan is not known. Our working hypothesis is that a change in the surface chemistry of the somatic antigen occurs during the transition of free-living rhizobia to bacteroids, permitting their correct release from the infection thread and survival within the nodule. The rhamnan (if still present on the bacteroid membrane) may protect the rhizobia within the symbiosome compartment from the plant cytoplasm, allowing bacteroidal persistence for extensive periods of time. Nevertheless, we cannot rule out the possibility that the somatic antigen acts as a signal molecule during symbiosis, as has been claimed for the interaction between Azorhizobium caulinodans and Sesbania rostrata (36). In fact, the rhamnan may have multiple functions, possibly depending upon the plant host, as ultrastructural analyses of nodules produced by NGRΩfixF show differences, such as variable bacteroid content, between different hosts (48; J. Kopciñska and W. Golinowski, unpublished data).
Based upon sequence homology and carbohydrate analysis, the roles of RmlB, RmlD, and WbgA seem to be to synthesize and, possibly, then polymerize rhamnose residues (Fig. 1). The exact function of FixF is more difficult to define, however. One possibility is that FixF may be involved in the transfer of dTDP-l-rhamnose to LPSs, forming the O antigens (48). Yet, FixF may have other functions, such as in the regulation of changes to somatic antigen structure, specific modifications of the somatic antigen core, or even activation of the polymerized rhamnan. It is also possible that FixF could possess several of these activities, which may explain why the symbiotic phenotype of NGRΩfixF is more drastic than that of NGRΔrmlB-wbgA (Table 3) and why there are important differences in the extracted polysaccharides from these two mutants (Fig. 2). Furthermore, direct comparisons of NGRΔrmlB-wbgA and NGRΩfixF in nodulation tests showed that the effects of two strains that are unable to produce rhamnan varied with the host.
The sixth ORF predicted to be involved in rhamnan production, rmlC, is thought to encode the third enzyme in the rhamnose synthetic pathway (a prediction based upon its homology [Table 1 and Fig. 1]). Mutation of rmlC has no effect on the appearance of rhamnan in LPS extracts or on NGR234 symbiosis, however. This suggests that either a second copy of rmlC exists in the NGR234 genome or that another epimerase provides the same function as RmlC. Of the fully sequenced rhizobia, Bradyrhizobium japonicum USDA110 does not contain a rmlC homologue, whereas Mesorhizobium loti MAFF303099 has one copy (35). The pSymB megaplasmid of Sinorhizobium meliloti 1021 possesses two rmlC homologues (17) and, although rmlC homologues were not detected in the partially sequenced megaplasmid of NGR234 (54), it is possible that another gene exists in the NGR234 genome. In fact, searching the draft genome sequence of ANU265 (an NGR234 derivative cured of the symbiotic plasmid and thus lacking rmlC) revealed the presence of another putative ORF encoding a dTDP-4-dehydrorhamnose 3,5-epimerase (data not shown).
Transcriptional control of rhamnan synthesis is complicated, with multiple regulatory proteins involved in the process. After flavonoid induction, the following signaling pathways up-regulate the fixF and rmlB-wbgA genes, which are absolutely required for rhamnan production (30, 35): NodD1 → SyrM2 → NodD2 → FixF, and NodD1 → TtsI → RmlB-WbgA.
Genes regulated this way are thought to be expressed after the bacteria have entered the plant but before they are released into cortical cells of the nodules. One subcellular location that matches these timing patterns is within developing infection threads. The different control mechanisms for induction of fixF and rmlB-wbgA may also explain the observed differences between the NGRΩfixF and NGRΔrmlB-wbgA nodulation phenotypes. Mutation of the intermediate regulators of FixF (NodD2 and SyrM2) does not block rhamnan synthesis, although the amount of rhamnan produced is lower in the mutant. It is possible that NodD2 and SyrM2 coordinate rhamnan synthesis with rhizobial release from infection threads, a critical point in the symbiotic interaction.
Perhaps the most surprising result of this study is that a mutation of y4gM affects the symbiotic properties of NGR234 in a manner similar to that of a mutation in fixF, although the production of rhamnan still occurs in NGRΩgM (Fig. 2). BacA of S. meliloti, which belongs to the same family of ABC transporters as y4gM, is essential for effective symbiosis with Medicago sativa and has been shown to affect the distribution of fatty acids in LPSs (15). With the S. meliloti model in mind, it is possible that the lipid A core of NGR234 changes following flavonoid induction in a process that is independent of FixF (23). As a result, both the constitutive and symbiotic species of lipid A accumulate, and y4gM helps transport the symbiotic subset to the exterior of the cells. Mutation of y4gM and, to a lesser extent, rmlC leads to the synthesis of vhm-R-LPSs, in addition to the rhamnan LPS. Although the precise functions of vhm-R-LPSs are not clear, the fact that they are only produced in induced cells suggests that they may play a symbiotic role. The apparent similarities in isolated LPSs from NGRΩgM and NGRΩrmlC need to be reconciled with the differences in nodulation phenotypes, however. Ultrastructural analyses of nodules formed by NGRΩgM showed that only a few plant cells contained bacteroids and that these few bacteroids were aptopic. Other work has shown that the lipid A core of LPS molecules is important for symbiosis, especially for the long-term survival of rhizobia within acidic compartments of plant cells (5, 28, 32).
It seems clear from this study that modified O antigens are required for effective symbiosis with many legumes. L. leucocephala was the only plant tested that did not show a variable response to any of the mutants. Yet, the exact function of the O antigen rhamnan remains elusive. Hopefully, the next challenges—to determine the structural changes to the lipid A core and the genetic mechanisms that control them—will shed light on both parts of this symbiotically important molecule.
Acknowledgments
We thank Aung Yin-Yin, Dora Gerber, Slobodan Relić, and Wong Chee-Hoong for their unstinting help with many different aspects of this work. We are very grateful to Peter Reeves of the School of Molecular and Microbial Biosciences of the University of Sydney (Australia) for his tireless help in sorting out the often-confusing nomenclature of genes that encode enzymes involved in polysaccharide metabolism.
This work was supported by grant MCB-9728564 from the National Science Foundation (to B. Reuhs), the Fonds National Suisse de la Recherche Scientifique (projects 31-30950.91, 31-36454.92, 31-63893.00, and 3100AO-104097/1), the Whistler Center for Carbohydrate Research, and the Université de Genève.
REFERENCES
- 1.Allaway, D., L. Cavalca, S. Saini, P. Hocking, E. M. Lodwig, M. E. Leonard, and P. S. Poole. 2000. Identification of a putative LPS-associated cation exporter from Rhizobium leguminosarum bv. viciae. FEMS Microbiol. Lett. 186:47-53. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Broughton, W. J., S. Jabbouri, and X. Perret. 2000. Keys to symbiotic harmony. J. Bacteriol. 182:5641-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broughton, W. J., C.-H. Wong, A. Lewin, U. Samrey, H. Myint, H. Meyer, z. A., D. N. Dowling, and R. Simon. 1986. Identification of Rhizobium plasmid sequences involved in recognition of Psophocarpus, Vigna, and other legumes. J. Cell Biol. 102:1173-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, G. R. O., B. L. Reuhs, and G. C. Walker. 2002. Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc. Natl. Acad. Sci. USA 99:3938-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Haeze, W., M. Gao, R. De Rycke, M. van Montagu, G. Engler, and M. Holsters. 1998. Roles for azorhizobial Nod factors and surface polysaccharides in intercellular invasion and nodule penetration, respectively. Mol. Plant-Microbe Interact. 11:999-1008. [Google Scholar]
- 7.Doerrler, W. T., F. Gibbons, and C. R. H. Raetz. 2004. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 279:45102-45109. [DOI] [PubMed] [Google Scholar]
- 8.Duelli, D. M., and K. D. Noel. 1997. Compounds exuded by Phaseolus vulgaris that induce a modification of Rhizobium etli lipopolysaccharide. Mol. Plant-Microbe Interact. 10:903-910. [Google Scholar]
- 9.Duelli, D. M., A. Tobin, J. M. Box, V. S. Kolli, R. W. Carlson, and K. D. Noel. 2001. Genetic locus required for antigenic maturation of Rhizobium etli CE3 lipopolysaccharide. J. Bacteriol. 183:6054-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Economou, A., W. D. O. Hamilton, A. W. B. Johnston, and J. A. Downie. 1990. The Rhizobium nodulation gene nodO encodes a Ca2+-binding protein that is exported without N-terminal cleavage and is homologous to haemolysin and related proteins. EMBO J. 9:349-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrhardt, D. W., R. Wais, and S. R. Long. 1996. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85:673-681. [DOI] [PubMed] [Google Scholar]
- 12.Fellay, R., M. Hanin, G. Montorzi, J. Frey, C. Freiberg, W. Golinowski, C. Staehelin, W. J. Broughton, and S. Jabbouri. 1998. nodD2 of Rhizobium sp. NGR234 is involved in the repression of the nodABC operon. Mol. Microbiol. 27:1039-1050. [DOI] [PubMed] [Google Scholar]
- 13.Fellay, R., X. Perret, V. Viprey, W. J. Broughton, and S. Brenner. 1995. Organization of host-inducible transcripts on the symbiotic plasmid of Rhizobium sp. NGR234. Mol. Microbiol. 16:657-667. [DOI] [PubMed] [Google Scholar]
- 14.Felle, H. H., E. Kondorosi, A. Kondorosi, and M. Schultze. 1998. The role of ion fluxes in Nod-factor signalling in Medicago sativa. Plant J. 13:455-463. [Google Scholar]
- 15.Ferguson, G. P., A. Datta, J. Baumgarter, R. M. Roop II, R. W. Carlson, and G. C. Walker. 2004. Similarity to peroxisomal-membrane protein family reveals that Sinorhizobium and Brucella BacA affect lipid-A fatty acids. Proc. Natl. Acad. Sci. USA 101:5012-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorhölter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Pühler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraysse, N., S. Jabbouri, M. Treilhou, F. Couderc, and V. Poinsot. 2002. Symbiotic conditions induce structural modifications of Sinorhizobium sp. NGR234 surface polysaccharides. Glycobiology 12:741-748. [DOI] [PubMed] [Google Scholar]
- 19.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 20.Gehring, C. A., H. R. Irving, A. A. Kabbara, R. W. Parish, N. M. Boukli, and W. J. Broughton. 1997. Rapid, plateau-like increases in intracellular free calcium are associated with Nod-factor-induced root hair deformation. Mol. Plant-Microbe Interact. 10:791-802. [Google Scholar]
- 21.Gibson, T. J., A. R. Coulson, J. E. Sulston, and P. F. Little. 1987. Lorist2, a cosmid library with transcriptional terminators insulating vector genes from interference by promoters within the insert: effect on DNA yield and cloned insert frequency. Gene 53:275-281. [DOI] [PubMed] [Google Scholar]
- 22.Golinowski, W., J. Kopciñska, and W. Borucki. 1987. The morphogenesis of lupine root nodules during infection by Rhizobium lupini. Acta Soc. Bot. Pol. 56:687-703. [Google Scholar]
- 23.Gudlavalleti, S. K., and L. S. Forsberg. 2003. Structural characterization of the lipid A component of Sinorhizobium sp. NGR234 rough and smooth form lipopolysaccharide. J. Biol. Chem. 278:3957-3968. [DOI] [PubMed] [Google Scholar]
- 24.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 25.Hanin, M., S. Jabbouri, D. Quesada-Vincens, C. Freiberg, X. Perret, J.-C. Promé, W. J. Broughton, and R. Fellay. 1997. Sulphation of Rhizobium sp. NGR234 Nod factors is dependent on noeE, a new host-specificity gene. Mol. Microbiol. 24:1119-1129. [DOI] [PubMed] [Google Scholar]
- 26.Jabbouri, S., M. Hanin, R. Fellay, D. Quesada-Vincens, B. Reuhs, R. W. Carlson, X. Perret, C. Freiberg, A. Rosenthal, D. Leclerc, W. J. Broughton, and B. Relić. 1996. Rhizobium species NGR234 host-specificity of nodulation locus III contains nod- and fix genes, p. 319-324. In G. Stacey, B. Mullin, and P. M. Gresshoff (ed.), Biology of plant-microbe interactions. International Society of Molecular Plant-Microbe Interactions, St. Paul, Minn.
- 27.Jones, J. D., and N. Gutterson. 1987. An efficient mobilizable cosmid vector, pRK7813, and its use in a rapid method for marker exchange in Pseudomonas fluorescens strain HV37a. Gene 61:299-306. [DOI] [PubMed] [Google Scholar]
- 28.Kannenberg, E. L., and R. W. Carlson. 2001. Lipid A and O-chain modifications cause Rhizobium lipopolysaccharides to become hydrophobic during bacteroid development. Mol. Microbiol. 39:379-391. [DOI] [PubMed] [Google Scholar]
- 29.Kim, J. S., B. L. Reuhs, M. M. Rahman, B. Ridley, and R. W. Carlson. 1996. Separation of bacterial capsular and lipopolysaccharides by preparative electrophoresis. Glycobiology 6:433-437. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi, H., Y. Naciri-Graven, W. J. Broughton, and X. Perret. 2004. Flavonoids induce temporal shifts in gene-expression of nod-box controlled loci in Rhizobium sp. NGR234. Mol. Microbiol. 51:335-347. [DOI] [PubMed] [Google Scholar]
- 31.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 32.Le Vier, K., R. W. Phillips, V. K. Grippe, R. M. Roop, and G. C. Walker. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287:2492-2493. [DOI] [PubMed] [Google Scholar]
- 33.Lewin, A., E. Cervantes, C.-H. Wong, and W. J. Broughton. 1990. nodSU, two new nod genes of the broad host range Rhizobium strain NGR234 encode host-specific nodulation of the tropical tree Leucaena leucocephala. Mol. Plant-Microbe Interact. 3:317-326. [DOI] [PubMed] [Google Scholar]
- 34.Lewin, A., C. Rosenberg, H. Meyer, C.-H. Wong, L. Nelson, J.-F. Manen, J. Stanley, D. N. Dowling, J. Dénarié, and W. J. Broughton. 1987. Multiple host-specificity loci of the broad host range Rhizobium sp. NGR234 selected using the widely compatible legume Vigna unguiculata. Plant Mol. Biol. 8:447-459. [DOI] [PubMed] [Google Scholar]
- 35.Marie, C., W. J. Deakin, T. Ojanen-Reuhs, E. Diallo, B. Reuhs, W. J. Broughton, and X. Perret. 2004. TtsI, a key regulator of Rhizobium species NGR234 is required for type III-dependent protein secretion and synthesis of rhamnose-rich polysaccharides. Mol. Plant-Microbe Interact. 17:958-966. [DOI] [PubMed] [Google Scholar]
- 36.Mathis, R., F. van Gijsegem, R. De Rycke, W. D'Haeze, E. van Maelsaeke, E. Anthonio, M. van Montagu, M. Holsters, and D. Vereecke. 2005. Lipopolysaccharides as a communication signal for progression of legume endosymbiosis. Proc. Natl. Acad. Sci. USA 102:2655-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Assay of β-galactosidase, p. 352-355. In J. H. Miller (ed.), Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Noel, K. D., J. M. Box, and V. J. Bonne. 2004. 2-O-methylation of fucosyl residues of a rhizobial lipopolysaccharides is increased in response to host exudate and is eliminated in a symbiotically defective mutant. Appl. Environ. Microbiol. 70:1537-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noel, K. D., D. M. Duelli, H. Tao, and N. J. Brewin. 1996. Antigenic change in the lipopolysaccharide of Rhizobium etli CFN42 induced by exudates of Phaseolus vulgaris. Mol. Plant-Microbe Interact. 9:180-186. [Google Scholar]
- 40.Perret, X., W. J. Broughton, and S. Brenner. 1991. Canonical ordered cosmid library of the symbiotic plasmid of Rhizobium species NGR234. Proc. Natl. Acad. Sci. USA 88:1923-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perret, X., C. Freiberg, A. Rosenthal, W. J. Broughton, and R. Fellay. 1999. High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol. Microbiol. 32:415-425. [DOI] [PubMed] [Google Scholar]
- 42.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pueppke, S. G., and W. J. Broughton. 1999. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host-ranges. Mol. Plant-Microbe Interact. 12:293-318. [DOI] [PubMed] [Google Scholar]
- 44.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 45.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. H. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 46.Relić, B., R. Fellay, A. Lewin, X. Perret, N. P. J. Price, P. Rochepeau, and W. J. Broughton. 1993. nod genes and Nod factors of Rhizobium species NGR234, p. 183-189. In R. Palacios, J. Mora, and W. E. Newton (ed.), New horizons in nitrogen fixation. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 47.Relić, B., X. Perret, M. T. Estrada-Garcia, J. Kopcinska, W. Golinowski, H. B. Krishnan, S. G. Pueppke, and W. J. Broughton. 1994. Nod factors of Rhizobium are a key to the legume door. Mol. Microbiol. 13:171-178. [DOI] [PubMed] [Google Scholar]
- 48.Reuhs, B. L., B. Relić, L. S. Forsberg, C. Marie, T. Ojanen-Reuhs, S. B. Stephens, C.-H. Wong, S. Jabbouri, and W. J. Broughton. 2005. Structural characterization of a flavonoid-inducible Pseudomonas aeruginosa A-band-like O antigen of Rhizobium sp. strain NGR234, required for the formation of nitrogen-fixing nodules. J. Bacteriol. 187:6479-6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reuhs, B. L., R. W. Carlson, and J. S. Kim. 1993. Rhizobuim fredii and Rhizobium meliloti produce 3-deoxy-d-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J. Bacteriol. 175:3570-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reuhs, B. L., D. P. Geller, J. S. Kim, J. E. Fox, V. S. K. Kolli, and S. G. Pueppke. 1998. Sinorhizobium fredii and S. meliloti produce structurally conserved lipopolysaccharides and strain-specific K-antigens. Appl. Environ. Microbiol. 64:4930-4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reuhs, B. L., S. B. Stephens, D. P. Geller, J. S. Kim, J. Glenn, J. Przytycki, and T. Ojanen-Reuhs. 1999. Epitope identification for a panel of anti-Sinorhizobium meliloti monoclonal antibodies and application to the analysis of K antigens and lipopolysaccharides from bacteroids. Appl. Environ. Microbiol. 65:5186-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 53.Stanley, J., D. N. Dowling, and W. J. Broughton. 1988. Cloning of hemA from Rhizobium sp. NGR234 and symbiotic phenotype of a gene-directed mutant in diverse legume genera. Mol. Gen. Genet. 215:32-37. [Google Scholar]
- 54.Streit, W. R., R. A. Schmitz, X. Perret, C. Staehelin, W. J. Deakin, C. Raasch, H. Liesegang, and W. J. Broughton. 2004. An evolutionary hot spot: the pNGR234b replicon of Rhizobium sp. strain NGR234. J. Bacteriol. 186:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 56.Viprey, V., A. Del Greco, W. Golinowski, W. J. Broughton, and X. Perret. 1998. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28:1381-1389. [DOI] [PubMed] [Google Scholar]
- 57.Wong, C.-H., C. E. Pankhurst, A. Kondorosi, and W. J. Broughton. 1983. Morphology of root nodules and nodule-like structures formed by Rhizobium and Agrobacterium strains containing a Rhizobium meliloti megaplasmid. J. Cell Biol. 97:787-794. [DOI] [PMC free article] [PubMed] [Google Scholar]