Abstract
The halophilic bacterium Chromohalobacter salexigens synthesizes and accumulates compatible solutes in response to salt and temperature stress. 13C-nuclear magnetic resonance analysis of cells grown in minimal medium at the limiting temperature of 45°C revealed the presence of hydroxyectoine, ectoine, glutamate, trehalose (not present in cells grown at 37°C), and the ectoine precursor, Nγ-acetyldiaminobutyric acid. High-performance liquid chromatography analyses showed that the levels of ectoine and hydroxyectoine were maximal during the stationary phase of growth. Accumulation of hydroxyectoine was up-regulated by salinity and temperature, whereas accumulation of ectoine was up-regulated by salinity and down-regulated by temperature. The ectD gene, which is involved in the conversion of ectoine to hydroxyectoine, was isolated as part of a DNA region that also contains a gene whose product belongs to the AraC-XylS family of transcriptional activators. Orthologs of ectD were found within the sequenced genomes of members of the proteobacteria, firmicutes, and actinobacteria, and their products were grouped into the ectoine hydroxylase subfamily, which was shown to belong to the superfamily of Fe(II)- and 2-oxoglutarate-dependent oxygenases. Analysis of the ectoine and hydroxyectoine contents of an ectABC ectD mutant strain fed with 1 mM ectoine or hydroxyectoine demonstrated that ectD is required for the main ectoine hydroxylase activity in C. salexigens. Although in minimal medium at 37°C the wild-type strain grew with 0.5 to 3.0 M NaCl, with optimal growth at 1.5 M NaCl, at 45°C it could not cope with the lowest (0.75 M NaCl) or the highest (3.0 M NaCl) salinity, and it grew optimally at 2.5 M NaCl. The ectD mutation caused a growth defect at 45°C in minimal medium with 1.5 to 2.5 M NaCl, but it did not affect growth at 37°C at any salinity tested. With 2.5 M NaCl, the ectD mutant synthesized 38% (at 37°C) and 15% (at 45°C) of the hydroxyectoine produced by the wild-type strain. All of these data reveal that hydroxyectoine synthesis mediated by the ectD gene is thermoregulated and essential for thermoprotection of C. salexigens.
A common strategy of osmoadaptation for eukaryotic and prokaryotic microorganisms involves the accumulation of small organic molecules by transport and/or biosynthesis. Such osmoregulatory compounds have been termed compatible solutes because they do not interfere with central metabolism, even if they accumulate to high concentrations (6). These compounds can be classified into different categories, such as polyols and heterosides, sugars, amino acids and their derivatives, N-acetylated diamino acids, betaines and tethines, and ectoines (ectoine [E] and hydroxyectoine [OH-E]) (5, 15, 16, 18, 54).
During recent years, a growing body of evidence has indicated that the role of compatible solutes goes beyond osmotic adjustment alone and includes the protection of cells and cell components from freezing, desiccation, high temperature, or oxygen radicals. Thus, the effect of compatible solutes is not only that they can be accumulated up to molar concentrations without inhibiting enzyme functions but also that they are able to stabilize proteins (42, 49, 55). Thermophilic and hyperthermophilic microorganisms that live in hot saline environments accumulate some compatible solutes that have not been found to play a role in mesophilic organisms, which led to the assumption that they are also involved in thermoprotection. Most interestingly, in some (hyper)thermophilic archaea, such as Pyrococcus furiosus, the concentration of the negatively charged compatible solute di-myo-inositol-phosphate varies as a function of the temperature but not of the salinity, indicating that this compound is not a compatible solute in the strict sense but is more like a thermoprotectant (39). On the other hand, the protective role of trehalose against several stress conditions, including salinity, desiccation, and temperature, has been broadly demonstrated for fungi, Saccharomyces cerevisiae, and bacteria (9, 24, 51, 55). Very recently, Holtmann and Bremer (26) reported that glycine betaine and several structurally related compounds serve as effective thermoprotectants for Bacillus subtilis, and they demonstrated that the osmoregulated Opu transporters are required for compound uptake at elevated growth temperatures. In contrast, glycine betaine did not have an influence on the desiccation tolerance of Escherichia coli (56). In vitro studies have suggested that the protective effect of hydroxyectoine against different types of stress is superior to that of ectoine or other compatible solutes. Thus, hydroxyectoine conferred protection to E. coli against osmotic and heat stress (35), and its capacity to provide desiccation tolerance to E. coli and Pseudomonas putida was comparable to that of trehalose (36, 37). On the other hand, Halomonas elongata and Streptomyces griseus accumulated hydroxyectoine in response to a temperature upshift, providing the first evidence that hydroxyectoine might function as a thermoprotectant in vivo (35, 57).
Ectoines (ectoine and its hydroxylated derivative, hydroxyectoine) are accumulated by halophilic/halotolerant representatives of bacteria belonging to the actinobacteria, firmicutes, and proteobacteria (16, 54). Ectoine was first discovered in the extremely halophilic phototrophic sulfobacterium Ectothiorhodospira halochloris (17), whereas hydroxyectoine was originally discovered in the actinomycin D producer Streptomyces parvulus (28). The ectABC gene cluster involved in the synthesis of ectoine has been isolated and characterized for a number of strains belonging to the above bacterial groups (10, 13, 21, 22, 32, 34, 46). In addition, the ectoine hydroxylase gene (thpD), responsible for the conversion of ectoine to hydroxyectoine, has been described for the actinomycin D producer Streptomyces chrysomallus as forming part of the thpABCD (ectABCD) gene cluster (22). The thpD gene was expressed in the heterologous host H. elongata (able to synthesize ectoine and hydroxyectoine itself), leading to more hydroxyectoine production (43). In contrast, all other ectABC clusters characterized so far do not have a thpD (ectD)-like gene downstream of ectC. To date, the gene(s) for hydroxyectoine biosynthesis in true halophilic bacteria has not been isolated.
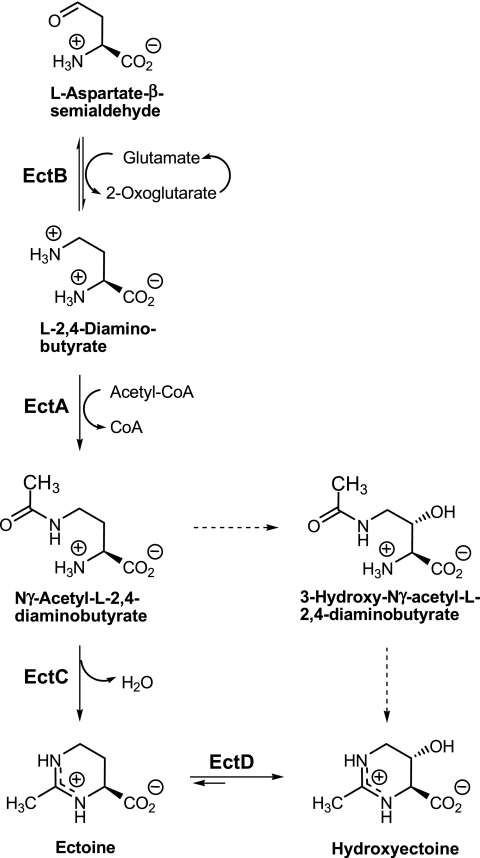
We are interested in the stress response of Chromohalobacter salexigens, a halophilic γ-proteobacterium of the family Halomonadaceae showing a very broad salinity range of growth (2). This microorganism can grow in minimal M63 medium with 0.5 to 3 M NaCl, with optimal growth at about 1.5 M NaCl (11). On the other hand, its temperature range in complex SW-10 medium is 15 to 45°C, with optimal growth at 37°C (2). Osmoadaptation is mainly achieved by de novo synthesis of ectoine and hydroxyectoine (13). In addition, C. salexigens accumulates externally supplied osmoprotectants, such as glycine betaine, which is taken up from the medium or synthesized from choline (11, 12). In a previous work, we suggested that C. salexigens can synthesize hydroxyectoine by two different pathways, either directly from ectoine or via an alternative pathway that converts Nγ-acetyldiaminobutyric acid to hydroxyectoine without the involvement of ectoine (8) (Fig. 1). In addition, we isolated and characterized the C. salexigens ectABC genes, which encode the proteins comprising the main route for ectoine synthesis in this microorganism. The ectB gene codes for the enzyme diaminobutyric acid transaminase, which catalyzes the conversion of aspartate semialdehyde to diaminobutyric acid; ectA encodes diaminobutyric acid acetyltransferase, responsible for the acetylation of diaminobutyric acid to Nγ-acetyldiaminobutyric acid; and ectC encodes ectoine synthase, which catalyzes the cyclic condensation of Nγ-acetyldiaminobutyric acid to ectoine (10). In this work, we report the isolation and characterization of the ectoine hydroxylase gene ectD, which is the main gene responsible for hydroxyectoine synthesis in C. salexigens. We show that whereas hydroxyectoine synthesis in this bacterium is both osmo- and thermoregulated, an ectD mutant strain is thermosensitive but not osmosensitive. Our data indicate that hydroxyectoine is crucial for cells to grow at the limiting temperature. This is the first time that a connection between hydroxyectoine accumulation and thermoprotection of a halophilic bacterium has been established firmly at the molecular level.
FIG. 1.
Biosynthetic pathway for ectoines from aspartate semialdehyde in C. salexigens DSM 3043T (8). The solid and dashed lines indicate established and proposed steps, respectively. The enzymes involved are l-diaminobutyric acid transaminase (EctB), l-diaminobutyric acid acetyltransferase (EctA), ectoine synthase (EctC), and ectoine hydroxylase (EctD).
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. Chromohalobacter salexigens (formerly Halomonas elongata DSM 3043) strains were routinely grown in complex SW-2 medium containing 2% (wt/vol) total salts (53). Escherichia coli was grown aerobically in complex LB medium (41). M63 (14), which contains 20 mM glucose as the sole carbon source, was used as minimal medium for C. salexigens and E. coli. The osmotic strength of M63 was increased by the addition of a 0.75 to 3.0 M final concentration of NaCl. Although C. salexigens can grow in M63 with 0.5 M NaCl, growth is extremely slow at this salinity, and cells take a very long time to reach exponential phase. Therefore, we used M63 with 0.75 M NaCl as the standard medium for a low salt concentration in all experiments. The pHs of all media were adjusted to 7.2 with KOH. Solid media contained 20 g of Bacto agar per liter (Difco). Cultures were incubated at 37°C (E. coli) or at temperatures ranging from 20 to 45°C (C. salexigens) in an orbital shaker at 200 rpm. When used, filter-sterilized antibiotics were added at the following final concentrations (μg ml−1): ampicillin (Ap), 150 for E. coli; chloramphenicol, 25 for E. coli; gentamicin (Gm), 20 for E. coli and 25 for C. salexigens; kanamycin (Km), 50 for E. coli and 75 for C. salexigens; rifampin (Rf), 25 for E. coli and C. salexigens; and streptomycin (Sm), 20 for E. coli and 50 for C. salexigens. When used, ectoine and hydroxyectoine (Bitop, Witten, Germany) were added to the media at a final concentration of 1 mM. Growth was monitored as the optical density of the culture at 600 nm (OD600) with a Perkin-Elmer Lambda 25 UV/Vis spectrophotometer.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or description | Source or reference |
|---|---|---|
| C. salexigens strains | ||
| DSM 3043T | Wild type | 2 |
| CHR61 | Spontaneous Rfr mutant of DSM 3043 | 13 |
| CHR62 | CHR61 ΔectABC::Tn1732; deficient in ectoine and hydroxyectoine synthesis; Rfr Kmr | 13; R. García-Estepa and C. Vargas, unpublished results |
| CHR136 | CHR61 ectD::Ω; deficient in hydroxyectoine synthesis; Rfr Smr Spr | This study |
| CHR137 | CHR61 ΔectABC::Tn1732ectD::Ω; deficient in ectoine and hydroxyectoine synthesis; Rfr Kmr Smr Spr | This study |
| E. coli strain | ||
| DH5α | supE44 Δ(lac)U169 φ80dlacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1; host for DNA manipulations | 23 |
| Plasmids | ||
| pKS(−) | Cloning vector; Apr | Stratagene |
| pHP45Ω | pBR322 derivative carrying the Ω cassette; Apr Smr Spr | 44 |
| pRK600 | Helper plasmid; Cmrtra | 29 |
| pJQ200-SK | Suicide vector; Gmrmob sac | 45 |
| pEctD16 | pVK102 derivative carrying the orf1-orf2-ectD-orf4 region of C. salexigens; Kmr | This study |
| PRectD1 | 2.9-kb PstI-EcoRI fragment from pEctD16 (containing orf2 ectD orf4) cloned into pKS; Apr | This study |
| pRectD4 | pRectD1 derivative with Ω cassette insertion within ectD; Apr Smr Spr | This study |
| pRectD5 | 4.9-kb PstI-SalI fragment from pRectD4 (containing orf2 ectD::Ωorf4) cloned into pJQ200-SK | This study |
Conjugal transfer of plasmids.
Plasmids were transferred from E. coli to C. salexigens by triparental mating on SW-2 medium, using pRK600 as a helper plasmid, as described by Vargas et al. (53).
Methods for nucleic acid manipulation.
Plasmid DNA was isolated from E. coli with a Wizard Plus SV miniprep kit (Promega), and genomic DNA was isolated with a Quantum Prep Aquapure genomic DNA kit (Bio-Rad). Restriction enzyme digestion and ligation were performed as recommended by the manufacturers (Amersham-Pharmacia Biotech and Promega). DNA sequencing was performed by Newbiotechniques (Seville, Spain).
To isolate a fragment of the gene encoding the ectoine hydroxylase from C. salexigens, a PCR strategy with degenerate oligonucleotides was designed. To design the primers for PCR, we performed an alignment of the amino acid sequences of the ectoine hydroxylase from S. chrysomallus, ThpD (kindly provided by Nicholas Grammel prior to publication), the product encoded by an ortholog of thpD immediately downstream of the ectABC cluster in Streptomyces coelicolor (accession number AL591322), and other proteins showing high similarity with these two, such as the l-proline 4-hydroxylase from Dactylosporangium sp. (BAA20094), the alpha-ketoglutarate-dependent hypophosphite dioxygenase from Pseudomonas stutzeri (AF061267), and a putative oxygenase from Streptomyces viridochromogenes (AF33038). After identification of well-conserved regions, a number of degenerate oligonucleotides were designed (up to 10) and used in different combinations in amplification reaction mixtures with 100 ng of chromosomal DNA from C. salexigens as a template, 3.5 mM MgCl2, and Taq DNA polymerase (Promega). The PCRs were performed in a Robocycler Gradient 96 device (Stratagene) under the following conditions: 3 min at 94°C, a first set of 12 cycles of 30 s at 94°C plus 1 min with a temperature gradient from 57 to 45°C plus 30 s at 72°C, a second set of 18 cycles with 30 s at 94°C plus 1 min at 45°C plus 30 s at 72°C, and finally, 5 min at 72°C. The PCR products generated were separated in a 2% agarose gel, extracted from the gel by using a QIAquick gel extraction test kit (QIAGEN), reamplified by PCR, and sequenced. The amino acid sequence of one of the resulting products (213 nucleotides) showed 49% identity with the corresponding region of the ThpD protein and therefore was assumed to be part of the ectoine hydroxylase from C. salexigens. The primers that yielded this product were P4, with the sequence 5′-ATGCC(GC)GG(GCT)TC(GC)CACAAG-3′, and P6, with the sequence 5′-GTTGTC(GC)CC(GC)GA(GC)CCGTG-3′.
To clone the complete C. salexigens gene for ectoine hydroxylase, the 213-bp PCR product was labeled with digoxigenin by using a nonradioactive labeling and detection kit (Roche) and was used as a probe against a wild-type C. salexigens genomic library constructed in E. coli HB101 (13). For colony hybridization, 3,000 single colonies were allowed to grow for 12 h at 37°C on LB plates with Km. After growth, plates were chilled for 1 h at 4°C and transferred to nylon membranes (Amersham-Pharmacia Biotech), as described by Sambrook and Russell (48), and membranes were squashed between two filter papers to remove cellular debris leading to background. Prehybridization was done with 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate with shaking for 2 h at 68°C. Hybridization, washes, and detection were done according to the instructions provided by the nonradioactive labeling and detection kit (Roche). Plasmid DNAs were isolated from 17 colonies giving a positive hybridization signal, digested with SalI, and hybridized with the digoxigenin-labeled 213-bp fragment. Fourteen of these cosmid clones shared a 4.7-kb SalI fragment that gave a positive hybridization signal. One of these clones was named pEctD16 and kept for further analysis.
To generate C. salexigens mutants affected in the ectoine hydroxylase gene, ectD, a 2,868-bp fragment from pEctD16 containing orf2, ectD, and 525 bp of orf4 was PCR amplified with Pfu Turbo DNA polymerase (Stratagene) by using two synthetic oligonucleotides (5′-CCTAGGCGGGCTGCAGGCAGGGTATT-3′ and 5′-CGGTGAGCACGAATTCGCCGGGCTGC-3′) that were modified (residues in bold) to introduce a PstI and an EcoRI site, respectively (underlined). The resulting PCR fragment was digested with PstI and EcoRI, and the 2,842-bp fragment generated was cloned into pKS to give plasmid pRectD1. pRectD1 was subsequently linearized with the enzyme Bsp119I (Fermentas, Germany), whose recognition site overlaps with residues 193 to 198 of the ectD sequence, made blunt ended by treatment with deoxynucleoside triphosphates and T4 DNA polymerase (Promega), and ligated to a 2-kb SmaI fragment from pHP45-Ω (44), containing the Ω interposon for insertional mutagenesis (Smr). The resulting plasmid was named pRectD4. To recombine the ectD mutation into the C. salexigens chromosome, a 4.9-kb PstI-SalI fragment from pRectD4 was cloned into the suicide vector pJQSK200 (Gmr) (45) to give plasmid pRectD5, which was mobilized into the C. salexigens wild-type strain and the ectoine-deficient mutant CHR62 (ΔectABC::Tn1732) by triparental mating. Mutant strains resulting from a double homologous recombination event were identified as Smr Gms colonies on SW-2 plates containing 10% sucrose. Two of these colonies were purified for further analysis and were named CHR136 (ectD::Ω) and CHR137 (ΔectABC::Tn1732 ectD::Ω). Insertion of the omega cassette in CHR136 and CHR137 was confirmed by PCR.
Extraction and determination of intracellular solutes by 13C-NMR spectroscopy.
C. salexigens was grown in 200 ml of M63 with 2.5 M NaCl at 45°C until late exponential phase (OD600 = 1 to 1.2). Cells were collected by centrifugation and washed with the same medium without any carbon source. The cell pellet was resuspended in 10 ml of extraction mixture (methanol:chloroform:water [10:5:4]) and extracted with gentle shaking for 30 min at 37°C. Cell debris was removed by centrifugation, and supernatants were extracted once with chloroform:water (1:1) and freeze dried. The solids were dissolved in D2O (0.6 ml). 13C-nuclear magnetic resonance (13C-NMR) spectra were recorded at 25°C on a Brucker AV500 spectrometer at 125 MHz. The chemical shifts are reported in ppm on the δ scale relative to tetramethylsilane. Signals were assigned by comparison with previously published chemical shift values (8, 13) and confirmed by comparison with 13C-NMR spectra of pure compounds.
HPLC analysis of ectoine and hydroxyectoine.
Cells for high-performance liquid chromatography (HPLC) analysis of ectoine and hydroxyectoine were extracted by using a modified Bligh and Dyer technique as described by Kraegeloh and Kunte (31). Samples of 20 μl of the water-soluble fraction containing compatible solutes were analyzed by isocratic HPLC on a Spherisorb S3 NH2 column (150 mm by 4.6-mm inner diameter; Waters), with 80% acetonitrile in water as the eluant, at a flow rate of 1 ml/min at 25°C. Ectoine and hydroxyectoine were monitored by their absorbance values at 210 nm by using a UV/Vis detector. The retention times of ectoine and hydroxyectoine were determined by using commercially available ectoine and hydroxyectoine samples.
Determination of protein content.
The amount of cell protein was determined by the method described by Smith et al. (52), using a bicinchoninic acid protein assay kit (Pierce). Cell suspensions (1 ml) were centrifuged at 13,000 rpm for 3 min, and the supernatants were removed. Cell pellets were dried overnight at 100°C and resuspended in 1 ml of demineralized water by shaking at room temperature for 30 min. After the addition of 2 ml of the bicinchoninic acid reagent to 100 μl of sample and incubation at 37°C for 30 min, the absorption of samples was measured at 550 nm in a Perkin-Elmer spectrophotometer (Lambda 25) and compared to that of protein standards containing bovine serum albumin in the concentration range from 0 to 600 μg/ml.
Computer analysis.
DNA sequences were assembled with the CAP program (27) available at http://bio.ifom-firc.it/ASSEMBLY/capdoc.html. Searches for open reading frames and homologies were performed at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/), using the ORF Finder and iterative PSI-BLAST (1) programs. Conserved domains within proteins were identified at NCBI's Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (38). Specific BLAST searches within the C. salexigens genome database were performed at http://genome.ornl.gov/microbial/csal/. All searches in the databases were repeated immediately before submission of this work. Primary structure analysis of proteins was performed by using ProtParam (20) at the EXPASY proteomic server (http://www.expasy.ch/tools/protparam.html). Protein sequences were formatted by using ReadSeq (http://searchlauncher.bcm.tmc.edu/seq-util/seq-util.html) and aligned with the CLUSTALW 1.8 algorithm (25) available at the Baylor College of Medicine Search Launcher (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html). For a publishable presentation, the output of the alignment made with CLUSTALW was used as input for the BOXSHADE 3.21 program at http://www.ch.embnet.org/software/BOX_form.html. Sequence identity between two proteins was estimated by performing a global alignment, using the ALIGN program at the GENESTREAM network server (http://www2.igh.cnrs.fr/bin/align-guess.cgi). Phylogenetic analyses were conducted using MEGA, version 3.1 (33).
Nucleotide sequence accession number.
The nucleotide sequence of the C. salexigens orf1-orf2-ectD-orf4 region has been deposited in the EMBL database with the accession number AM231629.
RESULTS
Accumulation of ectoine and hydroxyectoine during the growth cycle of C. salexigens.
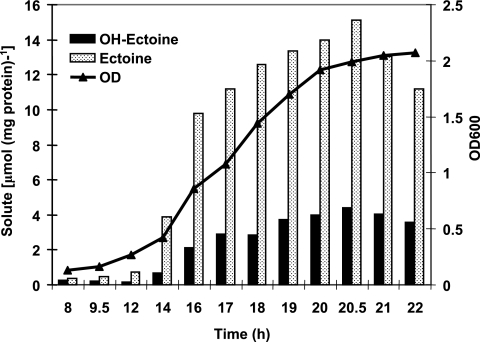
In previous works, we used 13C-NMR to show that ectoines are the main compatible solutes accumulated by C. salexigens cells grown in M63 at 37°C with 0.5 to 2.5 M NaCl (7, 10, 13). In this work, we used HPLC to quantify the cytoplasmic accumulation of these solutes during the growth cycle or in response to environmental stressors, such as increasing salinity or temperature. To assess the influence of growth phase on compatible solute levels, the intracellular ectoine and hydroxyectoine pools were determined in a C. salexigens wild-type strain grown in M63 at the optimal temperature (37°C) (2) and salinity (1.5 M NaCl) (11) (Fig. 2). Despite ectoine being by far the predominant solute at all growth stages, both compatible solutes followed similar patterns of accumulation, with low levels during early exponential phase (OD600 = 0.4) and a sudden increase at middle exponential phase (OD600 = 0.8). This was followed by increasing levels of solutes during late exponential growth, reaching a maximum after 20.5 h, which corresponded to the stationary phase (OD600 = 1.99). At this point, the concentrations of ectoine and hydroxyectoine were 15.12 and 4.4 μmol mg of protein−1, respectively. After this growth stage, the levels of both solutes started to decrease progressively. These results, which show that the accumulation of ectoines in C. salexigens is maximal during stationary phase, prompted us to perform all measurements in cultures grown to this phase of growth.
FIG. 2.
Correlation between growth phase and accumulation of ectoines by C. salexigens. The wild-type strain was grown in M63 minimal medium containing 1.5 M NaCl at 37°C, and the intracellular concentrations of ectoine and hydroxyectoine were determined by HPLC at different phases of growth. Experiments were repeated twice, and the data correspond to means, which on no occasion varied more than 5%. OD600, optical density at 600 nm.
Effect of temperature and salinity on accumulation of ectoine and hydroxyectoine by C. salexigens.
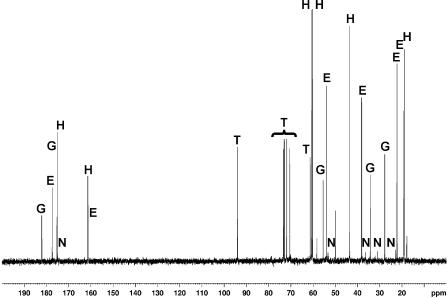
Since the compatible solute content of C. salexigens had never been measured in cells grown at high temperatures, we first used 13C-NMR to test the solute pattern of cells grown at 45°C in M63 with 2.5 M NaCl, which is the optimal salinity for growth at this temperature (see below). The 13C-NMR spectrum of the extract of C. salexigens grown under these conditions contained five sets of resonances, which were assigned to hydroxyectoine, ectoine, trehalose, glutamate, and the ectoine precursor, Nγ-acetyldiaminobutyric acid (Fig. 3). In addition, there was a sixth set of resonances (at 17.7, 49.7, and 58.2 ppm) that could not be assigned to any known compound. The finding that C. salexigens accumulates trehalose under high-temperature conditions was unexpected, since this compatible solute had never been detected in the wild-type strain grown at 37°C (7, 13). Although 13C-NMR is not a quantitative technique, it was evident that cells grown at 45°C had a proportion of hydroxyectoine that was much higher than that in cells grown at 37°C with the same salinity (7).
FIG. 3.
Natural-abundance 13C-NMR spectrum of major cytosolic solutes of the C. salexigens wild-type strain grown in M63 minimal medium with 2.5 M NaCl at 45°C. The major solutes were hydroxyectoine (H), ectoine (E), trehalose (T), glutamate (G), and Nγ-acetyldiaminobutyric acid (N).
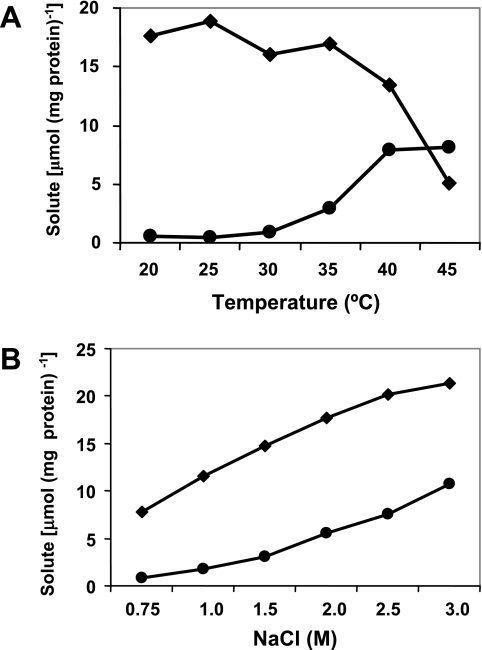
To determine the influence of temperature on the accumulation of ectoines by C. salexigens, the wild-type strain was grown in M63 with 1.5 M NaCl at temperatures ranging from 20 to 45°C (Fig. 4A). Ectoine was the predominant solute from 20 to 35°C, while the level of hydroxyectoine remained very low at these temperatures. From 35°C to 45°C, the content of ectoine decreased and that of hydroxyectoine increased, resulting in higher levels of hydroxyectoine than of ectoine in cells grown at 45°C. From 20°C to 45°C, the concentration of ectoine decreased 3.5-fold, whereas the concentration of hydroxyectoine increased 16-fold. The compatible solutes trehalose and glutamate could not be measured with the HPLC column used to quantify ectoines.
FIG. 4.
Effects of temperature (A) and salt stress (B) on the accumulation of ectoine (diamonds) and hydroxyectoine (circles) by C. salexigens. (A) Cells were grown in M63 minimal medium at the optimal salinity (1.5 M NaCl), and the concentrations of ectoines were determined by HPLC at temperatures ranging from 20 to 45°C. (B) Cells were grown in M63 minimal medium at the optimal growth temperature (37°C), and the concentrations of ectoines were determined by HPLC at salinities ranging from 0.75 to 3.0 M NaCl. The data are averages of three different replicates, which on no occasion varied more than 5%.
To quantify the influence of salinity on the accumulation of ectoines by C. salexigens, the wild-type strain was grown at 37°C in M63 with NaCl concentrations ranging from 0.75 to 3 M (Fig. 4B). Although higher levels of ectoine were found at all salinity levels, there was an evident increase in the contents of both compatible solutes with increasing salt concentrations. Cells grown with 3 M NaCl accumulated 21.3 μmol of ectoine mg of protein−1, a 2.75-fold higher level than that of cells grown with 0.75 M NaCl. The influence of salinity was more remarkable in the case of hydroxyectoine. Although the hydroxyectoine content remained low with 0.75 to 1.5 M NaCl and became more important above this salinity, cells grown with 3 M NaCl had 13.8-fold higher levels of hydroxyectoine than cells grown with 0.75 M NaCl. Taken together, these data indicate that the accumulation of hydroxyectoine in C. salexigens is up-regulated by salinity and temperature, whereas the accumulation of ectoine is up-regulated by salinity and down-regulated by temperature. In addition, hydroxyectoine becomes more important for cells grown under high-salinity and high-temperature conditions.
Cloning and DNA sequence analysis of the region carrying the ectD gene, which is involved in hydroxyectoine synthesis.
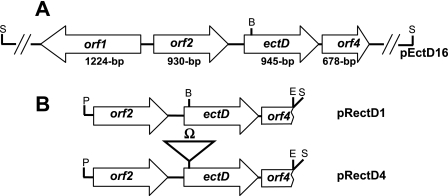
The cosmid clone pEctD16, containing the C. salexigens ectD gene, was isolated as described in Materials and Methods and then sequenced. Analysis of the 4,500-bp sequenced region with ORF Finder at the NCBI website revealed the presence of four open reading frames, with the first lying within the complementary strand (orf1) and the other three lying within the forward strand (orf234). They encode proteins with predicted molecular masses of 45.0 kDa (407 amino acids; Orf1), 34 kDa (309 amino acids; Orf2), 35.3 kDa (314 amino acids; Orf3 [EctD]), and 24.2 kDa (225 amino acids; Orf4) (Fig. 5). Each of these open reading frames is preceded by a potential ribosome binding site. Two more putative start codons were found upstream of ectD, but they are not preceded by ribosome binding sites. Inspection of the C. salexigens genome revealed the presence of an open reading frame downstream of orf4 lying within the complementary strand (not shown).
FIG. 5.
(A) Genetic organization of the sequenced 4,500 bp from plasmid pEctD16. (B) Generation of a construct carrying a mutated ectD gene. pRectD1 is a subclone of pEctD16 carrying the orf2-ectD-orf4 region. In pRectD4, the ectD gene was inactivated by the insertion of an Ω cassette, which carries resistance genes for streptomycin and spectinomycin, into its unique Bsp119I site (B).
Amino acid sequence homologies.
The amino acid sequences of the products encoded by the four open reading frames were compared with protein sequences deposited in the databases. These included the 3,347 proteins deduced from the sequence of the unfinished genome of C. salexigens, which were deposited in the databases while this work was in progress. The products encoded by orf1 and orf2 were very similar to γ-butyrobetaine hydroxylases (47) and to proteins annotated as members of the AraC-XylS family of transcriptional regulators (19), respectively. The product encoded by the fourth open reading frame contained two overlapping conserved motifs of the fumarylacetoacetate hydrolase and MhpD (2-keto-4 pentenoate hydratase) families of proteins, which are related to the catabolism of aromatic compounds (3, 4). orf1, orf2, and orf4 remain unassigned until experimental support confirms their functions.
The product encoded by the third open reading frame (ZP_00472926 in the annotated genome of C. salexigens) showed high similarity to the ectoine hydroxylase from S. chrysomallus (22), the putative ectoine hydroxylases from Streptomyces avermitilis and Nocardia farcinica, and two putative hydroxylases from “Methylobacter alcaliphilus” 20Z (“Methylomicrobium alcaliphilum” 20Z) (46) and S. coelicolor. Therefore, orf3 was designed ectD. We also found a homologous gene within the C. salexigens genome encoding a protein (ZP_00472926) which showed 51.9% identity to EctD. In addition, we found a high similarity between EctD and proteins from proteobacteria, firmicutes, or actinobacteria that had been annotated as l-proline-4-hydroxylases, proteins involved in the biosynthesis of mitomycin antibiotics, or phytanoil-coenzyme A (CoA)-dioxygenases. Despite the fact that the products of C. salexigens ectD and its homolog had themselves been annotated as phytanoil-CoA-dioxygenases (PhyH), the similarity between EctD and the human PhyH protein (40) was low. Many of the bacteria bearing homologs to C. salexigens EctD are either halophilic or halotolerant microorganisms. In order to check if they also carry the biosynthetic route for ectoine, we performed a search in the databases by using the ectA, ectB, and ectC genes as independent queries. Except for Bradyrhizobium japonicum, which only carries ectC, and Dactylosporangium sp., which could not be tested since its genome sequence is not available, all other bacteria carrying homologs to ectD also carried the ectABC genes for ectoine synthesis.
To gain insights into the relationships among EctD and its homologs, we performed an alignment with representative proteins, including the l-proline-4-hydroxylase from Dactylosporangium sp. (50) and the human phytanoil-CoA-hydroxylase, whose crystal structure and active sites have been determined recently (40). These enzymes are nonheme Fe(II)- and 2-oxoglutarate (2OG)-dependent oxygenases, which in general catalyze hydroxylation reactions. Figure 6A shows two partial alignments of the EctD-related proteins, corresponding to two evolutionarily very well-conserved regions containing the active sites of 2OG-dependent dioxygenases. Whereas the main residues of PhyH involved in binding of Fe(II) and 2OG were present in all enzymes, they were grouped into three clearly differentiated blocks, as shown in the cladogram derived from the complete alignment (Fig. 6B). The first block corresponds to PhyH, and the second one includes the l-proline-4-hydroxylase from Dactylosporangium and the putative l-proline-4-hydroxylase from B. japonicum. The third block contains the S. chrysomallus and N. farcinica ectoine hydroxylases and orthologous proteins, including EctD (and its homolog) from C. salexigens.
FIG. 6.
In silico analysis of EctD. (A) Partial alignment of EctD and related proteins. The amino acid sequence of the EctD protein cloned from Chromohalobacter salexigens (Csa1) was used as a query for PSI-BLAST. The sequences used were found in Streptomyces chrysomallus (Schr, [ThpI]), C. salexigens (Csa1 [EctD] and Csa2), Marinobacter aquaeoli (Maq1 and Maq2), Nocardia farcinica (Nfar), Brevibacterium linens (Blin), Bacillus clausii (Bcla), Nitrosococcus oceani (Noce), Sphingopyxis alaskensis (Sala), “Methylobacter alcaliphilus” (Malc), Bordetella parapertussis (Bpar), Alkalilimnicola ehrlichei (Aehr), Dactylosporangium sp. (Dact), Bradyrhizobium japonicum (Bjap), and humans (Huma). Identical and conserved residues are shown against black and gray backgrounds, respectively. The Fe(II)-binding and other residues involved in the active site of the human PhyH protein are indicated by arrows below the sequence. The 2-oxoglutarate-binding residues of PhyH are indicated by dots. Residues that are strictly conserved in all aligned sequences are indicated by asterisks below the PhyH sequence. Residues that are strictly conserved in all proposed ectoine hydroxylases are indicated by arrowheads above the ThpD sequence. The regions used to design the primers for isolation of the C. salexigens ectD gene are indicated by horizontal arrows above the ThpD sequence. (B) Cladogram of the alignment of the complete sequences of the proteins shown in panel A. Sequences in the neighbor-joining tree are identified by their species names and accession numbers. The ectoine hydroxylase family members are boxed to highlight their distinctness from other Fe(II)- and 2OG-dependent enzymes. Bar, 0.2 change per site.
The ectD gene is required for the main ectoine hydroxylase activity in C. salexigens.
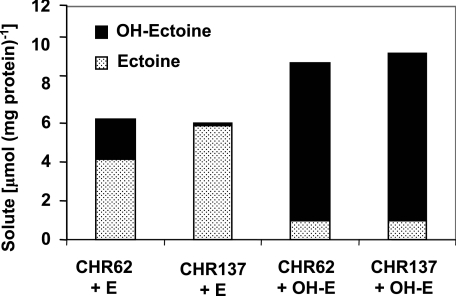
In a previous work, we demonstrated the existence of a reversible ectoine hydroxylase activity in C. salexigens by feeding the ectoine-deficient mutant CHR62 with ectoine or hydroxyectoine and determining its compatible solute content by 1H-NMR analysis (8). Although the CHR62 mutant, which accumulates low levels of diaminobutyric acid, was described as being affected in the ectA gene (8), we have found that the insertion of transposon Tn1732 actually caused a deletion covering ectABC (R. Garcia-Estepa and C. Vargas, unpublished data). To check the functionality of the ectD gene, we constructed an ectABC ectD double mutant (strain CHR137) by insertion of an omega cassette within ectD (Fig. 5B) followed by double recombination in the chromosome of strain CHR62. The parental strain CHR62 and the mutant CHR137 were grown at 37°C in M63 with 1.5 M NaCl in the presence of 1 mM ectoine or hydroxyectoine, and their compatible solute contents were measured by HPLC (Fig. 7). In the presence of 1 mM ectoine or 1 mM hydroxyectoine, CHR62 was able to accumulate both ectoine and hydroxyectoine, with E:OH-E ratios of 2:1 (in cells fed with ectoine) and 1:8.4 (in cells fed with hydroxyectoine). These data suggest that the conversion of hydroxyectoine to ectoine was very low under these conditions. Inactivation of the ectD gene drastically reduced hydroxyectoine production from ectoine in the ectABC ectD mutant strain CHR137. Thus, CHR137 grown in the presence of 1 mM ectoine accumulated 14-fold less hydroxyectoine than strain CHR62 grown under the same conditions, with an E:OH-E ratio of 39:1. However, strain CHR137 grown with ectoine was found to accumulate 7% of the hydroxyectoine accumulated by the parental strain CHR62 under the same conditions. When CHR137 was fed with 1 mM OH-E, the E:OH-E ratio was similar to that of strain CHR62 grown with OH-E (1:9.1). These data demonstrate that the product encoded by ectD is the main enzyme responsible for hydroxyectoine synthesis via ectoine hydroxylation.
FIG. 7.
ectD is required for the main ectoine hydroxylase activity in C. salexigens. Cells of the C. salexigens strains CHR62 (ΔectABC) and CHR137 (ΔectABC ΔectD) were grown at 37°C in M63 minimal medium with 1.5 M NaCl in the presence of 1 mM ectoine (E) or 1 mM hydroxyectoine (OH-E), and their compatible solute contents were determined by HPLC as described in Materials and Methods. The data are averages of three different replicates, which on no occasion varied more than 5%.
Hydroxyectoine synthesis mediated by ectD is essential for thermoprotection of C. salexigens.
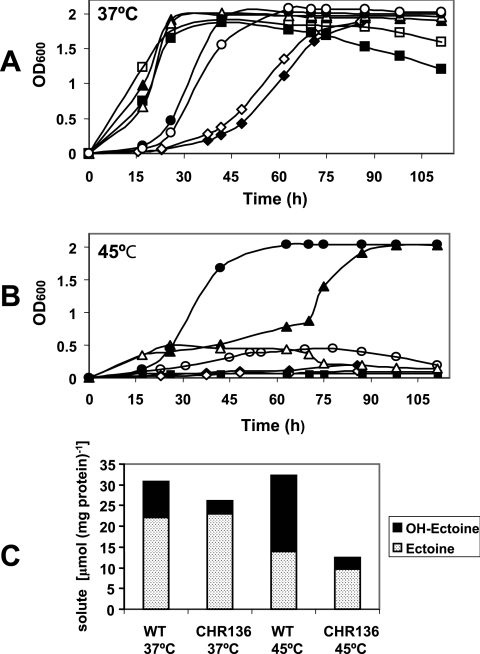
HPLC analysis of cytoplasmic extracts of C. salexigens cells grown at 45°C strongly suggested that hydroxyectoine may play a role in temperature protection. To test this hypothesis, a single ectD mutant was constructed (strain CHR136) after recombination of the mutated ectD gene in the chromosome of the wild-type strain (Fig. 5B). The growth of the wild type and the CHR136 mutant was compared at 37°C and 45°C in M63, with NaCl concentrations ranging from 0.75 to 3 M (Fig. 8). At 37°C (Fig. 8A), the wild-type strain grew optimally with between 0.75 and 1.5 M NaCl, although with 0.75 M NaCl, the stationary phase of growth was shorter than that with 1.5 M NaCl, and cultures entered death phase sooner. Above 1.5 M NaCl, and especially at 3 M NaCl, the growth curve of the wild-type strain showed an attenuated exponential phase, but it eventually reached stationary phase, with absorbance values comparable to those of cultures grown with 1.5 M NaCl. At this temperature, the growth curves of the ectD mutant CHR136 were parallel to those of the wild-type strain, regardless of the salt concentration of the culture medium. Likewise, at 37°C, the growth of strain CHR137 (ectABC ectD) was indistinguishable from that of its parental strain, CHR62 (ectABC), in M63 with 0.75 M NaCl (not shown).
FIG. 8.
ectD is involved in thermoprotection of C. salexigens. (A and B) Effects of temperature on the growth of C. salexigens at different salinities. Cells of the C. salexigens wild-type strain (closed symbols) and the ectD mutant strain CHR136 (open symbols) were grown at 37°C (A) and 45°C (B) in M63 minimal medium in the presence of 0.75 (squares), 1.5 (triangles), 2.5 (circles), and 3.0 (diamonds) M NaCl. (C) Intracellular concentrations of ectoine and hydroxyectoine of wild-type strain and CHR136 mutant. Cells were grown in M63 minimal medium with 2.5 M NaCl at 37°C or 45°C, and the solute content was determined by HPLC as described in Materials and Methods. The data are averages of three different replicates, which on no occasion varied more than 5%. OD600, optical density at 600 nm.
At 45°C (Fig. 8B), the C. salexigens wild-type strain was unable to grow at a low (0.75 M NaCl) or very high (3 M NaCl) salinity. Remarkably, the optimal salinity for growth of the wild type at 45°C was 2.5 M NaCl, with the cells showing a growth rate similar to that of cells grown at 37°C with the same salinity. In contrast, at 1.5 M NaCl, the growth of the wild type was much slower than that with 2.5 M NaCl or for cells grown at 37°C with the same salinity. Like the wild-type strain, the ectD mutant CHR136 was unable to cope with conditions of 45°C and 0.75 or 3 M NaCl. In addition, the growth of CHR136 was severely impaired at 1.5 M and 2.5 M NaCl, with absorbance values that never exceeded 0.5 optical density units at 600 nm.
The above data indicate that mutation in the ectoine hydroxylase gene ectD does not confer a salt-sensitive phenotype but does confer a temperature-sensitive phenotype. This correlated with the ectoine and hydroxyectoine contents of wild-type and CHR136 cultures grown at 2.5 M NaCl (Fig. 8C). At 37°C, the ectoine contents were similar for the wild type and the ectD mutant. At this temperature, the hydroxyectoine level of the ectD mutant strain was 2.6-fold lower than that of the wild type, although the mutant was able to synthesize 38% of the total hydroxyectoine produced by the wild-type strain. As expected, at 45°C the ectoine content of the wild-type decreased (1.6-fold), and the hydroxyectoine content increased (2.1-fold). At this temperature, the ectoine concentration in the ectD mutant decreased slightly (1.4-fold), and the hydroxyectoine level was drastically reduced, by 6.6-fold. In this case, the mutant was able to synthesize only 15% of the total hydroxyectoine produced by the wild-type strain. All of these data taken together demonstrate that hydroxyectoine synthesis mediated by ectD is involved in thermoprotection of C. salexigens.
DISCUSSION
In this work, we have approached the physiological and molecular characterization of the biosynthesis of the compatible solute hydroxyectoine in the halophilic bacterium Chromohalobacter salexigens. We have found evidence that ectoine hydroxylation is the main route for hydroxyectoine synthesis in C. salexigens. Our finding that hydroxyectoine accumulation is drastically reduced in ectD insertion mutants strongly indicates that ectD is the principal gene responsible for this route. Although we cannot totally exclude that insertion mutations in ectD might have a polar effect on the downstream gene orf4 and that the defects in hydroxyectoine accumulation might be attributed in part to the lack of the orf4-encoded product, this possibility seems remote. First, ectD and orf4 are separated by one intergenic region of 118 bp, suggesting that these genes are not cotranscribed. In addition, the orf4-encoded product has very well-conserved domains of enzymes involved in the degradation of aromatic compounds, strongly suggesting that it does not play any role in hydroxyectoine synthesis. Complementation studies with a plasmid carrying only ectD, which might have confirmed that ectD is the only gene responsible for the observed phenotype of the mutants, were not carried out, since in previous experiences with plasmids carrying the C. salexigens ectABC genes only a partial restoration of the wild-type phenotype was achieved (13).
Our finding that under high-salinity and -temperature conditions, hydroxyectoine levels exceed ectoine levels in C. salexigens, together with the temperature-sensitive phenotype of an ectD mutant strain, demonstrates that the compatible solute hydroxyectoine is responsible for thermoprotection of this halophilic bacterium. This is supported by previous data on the accumulation of hydroxyectoine in S. griseus (35) and H. elongata (57) in response to temperature. On the other hand, at 37°C and 45°C with 2.5 M NaCl, the ectD mutant strain accumulated 38% and 15%, respectively, of the total hydroxyectoine produced by the wild-type strain. These data indicate that the contribution of EctD to hydroxyectoine synthesis is greater at higher temperatures and therefore that ectD-induced ectoine hydroxylation is thermoregulated.
In addition to increased hydroxyectoine levels, we found that the C. salexigens response to elevated temperatures involves the accumulation of trehalose. This was unexpected, since trehalose had not been detected in a C. salexigens wild-type strain grown at 37°C (7, 13) or in the closely related organism H. elongata grown at 40°C (57). However, this finding corroborates the general assumption that cells tend to accumulate a cocktail of compatible solutes whose relative proportions may vary as a function of the environmental conditions, such as the absence of nutrients, the external salinity, or the temperature. The role of trehalose as a general stress protectant, including in response to heat stress, has previously been shown for both prokaryotic and eukaryotic microorganisms (9, 24, 51, 55). Because of the dramatic decrease in ectoine content at 45°C, the total pool size of ectoine and hydroxyectoine is decreasing at this temperature (Fig. 4A). Therefore, it might be possible that trehalose is a substitute for ectoine at high temperatures. Within the unfinished genome of C. salexigens, there are orthologs of the otsA and otsB genes, which encode the enzymes for trehalose synthesis from glucose. Since a C. salexigens ectD mutant strain is able to synthesize trehalose (Garcia-Estepa and Vargas, unpublished data), it is probable that trehalose allows residual growth at intermediate salinities. This will be confirmed by analyzing the phenotype of an otsA ectD double mutant, whose construction is in progress in our laboratory.
The C. salexigens wild-type strain cannot cope with extreme (low or high) salt concentrations and high temperatures. Whereas the temperature-sensitive phenotype at the low salt concentration (0.75 M NaCl) might be explained by the practical lack of hydroxyectoine at this salinity (Fig. 4B), the absence of growth at 45°C and 3 M NaCl might be due to the fact that the amount of hydroxyectoine (plus trehalose) accumulated by C. salexigens is not enough to protect the cells against these two environmental constraints. This is in agreement with our finding that the optimal salinity for growth of the wild type at 37°C is 1.5 M NaCl, whereas at 45°C it is 2.5 M NaCl, where a higher concentration of hydroxyectoine is accumulated in the cytoplasm. In this regard, it would be interesting to test the growth phenotypes of other microorganisms that produce higher hydroxyectoine levels, such as Marinococcus halophilus and Brevibacterium iodinum (18), at supraoptimal salt and temperature conditions.
Although the ectD-encoded ectoine hydroxylase is the main enzyme responsible for hydroxyectoine synthesis in C. salexigens, an ectD mutation does not totally suppress hydroxyectoine accumulation in this microorganism. There are at least two more possible routes for the biosynthesis of this compatible solute in C. salexigens, namely, a second putative ectoine hydroxylase encoded by the homolog found within the C. salexigens genome and alternative route branching of the ectoine precursor, Nγ-acetyldiaminobutyric acid (Fig. 1). In the absence of ectD and the alternative route, the homolog might be responsible for the 7% of hydroxyectoine (relative to the wild type) produced by strain CHR137 (ectABC ectD) at 37°C in M63 with 1.5 M NaCl and ectoine (Fig. 7). Alternatively, the residual hydroxyectoine synthesis observed in strain CHR137 might occur by a nonenzymatic conversion.
Although their functionality needs experimental support, our phylogenetic analysis suggests that whereas all proteins included in Fig. 6B belong to the superfamily of Fe(II)- and 2OG-dependent oxygenases, only the proteins boxed in this figure utilize ectoine as a substrate and therefore can be included in the ectoine hydroxylase subfamily. This hypothesis is supported by the presence of ectoine biosynthetic genes (ectA, ectB, and ectC) within the genomes of the bacteria carrying members of this family. Among all bacterial genera carrying members of the proposed ectoine hydroxylase family, species of Chromohalobacter (13), Streptomyces (28), Nocardia (18) (but N. farcinica has not been tested), and Brevibacterium (18) have been shown to synthesize ectoine and hydroxyectoine. In the haloalkalotolerant obligate methanotroph “Methylobacter alcaliphilus,” the synthesis of ectoine but not of hydroxyectoine has been reported (30). Nevertheless, it is possible that this microorganism can make hydroxyectoine under growth conditions different from those tested so far. Finally, and very interestingly, some of the microorganisms bearing orthologs of ectD are not typical for saline environments, as is C. salexigens, but are antibiotic-producing streptomycetes or even pathogenic bacteria, such as Bordetella parapertussis, Bordetella bronchiseptica, and N. farcinica (28, 35). In Streptomyces parvulus, the onset of ectoine and hydroxyectoine synthesis seems to correlate with the time of actinomycin D synthesis (28). Whether or not ectoines play a defensive role against antimicrobial agents and if this effect might function as a virulence factor for pathogenic bacteria are interesting questions that deserve to be investigated in the near future.
Acknowledgments
We express our gratitude to Nicholas Grammel for providing the sequence of the ThpD protein prior to publication and to Michael Galperin for his help in the initial alignment of hydroxylases that allowed us to clone the ectoine hydroxylase gene. We thank Rocío Rodriguez and Martín Romero (Centro de Investigaciones Científicas Isla de La Cartuja, CSIC, Sevilla, Spain) for technical assistance with HPLC analysis.
This research was financially supported by grants from the European Community (contract INCO-CT-2004-509115), the Spanish Ministerio de Educación y Ciencia (project BIO2005-06343-CO2-01), and Junta de Andalucía. Raúl García-Estepa and Mercedes Reina-Bueno were recipients of fellowships from Junta de Andalucía and Ministerio de Educación y Ciencia (Spain), respectively.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arahal, D. R., M. T. García, C. Vargas, D. Cánovas, J. J. Nieto, and A. Ventosa. 2001. Chromohalobacter salexigens sp. nov., a moderately halophilic species that includes Halomonas elongata DSM 3043 and ATCC 33174. Int. J. Syst. Evol. Microbiol. 51:1457-1462. [DOI] [PubMed] [Google Scholar]
- 3.Arai, H., T. Yamamoto, T. Ohishi, T. Shimizu, T. Nakata, and T. Kudo. 1999. Genetic organization and characteristics of the 3-(3-hydroxyphenyl)propionic acid degradation pathway of Comamonas testosteroni TA441. Microbiology 145:2813-2820. [DOI] [PubMed] [Google Scholar]
- 4.Arias-Barrau, E., E. R. Olivera, J. M. Luengo, C. Fernández, B. Galán, J. L. García, E. Díaz, and B. Minambres. 2004. The homogentisate pathway: a central catabolic pathway involved in the degradation of l-phenylalanine, l-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 186:5062-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bremer, E., and R. Krämer. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in bacteria, p. 79-97. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 6.Brown, A. D. 1976. Microbial water stress. Bacteriol. Rev. 40:803-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderón, M. I., C. Vargas, F. Rojo, F. Iglesias-Guerra, L. N. Csonka, A. Ventosa, and J. J. Nieto. 2004. Complex regulation of the synthesis of the compatible solute ectoine in the halophilic bacterium Chromohalobacter salexigens. Microbiology 150:3051-3063. [DOI] [PubMed] [Google Scholar]
- 8.Cánovas, D., N. Borges, C. Vargas, A. Ventosa, J. J. Nieto, and H. Santos. 1999. Role of N-γ-acetyldiaminobutyrate as an enzyme stabilizer and an intermediate in the biosynthesis of hydroxyectoine. Appl. Environ. Microbiol. 65:3774-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cánovas, D., S. A. Fletcher, M. Hayashi, and L. N. Csonka. 2001. Role of trehalose in growth at high temperature of Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:3365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cánovas, D., C. Vargas, M. I. Calderón, A. Ventosa, and J. J. Nieto. 1998. Characterization of the genes for the biosynthesis of the compatible solute ectoine in the moderately halophilic bacterium Halomonas elongata DSM 3043. Syst. Appl. Microbiol. 21:487-497. [DOI] [PubMed] [Google Scholar]
- 11.Cánovas, D., C. Vargas, L. N. Csonka, A. Ventosa, and J. J. Nieto. 1996. Osmoprotectants in Halomonas elongata: high-affinity glycine betaine transport system and choline-glycine betaine pathway. J. Bacteriol. 178:7221-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cánovas, D., C. Vargas, L. N. Csonka, A. Ventosa, and J. J. Nieto. 1998. Synthesis of glycine betaine from exogenous choline in the moderately halophilic bacterium Halomonas elongata. Appl. Environ. Microbiol. 64:4095-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cánovas, D., C. Vargas, F. Iglesias-Guerra, L. N. Csonka, D. Rhodes, A. Ventosa, and J. J. Nieto. 1997. Isolation and characterization of salt-sensitive mutants of the moderate halophile Halomonas elongata and cloning of the ectoine synthesis genes. J. Biol. Chem. 272:25794-25801. [DOI] [PubMed] [Google Scholar]
- 14.Csonka, L. N. 1982. A third l-proline permease in Salmonella typhimurium which functions in media of elevated osmotic strength. J. Bacteriol. 151:1433-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csonka, L. N., and W. Epstein. 1996. Osmoregulation, p. 1210-1223. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 16.da Costa, M. S., H. Santos, and E. A. Galinski. 1998. An overview of the role and diversity of compatible solutes in bacteria and archaea, p. 117-153. In T. Scheper (ed.), Advances in biochemical engineering/biotechnology, vol. 61. Springer-Verlag, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 17.Galinski, E. A., H. P. Pfeiffer, and H. G. Trüper. 1985. 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid. A novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. Eur. J. Biochem. 149:135-139. [DOI] [PubMed] [Google Scholar]
- 18.Galinski, E. A., and H. G. Trüper. 1994. Microbial behaviour in salt-stressed ecosystems. FEMS Microbiol. Rev. 15:95-108. [Google Scholar]
- 19.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, N.J.
- 21.Göller, K., A. Ofer, and E. A. Galinski. 1998. Construction and characterization of an NaCl-sensitive mutant of Halomonas elongata impaired in ectoine biosynthesis. FEMS Microbiol. Lett. 161:293-300. [DOI] [PubMed] [Google Scholar]
- 22.Grammel, N. 2000. Molekulargenetische und biochemische analyse der biosynthese von 2-mehyl-4-carboxy-3,4,5,6-tetrahydropyrimidin und seinem 5-hydroxyderivat, zwei salzstressinduzierbaren osmolyten in Streptomyces chrysomallus. Ph.D. thesis. Technische Universität Berlin, Berlin, Germany. [Online.] http://edocs.tu-berlin.de/diss/2000/grammel_nicholas.htm.
- 23.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 24.Hengge-Aronis, R., W. Klein, R. Lange, M. Rimmele, and W. Boss. 1991. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 173:7918-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holtmann, G., and E. Bremer. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J. Bacteriol. 186:1683-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, X. 1992. A contig assembly program based on sensitive detection of fragment overlaps. Genomics 14:18-25. [DOI] [PubMed] [Google Scholar]
- 28.Inbar, L., and A. Lapidot. 1988. The structure and biosynthesis of new tetrahydropyrimidine derivatives in actinomycin D producer Streptomyces parvulus. J. Biol. Chem. 263:16014-16022. [PubMed] [Google Scholar]
- 29.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusion into the chromosome of gram negative bacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 30.Khmelenina, V. N., M. G. Kalyuzhanaya, V. G. Sakharovsky, N. E. Suzina, Y. A. Trotsenko, and G. Gottschalk. 1999. Osmoadaptation in halophilic and alkaliphilic methanotrophs. Arch. Microbiol. 172:321-329. [DOI] [PubMed] [Google Scholar]
- 31.Kraegeloh, A., and H. J. Kunte. 2002. Novel insights into the role of potassium for osmoregulation in Halomonas elongata. Extremophiles 6:453-462. [DOI] [PubMed] [Google Scholar]
- 32.Kuhlmann, A. U., and E. Bremer. 2002. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 68:772-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 34.Louis, P., and E. A. Galinski. 1997. Characterization of genes for the biosynthesis of the compatible solute ectoine from Marinococcus halophilus and osmoregulated expression in Escherichia coli. Microbiology 143:1141-1149. [DOI] [PubMed] [Google Scholar]
- 35.Malin, G., and A. Lapidot. 1996. Induction of synthesis of tetrahydropyrimidine derivatives in Streptomyces strains and their effect on Escherichia coli in response to osmotic and heat stress. J. Bacteriol. 178:385-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manzanera, M., A. García de Castro, A. Tøndervik, M. Rayner-Brandes, A. R. Strøm, and A. Tunnacliffe. 2002. Hydroxyectoine is superior to trehalose for anhydrobiotic engineering of Pseudomonas putida KT2440. Appl. Environ. Microbiol. 68:4328-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manzanera, M., S. Vilchez, and A. Tunnacliffe. 2004. High survival and stability rates of Escherichia coli dried in hydroxyectoine. FEMS Microbiol. Lett. 233:347-352. [DOI] [PubMed] [Google Scholar]
- 38.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martins, L. O., and H. Santos. 1995. Accumulation of mannosylglycerate and di-myo-inositol-phosphate by Pyrococcus furiosus in response to salinity and temperature. Appl. Environ. Microbiol. 61:3299-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDonough, M. A., K. L. Kavanagh, D. Butler, T. Searls, U. Oppermann, and C. J. Schofield. 2005. Structure of human phytanoyl-CoA 2-hydroxylase identifies molecular mechanisms of Refsum disease. J. Biol. Chem. 280:41101-41110. [DOI] [PubMed] [Google Scholar]
- 41.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Müller, V., R. Spanheimer, and H. Santos. 2005. Stress response by solute accumulation in archaea. Curr. Opin. Microbiol. 8:1-8. [DOI] [PubMed] [Google Scholar]
- 43.Prabhu, J., F. Schauwecker, N. Grammel, U. Keller, and M. Bernhard. 2004. Functional expression of the ectoine hydroxylase gene (thpD) from Streptomyces chrysomallus in Halomonas elongata. Appl. Environ. Microbiol. 70:3130-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 45.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 46.Reshetnikov, A. S., V. Khmelenina, and Y. A. Trotsenko. 2006. Characterization of the ectoine biosynthesis genes of haloalkalotolerant obligate methanotroph “Methylomicrobium alcaliphilum 20Z”. Arch. Microbiol. 184:286-297. [DOI] [PubMed] [Google Scholar]
- 47.Ruetschi, U., I. Nordin, B. Odelhog, H. Jornvall, and S. Lindstedt. 1993. Gamma-butyrobetaine hydroxylase. Structural characterization of the Pseudomonas enzyme. Eur. J. Biochem. 213:1075-1080. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 49.Santos, H., and M. da Costa. 2002. Compatible solutes of organisms that live in hot saline environments. Environ. Microbiol. 4:501-509. [DOI] [PubMed] [Google Scholar]
- 50.Shibasaki, T., H. Mori, S. Chiba, and A. Ozaki. 1999. Microbial proline 4-hydroxylase screening and gene cloning. Appl. Environ. Microbiol. 65:4028-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simola, M., A. L. Hanninen, S. M. Stranius, and M. Makarow. 2000. Trehalose is required for conformational repair of heat-stress denatured proteins in yeast endoplasmic reticulum but not for maintenance of membrane traffic functions. Mol. Microbiol. 37:42-53. [DOI] [PubMed] [Google Scholar]
- 52.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Malia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 53.Vargas, C., M. J. Coronado, A. Ventosa, and J. J. Nieto. 1997. Host range, stability, and compatibility of broad host-range-plasmids and a shuttle vector in moderately halophilic bacteria. Evidence of intragenic and intergenic conjugation in moderate halophiles. Syst. Appl. Microbiol. 20:173-181. [Google Scholar]
- 54.Ventosa, A., J. J. Nieto, and A. Oren. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62:504-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welsh, D. T. 2000. Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol. Rev. 24:263-290. [DOI] [PubMed] [Google Scholar]
- 56.Welsh, D. T., and R. A. Herbert. 1999. Osmotically induced intracellular trehalose, but not glycine betaine accumulation promotes desiccation tolerance in Escherichia coli. FEMS Microbiol. Lett. 174:57-63. [DOI] [PubMed] [Google Scholar]
- 57.Wohlfarth, A., J. Severin, and E. A. Galinski. 1990. The spectrum of compatible solutes in heterotrophic halophilic eubacteria of the family Halomonadaceae. J. Gen. Microbiol. 136:705-712. [Google Scholar]