Abstract
Bacterial populations contain persisters, cells which survive exposure to bactericidal antibiotics and other lethal factors. Persisters do not have a genetic resistance mechanism, and their means to tolerate killing remain unknown. In exponentially growing populations of Escherichia coli the frequency of persister formation usually is 10−7 to 10−5. It has been shown that cells overexpressing either of the toxic proteins HipA and RelE, both members of the bacterial toxin-antitoxin (TA) modules, have the ability to form more persisters, suggesting a specific role for these toxins in the mechanism of persistence. However, here we show that cells expressing proteins that are unrelated to TA modules but which become toxic when ectopically expressed, chaperone DnaJ and protein PmrC of Salmonella enterica, also form 100- to 1,000-fold more persisters. Thus, persistence is linked not only to toxicity caused by expression of HipA or dedicated toxins but also to expression of other unrelated proteins.
A small number of cells in a bacterial population can survive treatment with lethal concentrations of bactericidal antibiotics. These survivors, which appear with a frequency of ∼10−6, are known as persisters (3). The nature of their tolerance to killing and the molecular mechanisms triggering the persister state remain unknown.
Persisters do not have a genetic resistance mechanism, and their progeny are fully sensitive to antibiotics (3, 14, 18). The only gene that has been linked to persistence in Escherichia coli has been hipA (4, 5, 15, 16, 19, 20, 23). A mutation in this gene, the hipA7 mutation, was identified because cells carrying this mutant gene formed persisters at a higher frequency (10−1 to 10−3) compared to the wild-type strain (16, 19, 23). The high frequency of persister formation is not restricted to cells with the chromosomal hipA7 mutation; it has also been shown for cells overexpressing the toxic wild-type HipA (4, 7, 16, 20). Cells expressing this protein form 10- to 1,000-fold more persisters tolerant to bactericidal antibiotics with different mechanisms of actions, such as ampicillin and ciprofloxacin (7).
It has been suggested that the hipA gene, encoding the toxic factor, together with hipB, encoding a putative antidote, constitutes one of the several toxin-antitoxin (TA) modules present in bacteria (8, 9). Similar to typical TA operons, like relBE and mazEF, hipB and hipA are organized in an operon with the gene encoding the antitoxin, hipB, located upstream of hipA, the toxin gene (4, 5, 8, 9).
It has been shown recently that, similar to HipA, cells overproducing the toxin RelE, another member of a TA module, form 10- to 10,000-fold more persisters (15). Both proteins, HipA and RelE, have the effect of slowing down or even stopping cell division, raising the possibility that toxins from the TA modules have a role in increasing the fraction of dormant or nongrowing cells in a population, the fraction, it has been suggested, that constitute the persister cells (2, 3, 14, 15, 18). The various roles in cell physiology of toxins RelE and MazF are most likely related to their specific activity in inhibiting translation by cleaving mRNA at particular sites (6, 21). It is unclear, however, whether the increase in the number of persisters observed when HipA (7) or RelE (15) is overexpressed from plasmids is a result of a specific dedicated role of these toxins in the mechanism of forming persisters. It is known that expression of a wide variety of recombinant proteins, even those that are not dedicated toxins, often has a detrimental effect on the growth of bacteria (11). Evidently, most of these proteins share no homology in sequence, gene organization, or function with the toxins of the TA modules, and thus their effects on cell physiology are probably of a different nature.
In this work, we asked whether the increase in frequency of persister formation is specific to cells overexpressing toxins from the TA modules or if cells expressing proteins that become toxic when produced from plasmids would also show high persistence.
MATERIALS AND METHODS
Strains and plasmids.
Plasmids pBAD18CM and pBAD22CM were used as cloning vectors for expression of E. coli genes thrB, dnaJ, mazF, and hipA and Salmonella enterica gene pmrC. pBADCM plasmids were prepared by replacing the ampicillin resistance gene with the chloramphenicol resistance gene. Genes to be cloned were PCR amplified using pairs of primers (Table 1) and high-fidelity AccuPrime Taq DNA polymerase (Invitrogen). DNA from E. coli strain MG1655 was prepared using a genomic DNA isolation kit (Sigma; catalog no. NA2120) and used as a template for amplification of hipA, mazF, dnaJ, and thrB. ppmrC plasmid (17) carrying the pmrC gene of Salmonella enterica serovar Typhimurium 14028s was used as a template for PCR amplification of pmrC. The amplified PCR products were cut with the appropriate restriction enzymes and cloned in the corresponding sites of the vector plasmids (Table 1).
TABLE 1.
Plasmids and primers used in this study
| Plasmid | Primersa | Source of template DNAb | Relevant features |
|---|---|---|---|
| pBAD22thrB | 5′-TTAGGAATTCGACATGGTTAAAGTTTATGCCC, 5′-CCTGAAGCTTGTGATCTTTCAGATTGTAGAGT | MG1655 | thrB gene in EcoRI-HindIII sites of pBAD22 |
| pBAD22dnaJ | 5′-CAATGAATTCAAGATGGCTAAGCAAGATTATTA, 5′-GCAGTCTAGAGGGGAGGTTAGCGGGTCAG | MG1655 | dnaJ gene in EcoRI-XbaI sites of pBAD22 |
| pBAD18pmrC | 5′-TGGAGAATTCAACATGTTAAAGCGCTT, 5′-TCAGAAGCTTCATTCGCTTAGTCTCCT | ppmrC (17) | pmrC gene in EcoRI-HindIII sites of pBAD22 |
| pBAD22mazF | 5′-AGGAGAATTCGTAATGGTAAGCCGATACGTA, 5′-ATAGAAGCTTGTTAGTAACACTACCCAATCAG | MG1655 | mazF gene in EcoRI-HindIII sites of pBAD22 |
| pBADCMhipA | 5′-GTGGGCATGCCTAAACTTGTCACTTGG, 5′-ACCCGAATTCTCACTTACTACCGTATTCTCG | MG1655 | hipA gene in NcoI-EcoRI sites of pBADMycHis; a stop codon (boldface) was introduced to avoid expression of Myc epitope and His tail |
Primers used for PCR amplification of the genes cloned in the corresponding plasmids. Restriction sites are underlined.
Template DNA used for PCR amplification of the genes.
Escherichia coli strain LMG194 (F− ΔlacX74 galE thi rpsL ΔphoA (PvuII) Δara714 leu::Tn10) was used as the host strain for the empty vector pBAD22CM and all the constructs described in Table 1.
Determination of the effect of protein overexpression on the different strains.
LMG194 cells carrying pBADCM plasmids were streaked on Luria-Bertani (LB) agar plates containing 0.2% glucose and 25 μg/ml chloramphenicol. Overnight cultures were grown from single colonies in RMG minimal medium (2% Casamino Acids, 1× M9 salts, 1 mM MgCl2, and 1% glycerol) supplemented with 25 μg/ml chloramphenicol (RMG-CM). Aliquots of 0.85 ml were mixed with 0.15 ml of sterile glycerol, snap-frozen in a dry ice-ethanol bath, and kept at −80°C.
For the protein expression experiments, cultures were started from these glycerol stocks. After an overnight growth in RMG-CM medium, cultures were diluted 100-fold into 30 ml of fresh medium in 125-ml Erlenmeyer flasks. Flasks were shaken at 160 rpm in a platform shaker (Innova 2300; New Brunswick Scientific) at 37°C. When cell densities of the cultures reached an optical density at 600 nm (OD600) of ∼0.15, 3- to 5-ml aliquots were transferred to 30- by 115-mm conical plastic tubes. Arabinose was added at different concentrations for an adequate time period (see below). Tubes were placed at an inclination of 45°, and shaking continued under the same conditions. Cell growth was determined by monitoring the OD600. The numbers of viable cells (CFU) in the cultures, before and after the addition of arabinose, were determined as follows. Ten-microliter aliquots were serially diluted in a 10-fold fashion. One hundred-microliter aliquots from these dilutions were plated on LB agar-chloramphenicol plates and kept overnight at 37°C. Due to the toxicity of the proteins expressed, a drop in the cell counts was expected. Therefore, it was important to carry out a wide range of dilutions in order to use the adequate ones to reliably determine CFU.
Determination of persistence frequency in cells overexpressing proteins.
To determine the number of persisters formed by cells expressing proteins from pBAD vectors, cells were grown and protein expression was induced as described above. We found that keeping these conditions was critical to obtain reproducible persistence frequencies between experiments. Similar observations about different factors influencing reproducibility of the persistence frequency have been reported previously (15). An antibiotic, ampicillin (Fisher Scientific BP1760) or ciprofloxacin (Bayer Co.), was added from stock solutions to a final concentration of 100 μg/ml or 0.4 μg/ml, respectively. Incubation continued for 4 h. Chloramphenicol (25 μg/ml) was kept throughout the experiments to ensure the stability of the plasmids. In order to determine viable-cell counts in the cultures after incubation with the antibiotics, 1-ml aliquots were removed and cells were spun and resuspended in fresh medium without antibiotics. Determination of viable cells was performed as described above. Persistence frequency was calculated by dividing the number of CFU/ml in the culture after incubation with the antibiotic by the number of CFU/ml in the culture before adding the antibiotic.
RESULTS AND DISCUSSION
Cells overexpressing toxins from TA modules show an increase in persistence frequency (4, 7, 15, 16, 20). Here we asked if expression of proteins that become toxic when ectopically produced also results in a higher frequency of persister formation. In order to test this, we chose to express in E. coli cells, in parallel to the toxins from the TA modules HipA and MazF, two unrelated proteins. One was protein DnaJ from the chaperone system DnaJ/DnaK/GrpE (12), which becomes toxic when overexpressed from a high-copy-number plasmid (http://ecoli.aist-nara.ac.jp/gb5/Resources/archive/archive.html). The second protein, PmrC from Salmonella enterica serovar Typhimurium 14028s, an enzyme that transfers phosphoethanolamine to lipid A, is also toxic to E. coli cells when expressed from a plasmid (17). In addition, ectopic expression of thrB was used as a control, since threonine B, a component of homoserine kinase, can be produced to high levels with no detrimental consequences for cell growth (13).
Due to the expected differential toxicity of the proteins, it was important to express them in a tightly regulated system. The PCR-amplified genes hipA, mazF, and dnaJ (from E. coli K-12 strain MG1655) and pmrC (from the ppmrC plasmid described in reference 17) were cloned under the control of the ParaBAD promoter into the pBADCM vector, carrying a chloramphenicol resistance determinant (see above). E. coli LMG194 cells unable to metabolize l-arabinose (10) were transformed with the obtained expression plasmids and were grown in RM minimal medium supplemented with 1% glycerol and chloramphenicol (25 μg/ml). In this expression system, the activity of the PBAD promoter could be tightly controlled by the concentration of arabinose, and the basal levels of expression of the cloned genes are negligible (10).
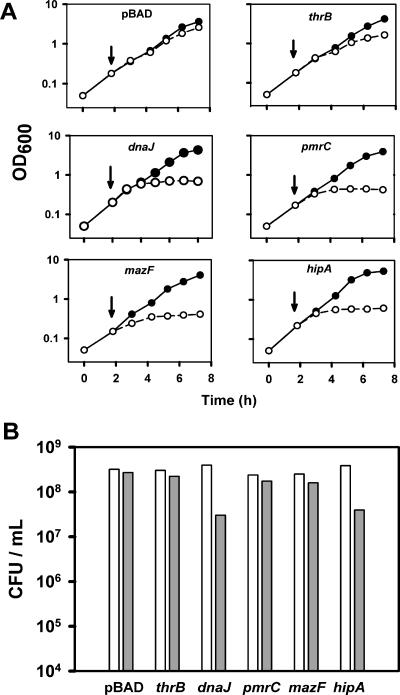
To test how expression of the cloned genes affects cell growth, arabinose at a final concentration of 0.2% was added to logarithmically growing cultures at 37°C. Overexpression of the control protein ThrB slowed down the growth rate of the cells in a relatively mild fashion. In contrast, expression of toxins HipA and MazF, as well as proteins DnaJ and PmrC, caused a complete growth arrest after some time at this saturating concentration of inducer (Fig. 1A). The toxic effect of the toxins HipA and MazF or the proteins DnaJ and PmrC on dividing cells could be reached at different levels of induction and, generally, could be bacteriostatic or bactericidal. This fact makes difficult the analysis and interpretation of any experiments comparing cells producing these proteins. In the case of TA module proteins, it has been reported that ectopic overexpression of MazF may have a lethal effect depending on the conditions and duration of the toxin expression (1, 22). Similarly, overproduction of HipA has been shown to cause occasional lysis of the cells (16). No information in this regard was available for the effect of overexpressing DnaJ or PmrC. Therefore, it was important to adjust the expression of the different proteins so that they would produce comparably mild inhibitory effects on the growth rate of the cells. In order to achieve this, logarithmically growing cultures of each strain were induced with different concentrations of arabinose. The densities (OD600) and numbers of viable cells (CFU) of the cultures were determined over time (not shown). Based on these data, induction conditions were chosen under which expression of the cloned proteins caused a minimal but evident decrease in the culture densities compared to the uninduced controls. Under these conditions, protein expression barely affected (strain with empty vector and strains expressing ThrB, PmrC, and MazF) or decreased only moderately (strains expressing DnaJ and HipA) the number of viable cells in the cultures (Fig. 1B). The level of protein expression in these cultures was examined by gel electrophoresis (not shown). A 34-kDa band corresponding to ThrB could be clearly identified, in agreement with the fact that ThrB can reach high expression levels without having any important effect on the growth rate or the number of cells (Fig. 1). In contrast, no bands could be detected for either DnaJ, PmrC, MazF, or HipA on Coomassie-stained gels. This suggests that the toxic effect of all these proteins is attained at low levels of induction. It should be noted that at low level of induction protein expression may vary between individual cells (24).
FIG. 1.
Effect of toxic proteins on the growth and viability of E. coli cells. A. Growth curves of LMG194 cells transformed with pBAD plasmids to induce expression of the indicated genes. Arrows show the moment of addition of arabinose. Solid circles, no arabinose; open circles, arabinose added. B. Number of live cells in strains producing toxic proteins after optimizing the concentration of inducer (see text). Arabinose concentrations were as follows: 0.2% for the empty vector and thrB; 0.05% for dnaJ; and 0.015% for pmrC, mazF, and hipA. White bars, no arabinose; gray bars, arabinose added.
The frequency of persister formation in cells expressing proteins under the conditions described above was determined. In order to do this, bactericidal antibiotics with unrelated mechanisms of action, ampicillin and ciprofloxacin, were used. Logarithmically growing cells were induced with adequate concentrations of arabinose for 1 (strains expressing ThrB, HipA, MazF, or PmrC or carrying an empty vector) or 2 h (DnaJ-producing strain). The induced cultures were then challenged with either 0.4 μg/ml of ciprofloxacin or 100 μg/ml of ampicillin for 4 h. In order to determine persistence frequency in the induced cultures, a fraction of the cells before and after the antibiotic treatment were washed with fresh medium and plated on LB agar-chloramphenicol plates containing 0.2% glucose to prevent any further production of the toxins. The frequency of persister formation was also determined for each strain grown in the absence of inducer. Figure 2 shows the ratios of the frequency of appearance of persisters in strains overproducing the cloned protein compared to the frequency in the absence of the inducer. Overexpression of the control protein ThrB, which showed only a mild negative effect on the growth of the cells, did not significantly increase the frequency of persisters. In contrast, strains expressing at limited induction levels members of the TA pairs MazF and HipA increased the frequency of formation of both ampicillin and ciprofloxacin persisters 100- to 10,000-fold. Remarkably, a similar increment of persistence was also detected for the strains expressing the chaperone DnaJ or even the foreign protein PmrC (100- to 1,000-fold), under comparably low levels of induction. These results show that, at least in the mid-log-phase cells used for these experiments, the increase in number of persisters is not unique to strains expressing HipA or other dedicated toxins. Instead, cells expressing proteins totally unrelated to the TA modules, which become toxic when ectopically expressed, also have the ability to form more persisters. It has been suggested that the formation of persisters during stationary phase is highly dependent on the hipBA locus, since its deletion causes a sharp drop in persister formation during this stage (15). However, to the best of our knowledge, no systematic effort to study the effect of other gene deletions on persistence of stationary-phase cells has been reported. Therefore, it is possible that deletions of genes other than hipA might also have the effect of diminishing persistence frequency.
FIG. 2.
Frequency of persister formation in strains expressing toxic proteins. Cells producing proteins under the conditions indicated in Fig. 1B were challenged with ampicillin (black bars) or ciprofloxacin (gray bars). The persistence frequency (f+) was estimated relative to that for cells challenged with the antibiotics but grown without arabinose (f−). A value of 1 would indicate no effect of protein overexpression on the frequency of persister formation.
One of the main questions about the phenomenon of persistence is whether bacterial cells have evolved a dedicated mechanism that allows them to survive lethal stresses including exposure to bactericidal antibiotics. Such a mechanism may potentially be based on a deliberate or sporadic expression of toxin proteins from the TA modules in a small fraction of cells to maintain them in a dormant or nongrowing stress-tolerant state (9, 15, 18). However, our findings suggest that limited levels of induction of unrelated proteins also increase persistence. Deleterious effects of such proteins will be attained at different intracellular concentrations, and the mechanism of their toxicity will likely have nonspecific targets. Given the low frequency of persisters in a wild-type population (∼10−6), it is conceivable that stochastic variations or illicit gene expression would lead to rare toxic levels of a broad variety of proteins in a small fraction of cells, increasing their chances to become persisters.
Acknowledgments
We are grateful to G. H. Araia for excellent technical assistance. The critical reading of the manuscript by A. S. Mankin is greatly appreciated.
This work was supported by Public Health Service grants AI-049214 and AI- 056575 from the National Institute of Allergy and Infectious Diseases to A.N.
REFERENCES
- 1.Amitai, S., Y. Yassin, and H. Engelberg-Kulka. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 186:8295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Lieber. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 3.Bigger, J. W. 1944. Treatment of staphyloccocal infections with penicillin. Lancet ii:497-500. [Google Scholar]
- 4.Black, D. S., A. J. Kelly, M. J. Mardis, and H. S. Moyed. 1991. Structure and organization of hip, an operon that effects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173:5732-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, D. S., B. Irwin, and H. S. Moyed. 1994. Autoregulation of hip, and operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 176:4081-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809-819. [DOI] [PubMed] [Google Scholar]
- 7.Falla, T. J., and I. Chopra. 1998. Joint tolerance to β-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob. Agents Chemother. 42:3282-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falla, T. J., and I. Chopra. 1999. Stabilization of Rhizobium symbiosis plasmids. Microbiology 145:515-516. [DOI] [PubMed] [Google Scholar]
- 9.Gerdes, K., S. K. Christensen, and A. Løbner-Olesen. 2005. Prokaryotic toxin-antitoxin stress loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 10.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann, F., and U. Rinas. 2004. Stress induced by recombinant protein production in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 89:73-92. [DOI] [PubMed] [Google Scholar]
- 12.Houry, W. A. 2001. Chaperone-assisted protein folding in the cell cytoplasm. Curr. Protein Pept. Sci. 2:227-244. [DOI] [PubMed] [Google Scholar]
- 13.Huo, X., and R. E. Viola. 1996. Functional group characterization of homoserine kinase from Escherichia coli. Arch. Biochem. Biophys. 330:373-379. [DOI] [PubMed] [Google Scholar]
- 14.Keren, I., N. Kaldalu, A. Spoering, Y. Wang, and K. Lewis. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13-18. [DOI] [PubMed] [Google Scholar]
- 15.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korch, S. B., T. A. Henderson, and T. M. Hill. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199-1213. [DOI] [PubMed] [Google Scholar]
- 17.Lee, H., F.-F. Hsu, J. Turk, and E. A. Groisman. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186:4124-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moyed, H. S., and S. H. Broderick. 1986. Molecular cloning and expression of hipA, a gene of Escherichia coli that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 166:399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131-140. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501-510. [DOI] [PubMed] [Google Scholar]
- 23.Scherrer, R., and H. S. Moyed. 1988. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J. Bacteriol. 170:3321-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siegele, D. A., and J. C. Hu. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 94:8168-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]