Abstract
The availability of complete genome sequences has highlighted the problems of functional annotation of the many gene products that have only limited sequence similarity with proteins of known function. The predicted protein encoded by open reading frame Rv3214 from the Mycobacterium tuberculosis H37Rv genome was originally annotated as EntD through sequence similarity with the Escherichia coli EntD, a 4′-phosphopantetheinyl transferase implicated in siderophore biosynthesis. An alternative annotation, based on slightly higher sequence identity, grouped Rv3214 with proteins of the cofactor-dependent phosphoglycerate mutase (dPGM) family. The crystal structure of this protein has been solved by single-wavelength anomalous dispersion methods and refined at 2.07-Å resolution (R = 0.229; Rfree = 0.245). The protein is dimeric, with a monomer fold corresponding to the classical dPGM α/β structure, albeit with some variations. Closer comparisons of structure and sequence indicate that it most closely corresponds with a broad-spectrum phosphatase subfamily within the dPGM superfamily. This functional annotation has been confirmed by biochemical assays which show negligible mutase activity but acid phosphatase activity with a pH optimum of 5.4 and suggests that Rv3214 may be important for mycobacterial phosphate metabolism in vivo. Despite its weak sequence similarity with the 4′-phosphopantetheinyl transferases (EntD homologues), there is little evidence to support this function.
The sequencing of complete genomes has highlighted the problems of assigning functional annotations to many of their gene products. For most gene products, no experimental data on either function or structure exist, and functions are assigned from bioinformatic analyses, generally on the basis of sequence similarities with other proteins that are functionally characterized. This becomes problematic when the sequence identity is lower than about 25%. Even when the general class of a protein can be established, its precise biochemical activity may be uncertain, and current estimates suggest that about 60% of gene products from a typical genome may be of unknown or uncertain function. In this environment, it is not surprising that misannotations occur and can be propagated through many genomes (14).
The predicted protein encoded by open reading frame Rv3214 in the genome of the H37Rv strain of Mycobacterium tuberculosis (4) was originally annotated as EntD, due to sequence similarities with Escherichia coli EntD, which is classified as a 4′-phosphopantetheinyl transferase and is involved in the synthesis of the siderophore enterobactin (33, 40). If confirmed, this activity would make Rv3214 a potential target for the design of new antituberculosis drugs, since the biosynthetic pathway leading to mycobactin, the siderophore used by M. tuberculosis for iron acquisition, is essential for the growth of the organism (6). Many of the M. tuberculosis genes coding for enzymes involved in mycobactin biosynthesis have been provisionally identified (32), but other genes remain unrecognized, as highlighted by the recent discovery that the gene product of Rv1347c, previously annotated as an aminoglycoside acetyltransferase, is a probable “missing” acyltransferase from mycobactin biosynthesis (3).
The 4′-phosphopantetheinyl transferases catalyze the conversion of acyl carrier proteins and peptidyl carrier proteins from their apo- to holoenzyme forms, usually in the context of multienzyme systems such as the polyketide synthases or fatty acid synthases (18, 46). This involves transfer of the 4′-phosphopantetheine moiety of coenzyme A onto a serine residue, with the free SH group created by this transfer then acting as a nucleophile for a further acylation reaction.
An alternative classification for Rv3214 has suggested, however, that its gene product belongs to the cofactor-dependent phosphoglycerate mutase (dPGM) family (9, 13). Phosphoglycerate mutase (EC 5.4.2.1) catalyzes the reversible conversion of 2-phosphoglycerate to 3-phosphoglycerate and is an essential component of the glycolysis and gluconeogenesis pathways. Cofactor-dependent and cofactor-independent forms are found; the cofactor-dependent form (dPGM) carries out this reaction via its cofactor 2,3-bisphosphoglycerate, whereas the cofactor-independent form (iPGM) is Mn2+ dependent (10, 13). The cofactor-dependent form is found in vertebrates, budding yeast, and eubacterial species, while the independent form is the only phosphoglycerate mutase present in higher plants, with some eubacteria possessing both forms of the enzyme (10).
The cofactor-dependent phosphoglycerate mutases are members of a large superfamily whose members share a common α/β fold and use a phosphohistidine as a catalytic intermediate (1, 8). Within this common framework, a variety of biochemical reactions can take place. Thus, the dPGMs can act not only as mutases but also as phosphatases (EC 3.1.3.13), by cleavage of one or other of the two phosphate groups of 2,3-bisphosphoglycerate, or as 2,3-bisphosphoglycerate synthases (EC 5.4.2.4), catalyzing the conversion of 1,3-bisphosphoglycerate to 2,3-bisphosphoglycerate. Other members of this superfamily include the prostatic acid phosphatase, phytase, glucose-1-phosphatase, and 6-phosphofructo-2-kinase phosphatase domain families (37).
In this context, the prediction of function is not trivial. For example, the broad-specificity phosphatase PhoE, from Bacillus stearothermophilus, was predicted from sequence data to be a member of the dPGM family and named YhfR. Assays of the expressed protein showed no mutase activity, however (28), and it was redesignated as a broad-specificity phosphatase through molecular modeling and functional assays (37). The solution of its three-dimensional structure (38) confirmed its characteristic dPGM fold and revealed a highly hydrophobic active site, consistent with an activity against larger hydrophobic substrates.
The predicted product of M. tuberculosis open reading frame Rv3214 has relatively low levels of sequence identity, typically 20 to 25% pairwise identity, with members of the dPGM superfamily and somewhat less (14%) with E. coli EntD, making sequence alone a poor discriminator. In this context, three-dimensional structure should provide a more reliable guide. Structures are available for a number of dPGM family enzymes, including the monomeric Schizosaccharomyces pombe dPGM (45), the dimeric human (47) and E. coli (1) dPGMs, and the tetrameric dPGMs from Saccharomyces cerevisiae (5, 36, 39) and M. tuberculosis (Rv0489) (25). In contrast, the structure of the Bacillus subtilis protein Sfp (35), the first for an EntD-like 4′-phosphopantetheinyl transferase, has an α/β fold completely different from that of the dPGMs and is monomeric but with a twofold internal repeat.
In order to address the uncertainty in the functional annotation of Rv3214 and consider its possible relevance as a drug target, we have determined its three-dimensional structure at 2.07-Å resolution. Both the structural details and our functional assays lead us to conclude that it is in fact a broad-specificity phosphatase, belonging to the dPGM family.
MATERIALS AND METHODS
Protein expression and purification.
The open reading frame corresponding to Rv3214 was amplified from genomic DNA from the H37Rv strain of M. tuberculosis by use of PCR and cloned into the E. coli expression vector pPROEX HTa. The Rv3214 gene product was then expressed as an N-terminally His-tagged protein and purified by nickel-affinity chromatography and gel filtration as previously described (48). The His tag was removed by cleavage with recombinant tobacco etch virus protease. Selenomethionine (SeMet)-substituted Rv3214 was also produced by use of the autoinduction method in the methionine prototroph BL21(DE3) and was purified with the same protocol.
Enzyme assays.
The assay for cofactor-dependent phosphoglycerate mutase activity was carried out in the forward direction as described previously (49) with 30 mM Tris-HCl (pH 7.0), 20 mM KCl, 5 mM MgSO4, 0.15 mM NADH, 0.2 mM 2,3-bisphosphoglycerate, 1 mM ATP, 0.08 U enolase, 0.5 U pyruvate kinase, 0.5 U lactate dehydrogenase, and concentrations of 3-phosphoglycerate in the range of 0.1 to 10.0 mM. All measurements were taken with a SpectraMAX-Pro 340 instrument (Molecular Devices) at 340 nm. To provide a positive control, the gene encoding the E. coli cofactor-dependent phosphoglycerate mutase was amplified from genomic DNA using PCR and isolated from E. coli cells using the reagent DNazol (Molecular Research Center, Inc). The gene was cloned into vector pDEST17 using the GATEWAY system (Invitrogen) and overexpressed in E. coli BL21(DE3) cells. The phosphoglycerate mutase enzyme was then purified as the His-tagged protein from E. coli using nickel affinity chromatography. Enzyme assays were carried out in parallel on Rv3214 and the control.
Phosphatase activity was measured by monitoring the release of p-nitrophenol from p-nitrophenyl phosphate (pNPP) at a range of pH values. The reaction, carried out at pH 5.5 and 37°C, was started by the addition of substrate (2.5 mmol), the mixture was incubated for 15 min, and then the reaction was stopped with the addition of 3 volumes of 1 M NaOH. The positive control, potato acid phosphatase (Sigma), was assayed at 37°C and pH 4.8. The amount of p-nitrophenol released was obtained from measurements of the absorbance at 405 nm after a standard curve that related the p-nitrophenol concentration to absorbance was calculated. For the assay of phosphatase activity with different substrates, the amount of inorganic phosphate released was assayed as previously described (44) over various substrate concentrations (0.1 to 10.0 mM). The reaction step was carried out at 37°C and pH 5.5, as before, and incubation with the molybdate reagent was then carried out for 25 min at 37°C. All absorbance results were corrected for enzyme-unrelated absorbance change, and all assays were carried out in triplicate.
Crystallization and data collection.
Rv3214 was crystallized in native and SeMet-substituted forms as described previously (48). Only the SeMet-substituted crystals, which were obtained by mixing equal volumes of protein solution (11 mg/ml protein, 50 mM Tris-HCl, 50 mM NaCl, 2 mM β-mercaptoethanol, pH 7.5) and precipitant (2.0 M Li2SO4, 6% 2-methyl-2,4-pentanediol, 0.1 M imidazole-HCl, pH 7.0), gave high-resolution diffraction. These were therefore used for the X-ray structural analysis. The crystals were monoclinic, space group P21, with the following unit cell dimensions: a, 44.36 Å; b, 79.03 Å; c, 52.85 Å; and β, 109.11°. Assuming the presence of two molecules of 22.0 kDa per asymmetric unit, the Matthews coefficient, VM, is 2.00 Å3 Da−1, consistent with a solvent content of about 37.7%.
A single SeMet-labeled crystal was soaked briefly in cryoprotectant solution comprising reservoir solution supplemented with 20% ethylene glycol and flash cooled in liquid nitrogen. Diffraction data were collected at 113 K at the peak wavelength of selenium anomalous scattering and at a second, remote wavelength on Beamline 8.2 at the Advanced Light Source (Lawrence Berkeley Laboratory, California). These data were processed using DENZO and SCALEPACK (27), giving the statistics shown in Table 1.
TABLE 1.
Unit cell, data collection, and phasing statistics
| Statistic | Value | Data collection value for indicated wavelengtha
|
|
|---|---|---|---|
| Peak | Remote | ||
| Space group | P21 | ||
| Unit cell dimensions | |||
| a (Å) | 44.36 | ||
| b (Å) | 79.03 | ||
| c (Å) | 52.85 | ||
| β (°) | 109.11 | ||
| Wavelength (Å) | 0.97972 | 0.91840 | |
| Resolution (Å) | 50-2.07 (2.15-2.07) | 50-2.6 (2.69-2.6) | |
| Completeness (%) | 97.8 (87.6) | 96.2 (75.5) | |
| Observed reflections | 151,474 | 67,903 | |
| Unique reflections | 20,637 | 10,306 | |
| <I/σ(I)> | 17.3 (2.3) | 10.1 (1.4) | |
| Rmergeb | 0.105 (0.489) | 0.157 (0.588) | |
| Phasing statistic | |||
| SOLVE figure of merit | 0.52 | ||
| Z-score | 28.35 | ||
| RESOLVE figure of merit | 0.57 | ||
Figures for the outermost shell are given in parentheses.
Rmerge = Σ|Ii − <I>|/ΣIi.
Structure determination and refinement.
The structure of Rv3214 was solved using the two-wavelength anomalous diffraction data. Initial phases to 2.6-Å resolution were obtained using SOLVE (43), which located six selenium sites in the asymmetric unit, and were extended to 2.2 Å with RESOLVE (42), which also built approximately 50% of the structure. The rest of the structure was manually built into the electron density by use of O (17). The initial rounds of crystallographic refinement were carried out using the experimental phases and strict noncrystallographic symmetry restraints in cycles of simulated annealing, energy minimization, and grouped B-factor refinement by use of CNS (2). Subsequent refinement, using the 2.07-Å resolution peak wavelength data set, was with individual isotropic B factors and no noncrystallographic symmetry restraints. The model was periodically examined in 2Fo-Fc and Fo-Fc difference electron density maps, rebuilt with Coot (7), and checked for geometrical correctness with PROCHECK (21). Water molecules were added to the structure by use of CNS and were carefully edited so that only those that made appropriate hydrogen-bonding contacts and had discrete spherical densities above 1.0 σ and 3.0 σ in 2Fo-Fc and Fo-Fc maps, respectively, were retained. Two sulfate ions were modeled into large spherical electron density peaks in equivalent positions in the two independent Rv3214 molecules. Refinement as water molecules gave B factors of less than 5 Å2 and left unexplained positive density around the central position, whereas refinement as sulfate ions gave average B factors that were only slightly higher than those of the surrounding protein structure.
Model completeness and quality.
Refinement statistics and other details of the final model are shown in Table 2. The model comprises residues 4 to 196 of each of the two independent molecules in the crystal asymmetric unit, with no interpretable electron density being found for residues 1 to 3 or 197 to 203, which are assumed to be disordered. A sulfate ion is also found in the active site of each molecule. The final model gives R and Rfree values of 20.9% and 22.4%, respectively, and conforms well with expected geometry; 92.7% of residues are found in the most favored regions of the Ramachandran plot, as defined in PROCHECK (21), with no outliers. The overall G factor is 0.3.
TABLE 2.
Refinement statistics
| Statistic | Value |
|---|---|
| Resolution range (Å) | 50-2.07 |
| Reflections (no.) for working set (test set) | 19,731 (1,038) |
| R factor (Rfree)a | 0.229 (0.245) |
| rms deviations from standard values | |
| Bond lengths (Å) | 0.023 |
| Bond angles (°) | 2.0 |
| Model details | |
| Protein atoms (no.) | 2,962 |
| Water molecules (no.) | 111 |
| SO42− ions (no.) | 2 |
| Avg B factor (Å2) | 26.6 |
| Residues in most favored region of Ramachandran plot (%) | 92.7 |
R = Σ∥Fobs|−|Fcalc∥/Σ|Fobs|, calculated for the 95% of data used in refinement. Rfree is the same as R but using only the 5% of data excluded from refinement.
Other methods.
Multiple sequence alignments were carried out using ClustalW (29) and ESPript (11). Figures were produced with Pymol (http://www.pymol.org).
Atomic coordinates.
Atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank (PDB) with accession code 2a6p.
RESULTS
Monomer fold.
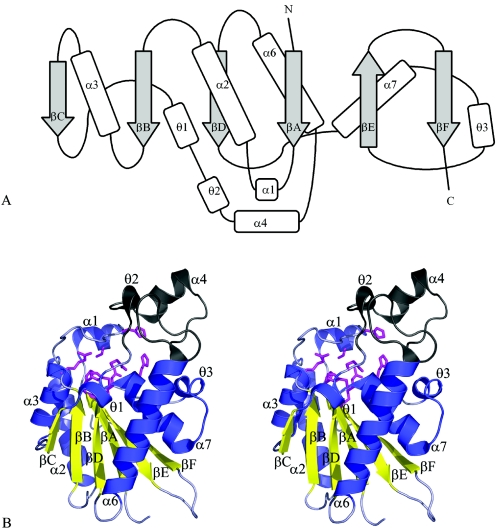
The Rv3214 monomer is folded around a central six-stranded β-sheet, in which the β-strands have the order βF-βE-βA-βD-βB-βC. All are parallel except for the penultimate strand, βE (Fig. 1A). This central sheet is surrounded by four α-helices, with α2 and α3 packed against one face and α6 and α7 against the other face (Fig. 1B). The active site is situated at the edge of the β-sheet, with key amino acid residues contributed by the C-terminal ends of strands βA, βB, βC, and βD and their following loops. A small subdomain is formed from residues 87 to 117 and contains one α-helix (α4) and several short, irregular helices.
FIG. 1.
Structure of Rv3214 from M. tuberculosis. (A) Topology diagram. Secondary structures, labeled to follow the nomenclature of Rigden et al. (38), comprise the following residues: βA, 7 to 13; α1, 16 to 22; α2, 33 to 50; βB, 55 to 60; α3, 62 to 73; βC, 77 to 80; θ1, 80 to 84; θ2, 88 to 92; α4, 95 to 104; α6, 118 to 137; βD, 141 to 146; α7, 147 to 160; θ3, 162 to 166; βE, 174 to 180; and βF, 187 to 193. (B) Stereoview of the Rv3214 monomer. The major parts of the molecule are shown in blue (helices) and gold (β-strands), with the small subdomain shown in gray and residues that contribute to the active site shown in magenta, in stick mode.
The Rv3214 dimer.
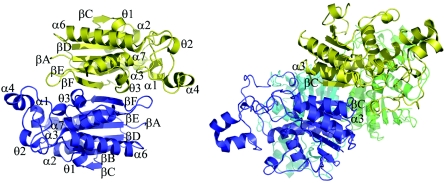
The two monomers in the asymmetric unit together form a discrete dimer, consistent with dynamic light-scattering measurements which indicate a dimer in solution. Dimerization involves structural elements from the C-terminal portion of the molecule: a few residues from helix α7, the following 310 helix, and the loop that connects to strand βE, the βE-βF loop, and the C-terminal β-strand βF. The βF strands of the two molecules pack in an antiparallel fashion with several β-type hydrogen bonds, effectively extending the central β-sheet through the whole dimer, and the βE-βF loop of each monomer extends over the active site of the other. Analysis with the Protein-Protein Interaction Server (http://www.biochem.ucl.ac.uk/bsm/PP/server) based on the principles described by Jones and Thornton (15, 16) shows that, for each molecule, a total of 990 Å2 of solvent-accessible surface (11.6% of the total) is buried in the dimer interface, implying a stable dimer. The interface is maintained by 10 hydrogen bonds and one salt bridge per monomer.
The dimer found here (Fig. 2) reveals a mode of oligomerization not previously seen in members of the dPGM family. The dimer formed by E. coli dPGM (1) makes use of strand βC, from the other end of the β-sheet, together with helix α3 and a portion of the small subdomain to create the dimer interface. The same interface is used in the tetrameric dPGMs from S. cerevisiae (36) and M. tuberculosis (25), with residues from helix α6 contributing to the dimer-dimer interface. In contrast, the S. pombe dPGM (30) and the phosphatase PhoE (38) are both monomeric. For both of these proteins, the monomeric state could be attributed to a large deletion from the small subdomain, which removes the region that participates in oligomerization in the E. coli, S. cerevisiae, and M. tuberculosis structures. In Rv3214, however, the same deletion occurs and a dimer is formed instead, using a different oligomerization surface. Although the catalytic activity of other dPGMs does not appear to depend on the oligomeric state, the fact that the βE-βF loop from one monomer forms a partial lid over the active site of the other monomer in Rv3214 (see below) suggests that dimer formation may in this case have some impact on function.
FIG. 2.
Oligomerization. (Left) The Rv3214 dimer, viewed down the approximate twofold axis relating the two monomers, shown in blue and gold. The C-terminal β-strands (βF) of the two monomers run in an antiparallel fashion, making several hydrogen bonds, and the βE-βF loop of each monomer extends over the active site of the other monomer. (Right) The tetrameric S. cerevisiae dPGM (5, 36, 39), shown for comparison. In this case, the βC strands and the small subdomain are used in oligomerization.
Structural comparisons.
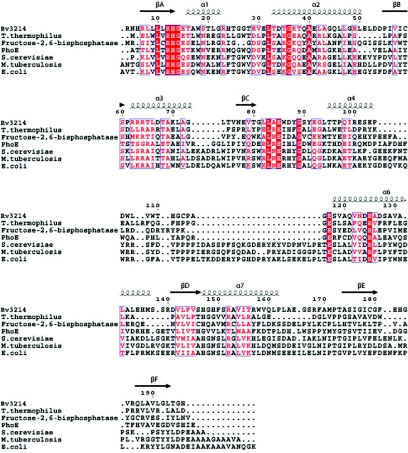
Searches of the current structural database using secondary-structure matching (19) (http://www.ebi.ac.uk/msd-srv/ssm/) show that there is significant structural homology across the whole dPGM superfamily. The closest structural match with Rv3214 is a functionally uncharacterized dPGM from Thermus thermophilus HB8 (PDB code, 1v37; Z-score, 8.0; 155 Cα aligned with a root mean square [rms] difference of 1.50 Å). The next closest matches, interestingly, are both phosphatases: the rat liver fructose-2,6-bisphosphatase (22) (Z-score, 7.6; 173 Cα aligned with an rms difference of 2.0 Å) and the broad-specificity phosphatase PhoE (38) from B. stearothermophilus (Z-score, 7.0; 181 Cα aligned with an rms difference of 2.25 Å). Weaker relationships are found with the dPGMs from S. cerevisiae (36), human (47), and M. tuberculosis (25) (Z-scores, ∼6.5; rms differences of ∼2.6 Å for 180 Cα atom positions). A structure-based sequence alignment of these proteins (Fig. 3) shows that the region of greatest sequence identity corresponds to strand βA and the following βA-α1 loop, where the conserved sequence motif LXRHGXXXW, characteristic of the dPGMs, includes the essential histidine. Other regions of strong conservation, including residues 80 to 90 at the start of the subdomain and the βE-α6 connection, all map to the surrounds of the active site, as described below.
FIG. 3.
Structure-based sequence alignment of the closest structural homologs of Rv3214. The sequences aligned are those of the dPGMs from T. thermophilus HB8, S. cerevisiae, M. tuberculosis, and E. coli, together with PhoE from B. stearothermophilus and rat fructose-2,6-bisphosphatase. Residue numbering and the location of secondary structure elements are shown for Rv3214.
More-detailed comparisons also place Rv3214 more clearly among the phosphatases than the true phosphoglycerate mutases. The latter possess a flexible C-terminal tail which is believed to protect the active site by folding over it when the protein becomes phosphorylated (1, 9). Proteolytic cleavage of this tail from the S. cerevisiae dPGM results in a 97% loss of mutase activity but an increase in phosphatase activity (41). Multiple sequence alignments show that although the C-terminal tail is mostly conserved for dPGMs, it is absent in both Rv3214 and PhoE (Fig. 3). Both Rv3214 and PhoE also share a major deletion of about 25 residues in the region between strand βC and helix α6 compared to what is seen with true mutases. The effect of this deletion is to make the subdomain considerably smaller and to remove the region that is involved in oligomerization in the S. cerevisiae, E. coli, and M. tuberculosis dPGMs (1, 25, 36). Only the first few residues of this subdomain (the 310 helix 88 to 92) are spatially conserved between Rv3214, PhoE, and the other dPGMs, with the rest showing structural variability and some flexibility; B factors are relatively high, especially for residues 98 to 105, which follow helix α4. The proximity of this flexible loop to the mouth of the active site may suggest that a gating effect which could have some effect on the activity of the enzyme can occur.
Active site.
A common feature of the dPGM family enzymes is that the catalytic reaction, whether mutase or phosphatase, is dependent on the transient phosphorylation of a histidine residue located at the end of strand βA, near the N terminus. This histidine is immediately preceded by an arginine, which helps stabilize the phosphate group (1), and is followed by a glycine, giving rise to the conserved sequence motif LXRHGXXXW.
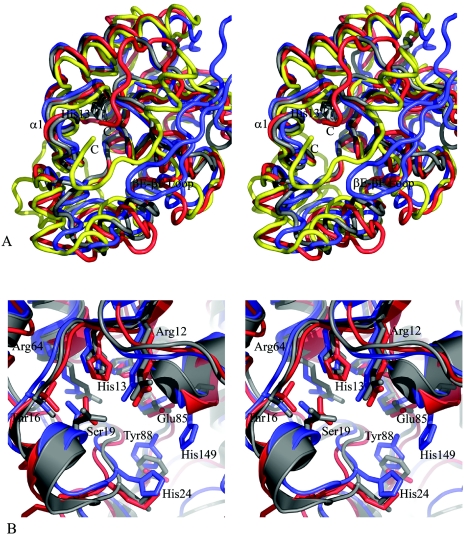
The key histidine residue in Rv3214, His13, is at the center of a deep, cup-shaped active site that is qualitatively similar to the active-site cleft of E. coli dPGM (1), although somewhat smaller and more enclosed. This cavity, which sits at the edge of the central β-sheet, is enclosed by helix α1 (residues 16 to 23), the small subdomain from 87 to 117, and the βE-βF loop (residues 180 to 186) from the other molecule in the dimer. Compared with E. coli dPGM, helix α1 is displaced ∼4 Å inwards, over the active site, as is the loop from residues 95 to 102. Moreover, the βE-βF loop from the other monomer also extends as a partial lid over the active site cavity and occupies a spatial position somewhat similar to that of the C-terminal residues in the E. coli dPGM (Fig. 4). Glu182 and His183 from this loop project into the active-site cavity but are too far from His13 (13.0 and 14.5 Å away, respectively) to directly contact a bound substrate. This makes it unlikely that this loop, and hence the Rv3214 dimer, has any impact on the catalytic reaction for small substrates, but it probably does influence the substrate selectivity of the enzyme. The active site still has a relatively large mouth region, which constricts around two large patches of positive charge to a smaller channel.
FIG. 4.
Comparison of Rv3214 structure with homologous proteins. (A) Stereoview showing a superposition of the Rv3214 structure (blue) onto those of PhoE (red), E. coli dPGM (gold), and the T. thermophilus dPGM homolog (gray). The view is looking down into the active site, with the essential histidine (His13 in Rv3214) shown in dark gray at the bottom. Differences in Rv3214 result from inward movement of several flanking helices and from the presence of the βE-βF loop (labeled) from the other monomer of the dimer; this helps enclose the active site, a role filled by the C-terminal regions of dPGM and PhoE. The T. thermophilus protein has no equivalent feature and a much more open active site. (B) Stereoview of a superposition of the active sites of Rv3214 (blue), PhoE (red), and the T. thermophilus enzyme (gray). Residue labels are for Rv3214.
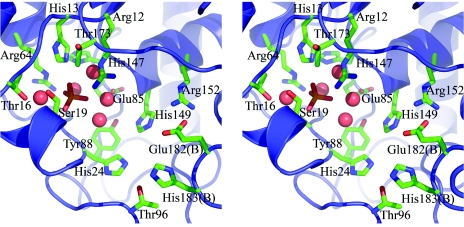
At the bottom of the active site are the residues Arg12, His13, Thr16, Ser19, His24, Thr25, Arg64, Glu85, Tyr88, His147, His149, and Thr173. Several residues also enclose the cavity from above, including Thr96 and Arg152 together with Glu182 and His183 from the βE-βF loop of the other molecule of the dimer. The surroundings of the essential His13 are thus almost exclusively polar, in sharp contrast to the situation for PhoE, in which a number of hydrophobic groups confer a preference for hydrophobic substrates (38). Two invariant residues that have been implicated in catalysis in the dPGMs, Glu85 and His147, form part of the floor of the active-site cavity. Glu 85 is situated ∼7 Å away from His13, but as in the other dPGMs, it is turned aside, with its carboxylate oxygens interacting with the N terminus of helix α7 (residues 147 to 161). His147 is very close to His13 (∼3.5 Å away), in a position where it could play a direct acid/base role in catalysis. Its orientation is identical to that of the corresponding His residues in other dPGMs, oriented by a hydrogen bond with another invariant residue, Ser60.
Both molecules of Rv3214 are found to contain a sulfate ion, bound deep in the active site between the side chains of Arg12 and His13 (Fig. 5). In molecule A, this sulfate makes probable hydrogen bonds with Arg12, His13, Thr25, and five water molecules. Although the hydrogen bonding is less clear in molecule B, because the oxygen positions are less defined, the positions of the sulfate ions differ by only 0.5 Å when the two molecules are superimposed. This sulfate ion is likely to model the position of a substrate phosphate group, as appears to occur in the structures of other dPGM family members, for example, the dPGM from S. cerevisiae (39). Also present in the active site are a number of very well defined water molecules which make multiple hydrogen bonds and whose positions correspond extremely closely between the two independent Rv3214 molecules. Altogether, 18 pairs of water molecules can be identified, with an rms difference in positions of only 0.24 Å.
FIG. 5.
Stereoview of the active site of Rv3214, shown for molecule A. The bound sulfate ion adjacent to Arg12 and His13 is shown in green, and water molecules are shown as large red spheres. Residues Glu182 and His183 are from the β6-β7 loop of the other molecule of the dimer (molecule B).
Activity assays.
Steady-state kinetic assays of phosphoglycerate mutase activity were carried out on both Rv3214 and, as a positive control, the E. coli cofactor-dependent phosphoglycerate mutase. Michaelis-Menten kinetics were obeyed by both enzymes, with the latter giving values for Km and kcat that were similar to published values for both the E. coli and the S. cerevisiae dPGMs (Table 3) . The threefold-lower value of Km for Rv3214 suggests a slightly increased affinity for the 3-phosphoglycerate substrate, but a striking reduction in kcat, over 5 orders of magnitude, and the corresponding reduction in kcat/Km indicate that Rv3214 has little or no phosphoglycerate mutase activity.
TABLE 3.
Kinetic constants for phosphoglycerate mutase assaya
| Kinetic constant | Result for:
|
|||
|---|---|---|---|---|
| Rv3214 | Positive control (E. coli dPGM) | E. coli dPGMb | S. cerevisiae dPGMb | |
| Km (μM) | 101.1 ± 0.27 | 271 ± 3 | 200 ± 27 | 510 ± 60 |
| kcat (s−1) | 7.1 × 10−4 ± 1.2 × 10−8 | 840 ± 0.003 | 330 ± 11 | 380 ± 10 |
| kcat/Km (M−1 s−1) | 0.7 | 3.09 × 106 | 1.6 × 106 | 7.4 × 105 |
All values were measured in triplicate.
See reference 10.
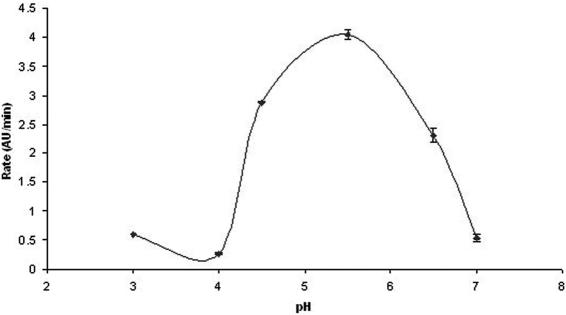
Given the significant structural similarity of Rv3214 to the broad-specificity phosphatase PhoE, Rv3214 was also assayed for acid phosphatase activity using pNPP as the substrate. This assay showed Rv3214 to have a pH optimum of 5.4 (Fig. 6), establishing it as an acid phosphatase. This compares with the known pH optimum of 6.2 for PhoE (37). In contrast with PhoE, the affinity of Rv3214 for pNPP is significantly higher (Km of 0.04 mM for Rv3214 compared with 3 mM for PhoE), but its specific activity against this substrate is comparable, being only fivefold lower (Table 4). Assays with the positive control, potato acid phosphatase, gave a specific activity of 0.12 μmol/min/mg, eightfold lower than the value of 1 μmol/min/mg quoted by the manufacturers.
FIG. 6.
pH-activity curve for Rv3214, when assayed for phosphatase activity, showing an optimum pH of 5.4. Error bars reflect the agreement from triplicate measurements. AU, absorbance units.
TABLE 4.
Results of phosphatase activity assays
| Substrate | Sp act (μmol/min/mg) for:
|
Km (mM) | ||
|---|---|---|---|---|
| PhoEa | Rv3214 | Positive control | ||
| 3-Phosphoglycerate | 27 | 0.00 | ||
| α-Naphthyl phosphate | 21 | 0.00 | ||
| p-Nitrophenyl phosphate | 16 | 2.6 ± 0.03 | 0.12 ± 0.02 | 0.04 |
| AMP | 5 | 0.00 | ||
| ADP | 2.4 × 10−3 ± 6 × 10−4 | 0.25 | ||
| 2,3-Bisphosphoglycerate | 0.00 | |||
| Phosphoenolpyruvate | 0.00 | |||
| Phytic acid | 5.8 × 10−2 ± 6 × 10−3 | 0.068 | ||
| Coenzyme A | 0.00 | |||
| Sucrose-6-phosphate | 1.5 × 10−2 ± 2 × 10−3 | 0.014 | ||
| Fructose-6-phosphate | 5.9 × 10−3 ± 6 × 10−4 | 0.3 | ||
| Maltose-1-phosphate | 0.00 | |||
| Glycerol-1-phosphate | 5.9 × 10−3 ± 4 × 10−4 | 0.16 | ||
| Ribose-5-phosphate | 0.00 | |||
| Dihydroxyacetone phosphate | 3.0 × 10−2 ± 3 × 10−3 | 1.62 | ||
| Phosphatidic acid | 0.00 | |||
See reference 37.
In order to further explore the substrate specificity of Rv3214, we also carried out phosphatase assays with a range of phosphorylated substrates (Table 4). This showed that Rv3214 has no phosphatase activity against 3-phosphoglycerate, 2,3-bisphosphoglycerate, or hydrophobic substrates such as α-napthyl phosphate, which is efficiently hydrolyzed by PhoE. On the other hand, Rv3214 does show phosphatase activity against a number of phosphorylated sugars, consistent with a preference for hydrophilic substrates and with the highly polar nature of the active-site cavity.
DISCUSSION
The crystal structure of the protein encoded by the open reading frame Rv3214 from M. tuberculosis clearly establishes that this protein belongs to the broad dPGM superfamily, which includes true mutases as well as a variety of phosphatase enzymes (13, 37). The original genomic annotation for this open reading frame product, however, was as EntD, a putative 4-phosphopantetheinyl transferase. This annotation was based on amino acid sequence similarities with the E. coli EntD, which has been shown biochemically to be able to transfer the 4-phosphopantetheinyl group from coenzyme A to the acyl carrier domain of the synthetase EntF during enterobactin biosynthesis (20, 40).
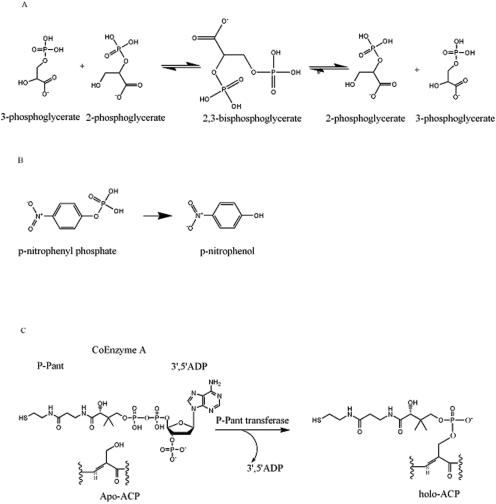
There are similarities evident in the chemical reactions carried out by these various proteins (Fig. 7). Both the phosphoglycerate mutases and the acid phosphatases operate through a phosphohistidine intermediate, involving the histidine of the conserved LXRHGXXXW motif close to the N terminus (His13 in Rv3214) (1, 8). For phosphatases such as PhoE, catalysis occurs through nucleophilic attack on the substrate phosphate by this histidine, with the resulting phosphohistidine later breaking down by attack from an essential acid (38). For PGM activity, the phosphate is transferred from the phosphohistidine to the nonphosphorylated hydroxyl of 2- or 3-phosphoglycerate to give 2,3-diphosphoglycerate, after which a conformational change allows the phosphate from the other hydroxyl to be transferred back to the histidine (1).
FIG. 7.
Reactions catalyzed by proteins of the dPGM family. Phosphoglycerate mutase activity (A) and phosphatase activity (B) are compared with the phosphopantetheinyl transfer reaction catalyzed by EntD (C) (20). P-Pant, phosphopantatheine; ACP, acyl carrier protein.
There exists a superficial similarity with the reaction catalyzed by phosphopantetheinyl transferases in that in these enzymes, the bond between the α- and β-phosphates of coenzyme A is cleaved with the formation of a covalent phosphopantetheinyl intermediate, followed by transfer of this group to a hydroxyl group on a target protein, such as EntF (20). While it is conceivable that Rv3214 could have such activity, it clearly does not belong to the phosphopantetheinyl transferase family to which E. coli EntD and B. subtilis Sfp belong. Rv3214 has a fold completely different from that of Sfp (35), and the sequence identities between Rv3214 and E. coli EntD found by pairwise sequence alignment do not match the residues that have recently been implicated in Sfp function (24) or the conserved motifs in the EntD family (20).
Instead, both the crystal structure of Rv3214 and the functional assays we have performed identify it as an acid phosphatase within the dPGM superfamily. The activity assays show that it does not have significant mutase activity but does have phosphatase activity with a pH optimum of 5.4. Structural homology also groups it with a subgroup of phosphatases within the wider dPGM family. Its closest structural homologues are the rat liver fructose-2,6-bisphosphatase (22) and the broad-specificity phosphatase PhoE (38) from B. stearothermophilus, together with a putative dPGM from Thermus thermophilus HB8 (PDB code 1v37). The latter has not yet been functionally characterized, but its close structural correspondence with Rv3214 and its lack of the C-terminal extension that seems to characterize true mutases (Fig. 3) lead to the prediction that it too functions primarily as a phosphatase. (We note here that the S. pombe dPGM also lacks a C-terminal tail and, although it still has mutase activity, its phosphatase activity is more pronounced [26]).
Comparisons of the active site of Rv3214 with that of PhoE suggest that it has a more restricted, and different, specificity. Both helix α1 and the residues of the “flap” subdomain fold inwards by 4 to 5 Å, compared with PhoE, enclosing the active- site cavity, and the β6-β7 loop from the other monomer also extends over it (Fig. 4A). The hydrophobic residues that line the active-site walls in PhoE and give it a preference for hydrophobic substrates (38) are also changed to polar groups in Rv3214 (Fig. 4B); Met21, Leu86, and Val153 in PhoE are replaced by His24, Tyr88, and His149 in Rv3214, and the positions of Trp109 and Trp171 in PhoE are occupied by Glu182 and His183 from the β6-β7 loop of the other monomer in Rv3214. These changes suggest that the broad specificity of PhoE, with its hydrophobic preference, is changed in Rv3214 to a more restricted specificity, i.e., for polar substrates. This is confirmed in the phosphatase assays that are summarized in Table 4, which show activity against a number of phosphorylated sugars and several other polar substrates. It is interesting to note that the putative T. thermophilus dPGM has a very open active site which resembles that of PhoE in the presence of hydrophobic residues flanking the catalytic center. His24, Tyr88, and His149 in Rv3214 are replaced by Leu19, Phe77, and Gly135 in the T. thermophilus enzyme and by Met24, Leu86, and Val153 in PhoE (Fig. 4B). Given that the T. thermophilus enzyme is the closest structural homolog to Rv3214 and PhoE, we suggest that it is also a phosphatase, but with a broad specificity and a substrate profile that resemble those of PhoE.
The results presented here demonstrate the value of three-dimensional structure in functional assignment. Although the assignment of Rv3214 to the dPGM family on the basis of relatively weak sequence identity proved correct, correcting an earlier misannotation as EntD, the crystal structure of Rv3214 provided conclusive proof of this. It also enabled Rv3214 to be grouped with the subfamily of phosphatases within the wider dPGM family, an assignment that was verified by our activity assays and thereby distinguished from the classical dPGM in M. tuberculosis, Rv0489.
The significance for M. tuberculosis biology is that there is growing evidence of the importance of phosphate metabolism, especially for survival and virulence after uptake by alveolar macrophages, which provide an acidic, phosphate-deficient environment. Thus, genome-wide transposon mutagenesis experiments identify a small number of genes that are essential for survival in macrophages, including a cluster of genes encoding a phosphate uptake system (34). In a phosphate-limiting environment, mycobacterial phosphatases may play a key role by liberating phosphate from host molecules or by recycling phosphate inside the cell. The recognition of Rv3214 as an acid phosphatase, which could operate in the acidic environment of macrophages, adds it to the repertoire of characterized phosphatases in M. tuberculosis, which also includes the serine/threonine and tyrosine protein phosphatases PtpA (23), PtpB (12), and PstP (31). In no case is a specific substrate specificity known, however; rather, it is likely that all show quite broad specificities.
Acknowledgments
We thank Minmin Yu for X-ray data collection at the Advanced Light Source (Berkeley, California) under the auspices of the International Structural Genomics Consortium and Shaun Lott for many helpful suggestions.
This work received funding support from the Health Research Council of New Zealand and from Centres of Research Excellence Funding to the Centre for Molecular Biodiscovery.
REFERENCES
- 1.Bond, C. S., M. F. White, and W. N. Hunter. 2001. High resolution structure of the phosphohistidine-activated form of Escherichia coli cofactor-dependent phosphoglycerate mutase. J. Biol. Chem. 276:3247-3253. [DOI] [PubMed] [Google Scholar]
- 2.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 3.Card, G. L., N. A. Peterson, G. A. Smith, B. Rupp, B. M. Schick, and E. N. Baker. 2005. The crystal structure of Rv1347c, a putative antibiotic resistance protein from Mycobacterium tuberculosis, reveals a GCN5-related fold and suggests an alternative function in siderophore biosynthesis. J. Biol. Chem. 280:13978-13986. [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 5.Crowhurst, G. S., A. R. Dalby, M. N. Isupov, J. W. Campbell, and J. A. Littlechild. 1999. Structure of a phosphoglycerate mutase:3-phosphoglyceric acid complex at 1.7 A. Acta Crystallogr. Sect. D 55:1822-1826. [DOI] [PubMed] [Google Scholar]
- 6.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. Barry. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 8.Fothergill-Gilmore, L. A., and P. A. M. Michels. 1993. Evolution of glycolysis. Prog. Biophys. Mol. Biol. 59:105-235. [DOI] [PubMed] [Google Scholar]
- 9.Fothergill-Gilmore, L. A., and H. C. Watson. 1989. The phosphoglycerate mutases. Adv. Enzymol. Relat. Areas Mol. Biol. 62:227-313. [DOI] [PubMed] [Google Scholar]
- 10.Fraser, H. I., M. Kvaratskhelia, and M. F. White. 1999. The two analogous phosphoglycerate mutases of Escherichia coli. FEBS Lett. 455:344-348. [DOI] [PubMed] [Google Scholar]
- 11.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 12.Grundner, C., H.-L. Ng, and T. Alber. 2005. Mycobacterium tuberculosis protein tyrosine phosphatase PtpB structure reveals a diverged fold and a buried active site. Structure 13:1625-1634. [DOI] [PubMed] [Google Scholar]
- 13.Jedrzejas, M. J. 2000. Structure, function and evolution of phosphoglycerate mutases: comparison with fructose-2,6-bisphosphatase, acid phosphatase and alkaline phosphatase. Prog. Biophys. Mol. Biol. 73:263-287. [DOI] [PubMed] [Google Scholar]
- 14.Johnston, J. M., V. L. Arcus, C. J. Morton, M. J. Parker, and E. N. Baker. 2003. Crystal structure of a putative methyltransferase from Mycobacterium tuberculosis: misannotation of a genome clarified by protein structural analysis. J. Bacteriol. 185:4057-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, S., and J. M. Thornton. 1996. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 93:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, S., and J. M. Thornton. 1995. Protein-protein interactions: a review of protein dimer structures. Prog. Biophys. Mol. Biol. 63:31-65. [DOI] [PubMed] [Google Scholar]
- 17.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 18.Kleinkauf, H. 2000. The role of 4′-phosphopantetheine in the biosynthesis of fatty acids, polyketides and peptides. Biofactors 11:91-92. [DOI] [PubMed] [Google Scholar]
- 19.Krissinel, E., and K. Henrick. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D 60:2256-2268. [DOI] [PubMed] [Google Scholar]
- 20.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 22.Lee, Y.-H., T. W. Olson, C. M. Ogata, D. G. Levitt, L. J. Banaszak, and A. J. Lange. 1997. Crystal structure of a trapped phosphoenzyme during a catalytic reaction. Nat. Struct. Biol. 4:615-618. [DOI] [PubMed] [Google Scholar]
- 23.Madhurantakam, C., E. Rajakumara, P. A. Mazumdar, B. Saha, D. Mitra, H. G. Wiker, R. Sankaranarayanan, and A. K. Das. 2005. Crystal structure of low-molecular-weight protein tyrosine phosphatase from Mycobacterium tuberculosis at 1.9-Å resolution. J. Bacteriol. 187:2175-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mofid, M. R., R. Finking, L. O. Essen, and M. A. Marahiel. 2004. Structure-based mutational analysis of the 4′-phosphopantetheinyl transferase Sfp from Bacillus subtilis: carrier protein recognition and reaction mechanism. Biochemistry 43:4128-4136. [DOI] [PubMed] [Google Scholar]
- 25.Muller, P., M. R. Sawaya, I. Pashkov, S. Chan, C. Nguyen, Y. Wu, L. J. Perry, and D. Eisenberg. 2005. The 1.70 Å X-ray crystal structure of Mycobacterium tuberculosis phosphoglycerate mutase. Acta Crystallogr. Sect. D 61:309-315. [DOI] [PubMed] [Google Scholar]
- 26.Nairn, J., N. C. Price, S. M. Kelly, D. Rigden, L. A. Fothergill-Gilmore, and T. Krell. 1996. Phosphoglycerate mutase from Schizosaccharomyces pombe: development of an expression system and characterisation of three histidine mutants of the enzyme. Biochim. Biophys. Acta 1296:69-75. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 28.Pearson, C. L., C. A. Loshon, L. B. Pederson, B. Setlow, and P. Setlow. 2000. Analysis of the function of a putative 2,3-diphosphoglyceric acid-dependent phosphoglycerate mutase from Bacillus subtilis. J. Bacteriol. 182:4121-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price, N. C., D. Duncan, and D. J. Ogg. 1985. Purification and preliminary characterization of phosphoglycerate mutase from Schizosaccharomyces pombe. Int. J. Biochem. 17:843-846. [DOI] [PubMed] [Google Scholar]
- 31.Pullen, K. E., H.-L. Ng, P.-Y. Sung, M. C. Good, S. M. Smith, and T. Alber. 2004. An alternate conformation and a third metal in PstP/Ppp, the M. tuberculosis PP2C-family Ser/Thr protein phosphatase. Structure 12:1947-1954. [DOI] [PubMed] [Google Scholar]
- 32.Quadri, L. E., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic genes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 33.Reichert, J., M. Sakaitani, and C. T. Walsh. 1992. Characterization of EntF as a serine-activating enzyme. Protein Sci. 1:549-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rengarajan, J., B. R. Bloom, and E. J. Rubin. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 102:8327-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reuter, K., M. R. Mofid, M. A. Marahiel, and R. Ficner. 1999. Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4′-phosphopantetheinyl transferase superfamily. EMBO J. 18:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rigden, D. J., D. Alexeev, S. E. Phillips, and L. A. Fothergill-Gilmore. 1998. The 2.3 Å X-ray crystal structure of S. cerevisiae phosphoglycerate mutase. J. Mol. Biol. 276:449-459. [DOI] [PubMed] [Google Scholar]
- 37.Rigden, D. J., I. Bagyan, E. Lamani, P. Setlow, and M. J. Jedrzejas. 2001. A cofactor-dependent phosphoglycerate mutase homolog from Bacillus stearothermophilus is actually a broad specificity phosphatase. Protein Sci. 10:1835-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigden, D. J., L. V. Mello, P. Setlow, and M. J. Jedrzejas. 2002. Structure and mechanism of action of a cofactor-dependent phosphoglycerate mutase homolog from Bacillus stearothermophilus with broad specificity phosphatase activity. J. Mol. Biol. 315:1129-1143. [DOI] [PubMed] [Google Scholar]
- 39.Rigden, D. J., R. A. Walter, S. E. Phillips, and L. A. Fothergill-Gilmore. 1999. Sulphate ions observed in the 2.12 Å structure of a new crystal form of S. cerevisiae phosphoglycerate mutase provide insights into understanding the catalytic mechanism. J. Mol. Biol. 286:1507-1517. [DOI] [PubMed] [Google Scholar]
- 40.Rusnak, F., M. Sakaitani, D. Drueckhammer, J. Reichert, and C. T. Walsh. 1991. Biosynthesis of the Escherichia coli siderophore enterobactin: sequence of the entF gene, expression and purification of EntF, and analysis of covalent phosphopantetheine. Biochemistry 30:2916-2927. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki, R., R. Sugimoto, and H. Chiba. 1966. Yeast phosphoglyceric acid mutase-modifying enzyme. Arch. Biochem. Biophys. 115:53-61. [DOI] [PubMed] [Google Scholar]
- 42.Terwilliger, T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. Sect. D 56:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. Sect. D 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner, W. L., and W. C. Plaxton. 2001. Purification and characterization of banana fruit acid phosphatase. Planta 214:243-249. [DOI] [PubMed] [Google Scholar]
- 45.Uhrinova, S., D. Uhrin, J. Nairn, N. C. Price, L. A. Fothergill-Gilmore, and P. N. Barlow. 2001. Solution structure and dynamics of an open beta-sheet, glycolytic enzyme, monomeric 23.7 kDa phosphoglycerate mutase from Schizosaccharomyces pombe. J. Mol. Biol. 306:275-290. [DOI] [PubMed] [Google Scholar]
- 46.Walsh, C. T., A. M. Gehring, P. H. Weinreb, L. E. Quadri, and R. S. Flugel. 1997. Post-translational modification of polyketide and nonribosomal peptide synthases. Curr. Opin. Chem. Biol. 1:309-315. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Y., Z. Wei, L. Liu, Z. Cheng, Y. Lin, F. Ji, and W. Gong. 2005. Crystal structure of human B-type phosphoglycerate mutase bound with citrate. Biochem. Biophys. Res. Commun. 331:1207-1215. [DOI] [PubMed] [Google Scholar]
- 48.Watkins, H. A., M. Yu, and E. N. Baker. 2005. Cloning, expression, purification and preliminary crystallographic data for Rv3214 (EntD), a predicted cofactor-dependent phosphoglycerate mutase from Mycobacterium tuberculosis. Acta Crystallogr. Sect. F 61:753-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White, M. F., and L. A. Fothergill-Gilmore. 1992. Development of a mutagenesis, expression and purification system for yeast phosphoglycerate mutase. Investigation of the role of active-site His181. Eur. J. Biochem. 207:709-714. [DOI] [PubMed] [Google Scholar]