Abstract
Enteric bacteria tumble, swim slowly, and are then paralyzed upon exposure to 390- to 530-nm light. Here, we analyze this complex response in Escherichia coli using standard fluorescence microscope optics for excitation at 440 ± 5 nm. The slow swimming and paralysis occurred only in dye-containing growth media or buffers. Excitation elicited complete paralysis within a second in 1 μM proflavine dye, implying specific motor damage, but prolonged tumbling in buffer alone. The tumbling half-response times were subsecond for onset but more than a minute for recovery. The response required the chemotaxis signal protein CheY and receptor-dependent activation of its kinase CheA. The study of deletion mutants revealed a specific requirement for either the aerotaxis receptor Aer or the chemoreceptor Tar but not the Tar homolog Tsr. The action spectrum of the wild-type response was consistent with a flavin, but the chromophores remain to be identified. The motile response processed via Aer was sustained, with recovery to either step-up or -down taking more than a minute. The response processed via Tar was transient, recovering on second time scales comparable to chemotactic responses. The response duration and amplitude were dependent on relative expression of Aer, Tar, and Tsr. The main response features were reproduced when each receptor was expressed singly from a plasmid in a receptorless host strain. However, time-resolved motion analysis revealed subtle kinetic differences that reflect the role of receptor cluster interactions in kinase activation-deactivation dynamics.
Escherichia coli chemotaxis has been the system of choice for elucidation of the design principles of transmembrane and intracellular signal transduction (13, 38, 41). The signal phosphorelay consists of a histidine kinase CheA, which in turn controls the phosphorylation state of CheY, the motor response regulator. CheA activity is coupled to and controlled by five membrane chemoreceptors (Aer, Tar, Tap, Trg, and Tsr) through a linker protein, CheW (12). Clockwise (CW) motor rotation increases in ultrasensitive fashion upon binding of phospho-CheY to sites on the rotor. This leads to shortened swimming runs and increased tumbling episodes until the CW rotation bias increases beyond 0.85, at which stage the bacteria tumble continuously. Three other proteins complete the signaling network: the receptor methyltransferase CheR, the receptor methylesterase CheB, activated via CheA-dependent phosphorylation, and CheZ, a facilitator of CheY autodephosphorylation. The system displays precise adaptation, restoring prestimulus rotational bias by stimulus-dependent feedback control of CheB activity (3, 13). The receptors, together with their modifying enzymes and the other Che signaling proteins, cluster at the cell poles (reference 34 and references therein). The clustering is important for both amplification and timing of the chemotactic signal (38, 41).
Avoidance responses to blue light, which can damage nucleic acids and proteins, are ubiquitous in plants, protozoa, and bacteria (15, 20, 26, 45-46). Early studies on Salmonella enterica serovar Typhimurium showed that high intensity light (390 to 530 nm; 200 mW/cm2) caused loss of motility preceded by tumbling, then slow smooth swimming (47, 49). More recent studies documented that E. coli hem mutants with elevated levels of the heme precursor protoporphyrin tumbled in response to 390- to 450-nm light at lower intensities. Reactive oxygen radicals generated by the photosensitization of protoporphyrin were thought to cause this tumbling response (51, 52).
Our interest in these studies was aroused for two reasons. We have a longstanding program in time-resolved analysis of chemotactic signaling (19, 24, 25, 28-32), and light stimuli provide an important addition to this program. Our recent focus has been on motor responses triggered by repellents. Protons and leucine, the repellents released by flash photolysis of caged derivatives, produce a mixed response because they simultaneously activate and inhibit different members of the chemoreceptor family (28). We hoped that, in contrast, blue light might provide a pure negative stimulus. Second, the recent discovery of the aerotaxis receptor (Aer), which binds a flavin adenine dinucleotide (FAD) prosthetic group via a PAS domain (7, 40), suggested a possible target for blue-light responses. Here we report that wild-type Escherichia coli responds to blue-light pulses administered via standard fluorescence microscope optics. We demonstrate that this response consists of two components, a long-lived response processed via Aer and a response processed via Tar, one of the abundant receptors, that is short lived due to methylation-dependent adaptation. The other abundant chemoreceptor, Tsr, also mediates aerotactic responses, in addition to Aer (40). Interestingly, E. coli strains containing Tsr alone did not respond to blue light.
MATERIALS AND METHODS
Media and chemicals.
Difco brand Luria broth (LB) tablets and Bacto agar were from Becton Dickinson (Franklin Lakes, NJ). Ampicillin, chloramphenicol, isopropyl-β-d-thiogalactopyranoside (IPTG), sodium salicylate, amino acids, thiamine, sodium lactate, proflavine hemisulfate, and resazurin were purchased from Sigma Chemical Co. (St. Louis, MO).
Photolabile N-1-(2-nitrophenyl)ethoxycarbonyl derivatives of l-serine and l-aspartate have been described previously (31, 32). (μ-Peroxo)(μ-hydroxo) bis[bis(bipyridyl)cobalt(III)] (HPBC) nitrate was a gift from Mathias Lubben (Ruhr-Universitat, Bochum, Germany) (33). 8-Hydroxypyrene-1,3,6-Tris-sulfonic acid (HPTS) and HPBC-perchlorate were purchased from Molecular Probes (Eugene, OR).
Strains and plasmids.
Strains used in this study were isogenic derivatives of E. coli K12 strain RP437 (37). Relevant markers were: RP8611 [Δtsr-7028 Δ(tar-tap)5201 Δtrg-100] (4); UU1250 [Δaer-1 Δtsr-7028 Δ(tar-tap)5201 Δtrg-100] (5); VS100 [ΔcheY] (42); RP8606 [Δ(tar-tap)5201 Δtrg-100] (22); UU1615 [Δaer-1 Δ(tar-tap)5201 Δtrg-100] (22); UU1623 [Δtsr-7028 Δtap-3654 Δtrg-100] (22); and UU1624 [Δaer-1 Δtsr-7028 Δtap-3654 Δtrg-100] (22).
The Tsr and Aer plasmids used in this study were derivatives of pCJ30 (7), an IPTG-inducible expression vector derived from pBR322 that confers ampicillin resistance: pPA56 (Tsr[290-470]) (4); pJC3 (wild-type Tsr) (5); pSB20 (wild-type Aer) (7); and pSB20 derivatives encoding mutant Aer proteins (7, 22). The wild-type Tar expression plasmid pLC113 (5) and the wild-type Aer expression plasmid pKG117 (16) were derived from a salicylate-inducible expression vector that confers chloramphenicol resistance (pLC112) (5).
Growth media.
Cultures for behavioral experiments were grown at 30°C in LB or H1 minimal medium (1) supplemented with 0.1 mM required amino acids (threonine, leucine, histidine, and methionine) and thiamine (1 mg/ml). Cells were harvested at mid-exponential phase by centrifugation, washed three times, and resuspended in potassium phosphate-EDTA motility buffer (29) containing 5 mM lactate, as respiratory substrate, and 100 μM methionine to maintain swim-tumble bias. Plasmid-containing cultures were grown in antibiotic-containing medium (ampicillin at 100 μg/ml; chloramphenicol at 25 μg/ml) and induced with either IPTG (50 μM) or sodium salicylate (0.7 μM) at early exponential phase (optical density at 600 nm = 0.15 to 0.2). Typically, cells displayed a wild-type run/tumble swimming pattern after induction for 120 to 180 min for Aer and 60 to 90 min for Tsr and Tar.
Microscopy.
The setup for temporal assays of the blue-light response is detailed in Fig. 1. Samples (8 μl) of cell suspensions were observed in coverslip chambers, constructed as described previously (29). The coverslips were acid cleaned and stored in double-distilled water prior to use. The mercury (Hg) lamp was used for routine experiments, since Hg has a 436-nm emission line. The xenon lamp was used for measurement of action spectra, since it has relatively uniform, bright radiation over the 400- to 500-nm range. The power of the monitoring and excitation light beams was measured by a thermopile-based energy meter (PEM-001; Melles Griot, Carlsbad, CA). The diaphragms were used to demarcate a defined image area under conditions of Kohler-transmitted or epi-illumination. The demarcated areas in the recorded images were measured, with the dimensions determined with the aid of a micrometer, together with the energy of the beam emerging from the field diaphragm or objective. The measurements were used to obtain the luminance power per unit sample area. The power of the infrared monitoring beam was 4.5 mW/cm2. The power of the 440 (±5)-nm band-pass-filtered excitation beam from the Hg lamp was 6.8 ± 1.8 mW/cm2. The Xe and Hg lamp intensities were comparable at this wavelength. The relative intensities of the Xe lamp excitation at other wavelengths were determined by measurement of HPTS fluorescence and normalization of the emitted fluorescence with the HPTS absorption spectrum at pH 7.0 (25).
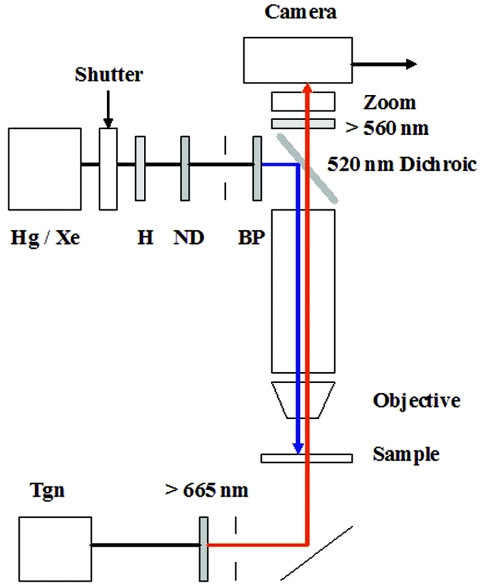
FIG. 1.
Microscope setup for blue-light assays. Cell samples (10 μl) were applied to bridged coverslips placed on the stage of a Nikon Optiphot microscope and observed under phase contrast (40×, 0.85 numerical-aperture fluorite objective), infrared illumination from a tungsten (Tgn) lamp (12 V, 50 W) selected by a 665-nm long-pass filter and reflected up towards the sample by a silvered mirror. A 100-W mercury (Hg) or xenon (Xe) lamp was used to stimulate the cells. The light pulse was heat filtered (H) and its duration was controlled by an electronic shutter (see reference 25 for details). Diaphragms, positioned after the filters, were used to obtain Kohler illumination for both the monitoring and excitation beams. A 520-nm dichroic mirror reflected the excitation beam down onto the sample, while a 560-nm long-pass barrier filter blocked the beam from the camera. Image magnification was adjusted by a zoom lens positioned in front of the camera. The wavelength of the excitation beam was controlled by band pass (BP) (440 ± 5 nm, 440 ± 15 nm, 450 ± 25 nm, 410 ± 5 nm, 470 ± 5 nm, 500 ± 5 nm) and attenuated by neutral density (ND) filters (1/2, 1/4, 1/8) positioned in series before the dichroic mirror. All filters were purchased from Omega Optical (Brattleboro, VT). The study of the E. coli hem mutant light responses used similar optics with two differences: a 50-W rather than 100-W Hg lamp was used for the excitation beam, with a broader (396- to 450-nm) band-pass filter (51).
A MAC-180 charge-coupled-device camera (Motion Analysis Inc., Santa Rosa, CA) was used for image acquisition either on videotape or directly to a computer using the LG-3 frame grabber card from Scion Corporation (Frederick, MD). A custom-made trigger box controlled the shutter via electronic circuitry (25, 31) to set the delay of the light pulse relative to the initiation of image acquisition and its duration. Alternatively, it was used to trigger discharge of a flash lamp (22) for photorelease of l-serine, l-aspartate, or molecular oxygen from photolabile derivatives.
Motion analysis.
Video records were digitized offline and analyzed using instrumentation and software that have been described previously (25). In brief, cell centroids were computed from the digitized outlines and connected frame to frame to form paths of the cell trajectories. Trajectories of poorly motile or stationary objects were filtered out. Motile behavior was expressed as population linear speed (SPD; in μm/s) or absolute angular speed (RCD; in degrees/s). Each time series was typically averaged over 1,000 paths merged from several video sequences. For each condition, we checked that the population histograms did not deviate from a unimodal distribution. The relationship between population RCD and the mean motor clockwise/counterclockwise bias has been documented (31). RCD values also depend on frame rate (31). Digitization was at 15 frames/second (fps) or 30 fps where specified. The values for smooth-swimming mutant strains UU1250 and VS100 were 207 ± 22 and 213 ± 18 degrees/second at 15 fps and 353 ± 15 and 334 ± 19 degrees/second at 30 fps, respectively. The corresponding values for continuously tumbling cell populations were 750 ± 25 and 1,107 ± 37 at 15 and 30 fps, respectively. The RCD increase upon turn-on (shutter opened) of blue light or adaptation upon turn-off (shutter closed) was fit by an expression of the form (a − b) [e(−kt)] + b, where k is the rate and a and b are final and initial RCD values, respectively. The RCD decrease upon blue light off or adaptation from its turn-on was fit by an equation of the form a − {b[e(−kt)]}, where k, a, and b are as defined above. The best fits were determined by eye.
RESULTS
Blue-light-induced paralysis of E. coli.
When excited by 440 (±5)-nm epi-illumination, E. coli organisms tumbled vigorously, then gradually slowed down and became paralyzed in rich media (e.g., LB), as observed earlier with high-intensity, broadband light. Importantly, however, we found that in transparent media (e.g., H1 or motility buffer), motility was unimpaired. This allowed the tumble response to be observed indefinitely and reset either by light turn-off or adaptation, as detailed below. Henceforth, these excitation conditions will be referred to as “blue-light” excitation.
We studied two smooth-swimming mutant strains, VS100 (ΔcheY) and UU1250 (Δaer Δtar Δtsr Δtrg Δtap), to identify the factors for tumbling and paralysis. In LB, the strains did not tumble but were paralyzed by blue light. In buffer, blue light had no effect. We wanted to check that the extreme counterclockwise rotation bias of the mutant strains did not mask the response and to establish that phosphorylated CheY was required rather than another molecule phosphorylated by CheA. To do so, we expressed the Tsr fragment, Tsr[290-470], which constitutively activates (4) CheA. This manipulation will increase phosphorylated CheY levels in strain UU1250 and was used to adjust its swim-tumble bias to normal values. The bias-adjusted UU1250 strain did not respond to blue light. The induction levels used for VS100 were fivefold greater than those used to restore the motile bias of UU1250. VS100 still did not tumble in the absence or presence of blue light. This showed that both the transmembrane chemoreceptors and CheY were needed for the blue-light tumble response.
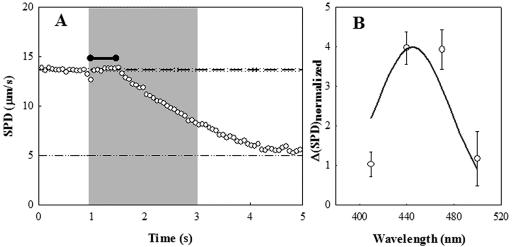
We reasoned, based on earlier studies (18, 47), that blue-light-induced paralysis was caused by flavin and other dyes present in the proteolytic digests of animal tissue that make up rich media. Extrinsic chromophores have long been known to be responsible for photo-oxidation of protein histidine residues (43). To test this supposition, we compared blue-light-induced paralysis of VS100 in broth with that in buffer containing proflavin. In 0.1 μM proflavin, the cells became gradually immotile in a fashion similar to that in broth under blue light. In 1 μM proflavin, they were rapidly paralyzed. An initial lag of 0.5 seconds was followed by an exponential decay in SPD with a half time of 1.2 seconds (Fig. 2A). The action spectrum for this effect over the 400- to 500-nm range could be superimposed on the known excitation spectrum of proflavin (http://omlc.ogi.edu/spectra/PhotochemCAD/html/proflavin(pH7).html) (Fig. 2B). This gave us confidence in the procedure used to calibrate the light intensity at different wavelength bandwidths.
FIG. 2.
(A) Proflavin-induced motor damage. Motility impairment of the cheY-deletion strain by 440 (±15)-nm light in the presence of 1 μM proflavine. The gray zone indicates the period of exposure to blue light. Reference lines indicate prestimulus mean SPD (dashed) and its frame-to-frame standard deviation (dotted) of the speed. The dashed-dotted line indicates the nonzero value for immotile bacteria due to Brownian motion. The black horizontal bar indicates the latency prior to speed impairment. (B) Action spectrum (410 to 500 nm) of the impairment. The continuous line denotes the adsorption spectrum of proflavine. The drop in speed (ΔSPD) was normalized by the light intensity to obtain a relative measure of the speed decrease (see text for details).
Blue-light-induced sensory responses of E. coli.
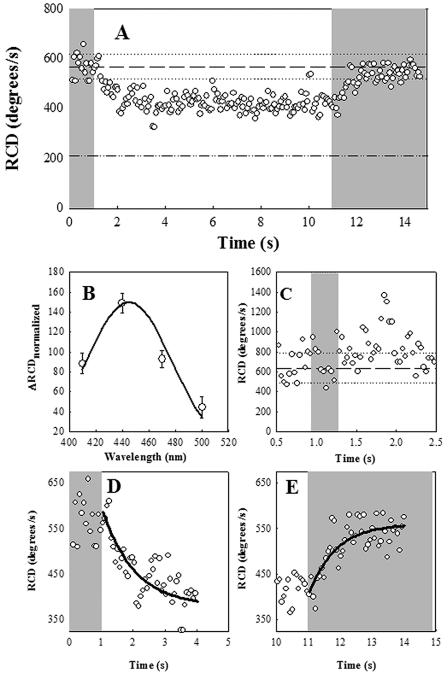
Wild-type strain RP437 responded to blue light by increased tumbling in defined, chromophore-free solutions. Sensory adaptation to continuous blue light took minutes (see below), being negligible during the first 20 seconds, in contrast to the rapid, subsecond responses to the onset and cessation of blue light. Thus, we could rapidly switch tumbling rates between high and low RCD values by opening and closing the shutter, respectively, at 10-second intervals (Fig. 3A). The action spectrum of the swimming response to blue light was consistent with the chromophore being a flavin (Fig. 3B) rather than a protoporphyrin (see Fig. 2 in reference 52). The response amplitude and rate of onset decreased upon four- and eightfold attenuation of the excitation beam intensity by neutral-density filters (not shown). However, increase of the excitation light intensity by increasing the 440-nm filter bandwidth from 10 to 50 nm did not affect response kinetics or amplitude (data not shown). This finding implies saturation of some process prior to CheA activation, because the peak population RCD was lower than that for continuous tumbling. Shorter light pulses (300 milliseconds) evoked brief tumbling with a delay of 0.55 s after shutter closure (Fig. 3C). Even-shorter pulses (30 milliseconds) did not elicit a response (not shown). The rates for the tumble-to-swim and the swim-to-tumble transitions obtained for the 10-second pulse from single exponential fits were 1 s−1 and 1.2 s−1, respectively (Fig. 3D and E).
FIG. 3.
Wild-type (RP437) E. coli response to blue light. Gray zones indicate exposure to blue light. (A) Motile response to turn-off and turn-on of blue light after a 10-second interval. Reference lines indicate prestimulus mean RCD (dashed) and its frame-to-frame standard deviation (dotted) for the 1 second prior to shutter closure. The value for smooth-swimming mutant E. coli is given by the dashed-dotted line. (B) Action spectrum. Open circles denote relative responses ± standard errors. Continuous line denotes superimposed absorption spectrum of proflavin. (C) Response to 0.3-s blue-light pulse. Reference lines indicate the prestimulus mean RCD and its ± frame-to-frame standard deviation as in panel A. (D) Smooth-swimming response on turn-off of blue light. (E) Tumbling response to turn-on of blue light. Lines in panels D and E are best exponential fits.
Roles of different receptors in the blue-light response.
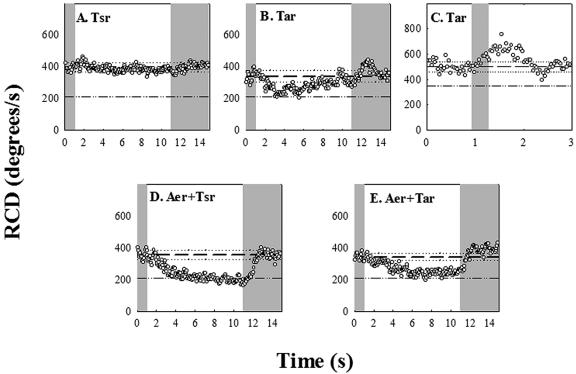
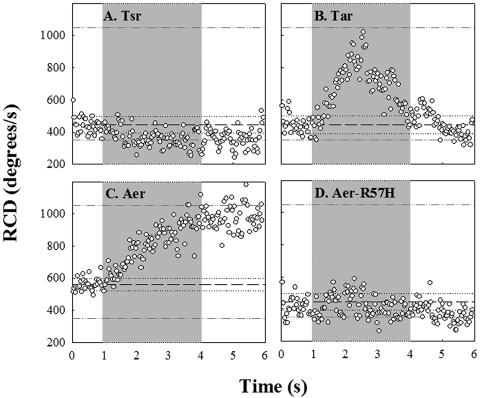
Isogenic strains lacking various combinations of receptor genes responded differently to blue light. A strain (UU1615) containing only Tsr, the high-abundance serine receptor, showed little response (Fig. 4A). In some experiments with this strain, we observed a slight decrease in population RCD (e.g., Fig. 4A), but this was not routinely observed. To our surprise, a strain (UU1623) containing only Tar, the high-abundance aspartate and maltose receptor, tumbled in response to blue-light turn-on and swam smoothly upon blue-light turn-off. In contrast to the wild-type response, however, these Tar-mediated responses were short lived, decaying rapidly with 0.6 s−1 and 0.15 s−1 rates, respectively (Fig. 4B). These adaptation rates are similar to those obtained for Tar-mediated responses of comparable strength to aspartate photorelease (25). The Tar-mediated CW response was strongest at 440 nm. However, rapid adaptation precluded measurement of the weaker, short-lived, peak response amplitudes at other wavelengths, hence determination of the action spectrum. A better measure of the response kinetics was obtained upon application of a 0.3-s pulse (Fig. 4C). Under these conditions the time for the CW response to reach peak amplitude was the same as that determined for wild-type E. coli. In addition, an adaptive undershoot of comparable magnitude was obtained, as expected for an impulse response that exhibits precise adaptation. The peak frequency, namely, the reciprocal of the dominant periodicity, reveals the intrinsic excitation and adaptation time constants for the response. This was 0.4 ± 0.05 Hz (Fig. 4C), comparable to the peak frequency of 0.25 Hz determined for impulse responses to 0.1-s pulses of l-aspartate addition or removal, applied by iontophoresis (11).
FIG. 4.
Chromosomal-deletion mutants. The gray zones indicate exposure to blue light, and the reference lines indicate the RCD values prior to shutter closure and the value for smooth-swimming populations, as in Fig. 3A. (A) UU1615; (B) UU1624; (C) UU1624 response to a 0.3-second pulse (lines denote prestimulus RCD [dashed] plus frame-to-frame standard deviations [dotted]); (D) RP8606; (E) UU1623.
The dramatically longer duration of the blue-light response time in wild-type than in the Tar-only strain could be due to the low-abundance receptors. We focused on the obvious suspect, Aer. The RP8611 strain expressing the Aer receptor alone was smooth swimming and did not respond. We adjusted its bias by transformation with pPA56 and expression of Tsr[290-470], but the bias-adjusted strain also failed to respond (data not shown). Conceivably, the lack of response in the bias-adjusted strain was an artifact, since Tsr[290-470] fragments may disrupt Aer-CheA association by binding more strongly to the kinase. To explore Aer-dependent responses under more physiological conditions, we studied strains expressing Aer in combination with either Tsr (RP8606) or Tar (UU1623). In contrast to RP8611, these strains had measurable tumble rates (RCD > 200 degrees/s) in the absence of blue light. In the 10-second pulse experiments, the RCD of the Tsr-plus-Aer strain increased upon blue-light turn-on and reverted to its unstimulated value upon blue-light turn-off, with amplitude and kinetics comparable to the wild-type time response (Fig. 4D). The Tar-plus-Aer strain behaved similarly (Fig. 4E). However, when the blue light was switched on after a 10-second off period, the RCD in the Tar-containing strain increased to a value slightly greater than just before turn-off, implying some adaptive recovery (Fig. 4E). Nevertheless, in both cases the responses were sustained, similar to that of the wild type and in marked contrast to the short-lived responses of the Tar-only strain. Thus, Aer expressed in combination with either Tsr or Tar reconstituted the wild-type response (Fig. 4D and E). The roles of the other low-abundance receptors Tap and Trg, if any, were not examined.
Blue-light responses mediated by Aer alone.
As noted previously, the receptorless UU1250 strain had a smooth-swimming phenotype and did not respond to blue light. Single receptor types were expressed from plasmid vectors in UU1250, with the induction conditions tuned to obtain roughly wild-type swimming patterns. This enabled us to examine blue-light responses mediated by Aer, which requires expression levels comparable to those of high-abundance receptors to adequately activate the CheA kinase (22).
The Tsr-only strain (pJC3/UU1250) did not tumble in response to blue-light turn-on. In some, but not all, experiments, the cells swam more smoothly in response (Fig. 5A), similarly to UU1515, the chromosomal Tsr-only strain (Fig. 4A). The Tar-only strain (pLC113/UU1250) responded and adapted to blue light (Fig. 5B), similarly to UU1624, the chromosomal Tar-only strain (Fig. 4B and C). Both strains gave rapid-saturation smooth-swimming responses, as reported previously, to photorelease of 0.1 mM serine (31) or aspartate (25), respectively (data not shown).
FIG. 5.
Single-receptor overexpression. The gray zones indicate exposure to blue light and the reference lines the mean RCD values (dashed) ± their standard deviations (dotted) prior to opening of the shutter. The dashed-dotted lines denote values for continuously “tumbly” and smooth-swimming populations. (A) pJC3/UU1250; (B) pLC113/UU1250; (C) pKG117/UU1250; (D) pKG117-R57H/UU1250.
We expressed Aer in UU1250 from two different vectors (pSB20 and pKG117) that carried different promoters and antibiotic resistance markers, to exclude possible plasmid-related perturbations of cellular metabolic state. In all cases, the UU1250 transformants expressing Aer at optimal levels for aerotactic responses also responded to blue-light turn-on by tumbling continuously (Fig. 5C). This result demonstrates that Aer can activate CheA independently of other receptors, which in turn suggests that the failure of chromosomally encoded Aer protein (in RP8611) to produce a blue-light response is due to its low expression level (50). In addition, if Aer binds or activates CheA less efficiently than Tsr, induction of the Tsr[290-470] CheA binding domain may sequester CheA from Aer. Even though switch transitions limit single-motor response, tumble signal processing times are sensitive to CheY∼P turnover since reversal of any one of several motors may trigger flagellar-bundle breakup (30). The comparable response times for the Aer- and Tar-mediated responses imply that the two receptors activate CheA to similar extents. However, recovery of Aer-only cell populations (e.g., pGK117/UU1250) to their basal RCD values upon blue-light turn-off was notably slower than in the Tar-only strains (Table 1).
TABLE 1.
Comparison of responses to blue-light stimuli
| Strain | Response ratea |
|||
|---|---|---|---|---|
| kon(s−1) | koff(s−1) | konad(s−1) | koffad(s−1) | |
| RP437 (wild type) | 1.2 | 1.0 | 0.028 | ND |
| RP8611 (Aer+) | − | − | − | − |
| UU1615 (Tsr+) | − | − | − | − |
| UU1623 (Tar+) | 1.0 | 0.6 | 0.6 | 0.15 |
| RP8606 (Aer+ Tsr+) | 1.0 | 0.5 | 0.025 | ND |
| UU1624 (Aer+ Tar+) | 1.6 | 0.6 | ND | ND |
| UU1250/pJC3 (Tsr+) | − | − | − | − |
| UU1250/pLC113 (Tar+) | 1.1 | ND | 0.7 | ND |
| UU1250/pKG117 (Aer+) | 0.6 | 0.2 | 0.02 | ND |
kon, Blue light turn-on; koff, blue-light turn-off; konad, adaptation to blue-light turn-on; koffad, adaptation to blue-light turn-off. −, no response. ND, not determined.
We sought to compare the kinetics of the Aer response to blue light to those of its response to oxygen, but have not been successful thus far in using the available caged-oxygen compounds (33, 53) for this purpose. The Aer-only UU1250 strains partially restored swarming in semisolid agar. In addition, they were resistant to oxygen depletion in liquid media, consistent with the induction by Aer of anaerobic respiration pathway components at low oxygen tension (39). In brief, the strains remained vigorously motile in the bridged coverslip chambers for tens of minutes after oxygen depletion as monitored by resazurin (51), whereas UU1250 strains expressing Tar or Tsr became immediately immotile. These observations argue that the expressed Aer protein possessed the expected oxygen sensitivity.
If blue-light sensing by Aer involves the same components used for aerosensing, we would expect some aerotaxis-defective Aer lesions to abrogate blue-light responses as well. To test this idea, we examined the ability of mutant Aer proteins (R57H, M112V, S261P, and Q263L) to mediate blue-light responses in strain UU1250. All mutant Aer proteins conferred aberrant motile bias. Aer-M112V/UU1250 and Aer-Q263L/UU1250 cell populations constantly tumbled, even in the absence of inducer, whereas Aer-S261P/UU1250 remained smooth swimming at all inducer levels. The “tumbly” mutant strains did not respond to cessation of blue light after many minutes of continuous exposure, while the smooth-swimming strain did not tumble in response to blue-light stimulation. The Aer-R57H strain, which is defective in FAD binding (data not shown), became “tumbly” upon induction. However, no tumbling response to blue light was evident even at shortened induction times, with RCD values in the normal-to-smooth-swimming range (Fig. 5D). These mutant data, albeit limited, suggest that Aer-mediated responses to oxygen and to blue light share common structure-function determinants. In particular, FAD binding may be essential, but is not sufficient, for either response.
The response and adaptation rate data for all experiments are compiled in Table 1. The two most noticeable findings are that (i) strains containing either chromosomally or plasmid-expressed Tar adapt on the second time scale, whereas strains expressing Aer do not and (ii) response rates to blue-light turn-on/turn-off for Aer-containing strains, particularly those expressing plasmid-encoded Aer, are slower than the wild type.
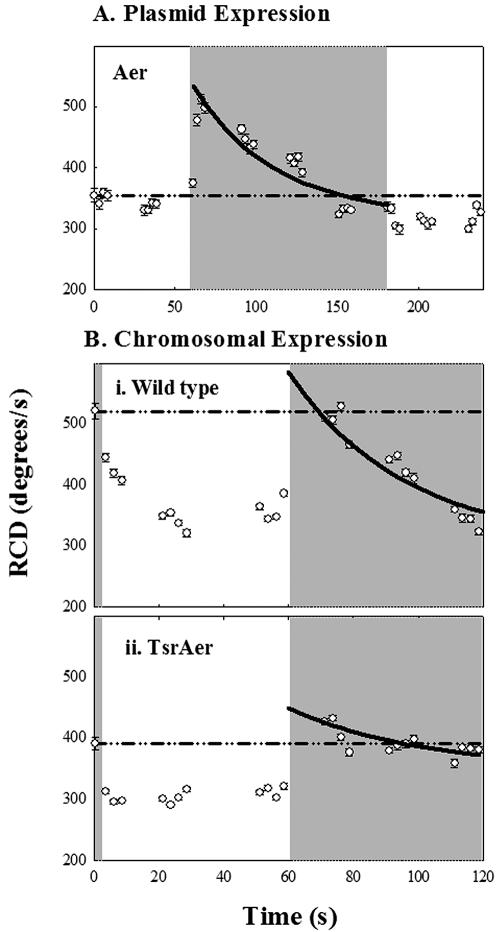
Adaptation kinetics.
We carried out observations over 1 to 2 min of continuous blue-light exposure to measure adaptation rates for strains expressing Aer. Aer was induced in strain pKG117/UU1250 at levels producing a close-to-wild-type swim-tumble bias before blue-light stimulation. Adaptation to blue-light turn-on was complete in about 2 minutes. Subsequent turn-off of blue light triggered a modest smooth-swimming response (Fig. 6A). Adaptation was also measured for the wild-type strain (RP437) (Fig. 6B, upper panel) and the chromosomal-deletion mutant RP8606 (Aer plus Tsr) (Fig. 6B, lower panel). These rates were all an order of magnitude slower than adaptation rates for the corresponding Tar-mediated response to blue light (Fig. 4B and C and 5B; Table 1).
FIG. 6.
Recovery kinetics of the Aer-mediated response. The gray zones indicate exposure to blue light. Symbols denote mean RCD values averaged over 2.5 seconds (38 frames) ± standard errors. The dashed-dotted reference lines indicate the mean RCD values for the first second in all sequences. Solid lines denote single exponential fits to the adaptation phase of the response to turn-on of blue light. (A) Recovery in pKG117/UU1250 of a nonsaturation response to a 2-minute step of blue light. (B) Recovery of (i) RP437 and (ii) RP8606 populations to a 1-minute step of blue light.
DISCUSSION
In this work we established conditions for utilizing blue light as a time-resolved repellent stimulus. We have found that the motor response requires either the chemoreceptor Tar or the aerotaxis receptor Aer. We have undertaken a preliminary kinetic analysis of the sensing and recovery mechanisms. As discussed below, our analysis provides important clues regarding signal generation and integration by the different receptors.
Blue-light effects on motile behavior.
Light, analogous to pH, oxygen, and temperature, is likely to have multiple effects on cellular physiology. Therefore, we first compare our observations with those of earlier studies to enumerate the different phenomena involved. In the early studies, critical illumination from a 75-W xenon lamp focused through a high-numerical-aperture (1.2) oil immersion condenser was used to excite the bacteria over a greater bandwidth (390 to 530 nm) and threefold-greater intensity than used here. The monitoring-beam power (3 mW/cm2) was similar but utilized higher-energy (>530 versus >665 nm) radiation (47). The action spectrum implicated a flavin (35), while proflavin caused paralysis (47); this was found by us as well. “Intrinsic” motor damage (47, 49) was avoided by the more benign radiation we used to monitor and excite the bacteria. In summary, it is likely that the responses we studied were observed previously but that the coarser optics employed in those pioneering studies (47, 49) hindered analysis of the tactic response separate from motor damage. Furthermore, we have benefited from the time-resolved motion analysis tools and extensive molecular knowledge about E. coli motility that are now available.
The light intensity used for the hem mutant studies was 10 μmol/m2s (52). The energy of 1 440-nm photon is 4.5 × 10−19 joules. Therefore, the intensity converts to (4.5 × 10−19) × (10 × 10−6 × 10−4 × 6.3 × 1023) = 0.28 mW/cm2, about 20-fold lower than what we employed. The difference is puzzling, since both studies used similar microscope optics, but is perhaps explained by their use of a low-transmission objective, in addition to the weaker lamp. In any case, the hem mutant responses are distinct from what we observed, because (i) their action spectrum is consistent with a protoporphyrin (405-nm peak absorption), (ii) they occur in the presence of either Tar or Tsr, and (iii) they take about 10 seconds to develop (52).
Motor paralysis.
E. coli has a significant store of endogenous ATP, and de-energization takes 1 to 2 min to complete upon addition of uncoupler (27). The fact that motility is abolished within a few seconds (Fig. 2) makes a kinetic argument for specific action of the dye on the motor itself. Studies of Streptococcus on the photodynamic effects of light in the presence of proflavin and other dyes drew a similar conclusion (18). Histidine residues have not been implicated as essential for proton transfer in the flagellar motor. However, their photo-oxidation could block transport by perturbing the molecular conformation of the proton channel proteins. Interestingly, there are two histidines close to Asp32, a residue critical for motor function, in the proton channel protein MotB (14).
Blue-light repellent responses.
A number of known photoreceptor families respond to blue light (15, 20). The archaebacterial retinal-containing sensory rhodopsins trigger motile repellent responses, while phototropin mediates a negative trophic response in plants. The phototropins, cryptochromes, and BLUF domains all contain a flavin as chromophore, while the activation of xanthopsins (e.g., photosensitive yellow proteins) and phytochromes is based on chromophore isomerization, like the rhodopsins. The responses we observe in E. coli may be mediated by homologous members of these photoreceptor families. In addition, heme-based oxygen sensors are present in E. coli (21). Their photosensitization may be relevant under certain conditions for its blue-light sensitivity. Our data require that photon adsorption at the unknown site perturb the conformation of either the chemoreceptor Tar or the aerotaxis receptor Aer, when it is present in abundance or together with Tar or Tsr, to effect motor responses.
Tar- and Aer-dependent blue-light responses.
Our data reveal an intriguing division of labor as regards “energy sensing,” a term that encompasses responses to pH, membrane potential, proton motive force, electron transport, and the phosphate (ATP/[ADP + Pi]) ratio (48). Aer and Tsr are oxygen sensors (40), while Tsr and Tar are pH sensors (reference 29 and references therein). The paired receptor responses interact to ensure migration to optimal oxygen tension and pH, respectively. Tar and Aer are now revealed to mediate responses to blue-light stimuli. Thus, a different set of receptors, albeit containing a common component (Aer), is utilized to respond to blue light versus oxygen. The parameter sensed by Tar to monitor blue-light levels must be different from that monitored by Tsr to respond to oxygen.
Tar does not have a chromophore and so must effect motor responses by monitoring a parameter perturbed by absorption of blue photons elsewhere. A reasonable suggestion, supported by evidence, is that blue light perturbs electron transport (50). Perturbation of electron transport will affect other energy parameters, such as pH. Irrespective of what exactly is sensed by Tar, the kinetics argued that the motor responses it triggers utilize both the CheA activation and methylation-dependent adaptation machinery used for chemotactic stimuli (Fig. 5B). Aer contains an FAD binding PAS domain that determines its response to oxygen (8, 50) Perhaps blue light directly reduces the Aer FAD cofactor or the free FAD in the cytoplasmic pool (23) that then binds to and triggers CheA-activating conformational changes within Aer. Alternatively, Aer residues unrelated to its FAD binding domain could, like Tar, monitor perturbation of a membrane parameter caused by blue-light adsorption elsewhere. All possibilities are consistent with the excitation times measured for the Aer-mediated blue-light response.
The dramatic difference in the adaptive recovery kinetics between the Aer- and Tar-mediated responses to blue light supports recent work demonstrating that methylation is not involved in Aer signal-processing biochemistry (9). Most simply, the FAD redox potential or the activated Aer conformation may be reset by a variety of mechanisms used for energy homeostasis. Alternatively, recovery could occur at later steps in the signaling pathway, with CheY∼P levels and/or motor bias being restored rather than CheA activity. Numerous “methylation-independent” adaptation mechanisms have been proposed. These include (i) restoration of CheY∼P levels by CheZ-mediated acceleration of CheY∼P dephosphorylation (2, 10, 17), (ii) other covalent modifications of CheY (6), or (iii) restoration of prestimulus motor bias by metabolites (e.g., fumarate [36]) that allosterically counteract CheY∼P. At this time there is little to argue strongly for or against any of these possibilities.
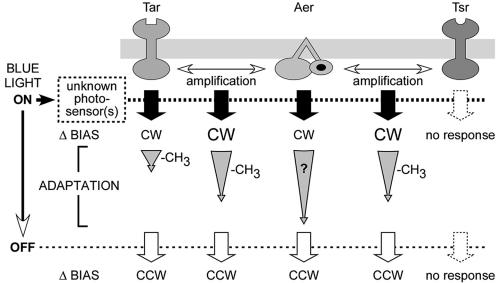
Receptor interactions during blue-light responses.
Our main findings are summarized in Fig. 7. Tar and Aer respond to blue light; Tsr does not. Recovery kinetics for the Tar response were on time scales expected for sensory adaptation based on receptor methylation. The Aer response was measured under two conditions: at chromosomal levels when expressed together with a major receptor and singly when expressed from a plasmid at levels sufficient to obtain a measurable swim-tumble bias. Excitation responses were more rapid in the former case (Table 1), which suggests that the major receptors enhance the Aer response. This collaborative signaling effect is most likely due to formation of mixed trimers of dimers between Aer and the other transducers (22, 44).
FIG. 7.
Chemoreceptor activation and adaptation by blue light. The Aer response is enhanced by the presence of either Tar or Tsr, but its adaptive recovery upon sustained exposure to blue light remains slow by comparison to the rapid adaptation, presumably driven by methylation, exhibited by Tar (see text for details).
The Aer contribution dominates the wild-type response to a temporal blue-light stimulus due to its slow adaptive recovery. The methylation state of its receptor homologs could affect this recovery if mixed complexes existed. Consistent with this idea, a slight overshoot was observed in the 10-second-pulse experiments for the Tar-plus-Aer strain, but not the wild-type or the Tsr-plus-Aer strain (Fig. 5). Tar responds and adapts rapidly to blue light and thus would be more highly methylated after a sustained 10-second absence of light than Tsr, which does not respond or may do so in an antagonistic fashion if the response is due to pH perturbation. The adapted, more highly methylated Tar molecules may act via cooperative interactions to partially reactivate neighboring Aer receptors. Culture-to-culture variability will need to be determined to assess whether the modest differences are meaningful.
Blue light as a tool for time-resolved analysis.
Temporal assays of increasing precision and temporal resolution have provided critical insights into sensory signal-processing mechanisms. Blue light provides a noninvasive stimulus whose strength may be precisely calibrated and which may be used to study motile responses in or on semisolid or solid substrates, as well as in solution. This work illustrates the advantages of its use relative to photolabile (“caged”) compounds, namely, (i) step-up or step-down stimuli, (ii) impulsive or step stimuli, and (iii) arbitrarily long- or short-duration stimuli. In particular, it should be an important tool for timing methylation-independent adaptive recovery, since other stimuli that initiate such responses (e.g., oxygen or phosphotransferase system carbohydrates) are difficult both to measure and to control.
Acknowledgments
We thank Osman Akcakir and Julie Moinnes for assistance with some of the experiments.
This work was supported by research grants GM19559 and GM62940 (to J.S.P.) and GM49319 (to S.K.) from the National Institutes of Health.
REFERENCES
- 1.Adler, J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77-91. [DOI] [PubMed] [Google Scholar]
- 2.Almogy, G., L. Stone, and N. Ben-Tal. 2001. Multi-stage regulation, a key to reliable adaptive biochemical pathways. Biophys. J. 81:3016-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alon, U., M. G. Surette, N. Barkai, and S. Leibler. 1999. Robustness in bacterial chemotaxis. Nature 397:168-171. [DOI] [PubMed] [Google Scholar]
- 4.Ames, P., and J. S. Parkinson. 1994. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 176:6340-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barak, R., and M. Eisenbach. 2001. Acetylation of the response regulator, CheY, is involved in bacterial chemotaxis. Mol. Microbiol. 40:731-743. [DOI] [PubMed] [Google Scholar]
- 7.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bibikov, S. I., A. C. Miller, K. K. Gosink, and J. S. Parkinson. 2004. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J. Bacteriol. 186:3730-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blat, Y., and M. Eisenbach. 1996. Mutants with defective phosphatase activity show no phosphorylation-dependent oligomerization of CheZ. The phosphatase of bacterial chemotaxis. J. Biol. Chem. 271:1232-1236. [DOI] [PubMed] [Google Scholar]
- 11.Block, S. M., J. E. Segall, and H. C. Berg. 1982. Impulse responses in bacterial chemotaxis. Cell 31:215-226. [DOI] [PubMed] [Google Scholar]
- 12.Boukhvalova, M. S., F. W. Dahlquist, and R. C. Stewart. 2002. CheW binding interactions with CheA and Tar. Importance for chemotaxis signaling in Escherichia coli. J. Biol. Chem. 277:22251-22259. [DOI] [PubMed] [Google Scholar]
- 13.Bourret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 14.Braun, T. F., and D. F. Blair. 2001. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H(+) channels in the stator complex. Biochemistry 40:13051-13059. [DOI] [PubMed] [Google Scholar]
- 15.Briggs, W. R., and J. L. Spudich (ed.). 2005. Handbook of photosensory receptors. Wiley Interscience, Indianapolis, Ind.
- 16.Buron-Barral, M., K. K. Gosink, and J. S. Parkinson. Loss- and gain-of-function mutations in the F1-HAMP region of the Escherichia coli aerotaxis transducer Aer. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 17.Cantwell, B. J., R. R. Draheim, R. B. Weart, C. Nguyen, R. C. Stewart, and M. D. Manson. 2003. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J. Bacteriol. 185:2354-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conley, M. P., and H. C. Berg. 1984. Chemical modification of Streptococcus flagellar motors. J. Bacteriol. 158:832-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo, M., L. Chang, K. Lipkow, R. D. Vale, and S. Khan. Unpublished data.
- 20.Gauden, M., S. Yeremenko, W. Laan, I. H. van Stokkum, J. A. Ihalainen, R. van Grondelle, K. J. Hellingwerf, and J. T. M. Kennis. 2005. Photocycle of the flavin-binding photoreceptor AppA, a bacterial transcriptional antirepressor of photosynthesis genes. Biochemistry 44:3653-3662. [DOI] [PubMed] [Google Scholar]
- 21.Gilles-Gonzalez, M. A., and G. Gonzalez. 2005. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 99:1-22. [DOI] [PubMed] [Google Scholar]
- 22.Gosink, K. K., M. Buron-Barral, and J. S. Parkinson. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in E. coli. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 23.Hodgson, A. V., and H. W. Strobel. 1996. Quantitation of FAD-dependent cytochrome P450 reductase activity by photoreduction. Anal. Biochem. 243:154-157. [DOI] [PubMed] [Google Scholar]
- 24.Jasuja, R., Y. Lin, G. P. Reid, D. R. Trentham, and S. Khan. 1999. Response tuning in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 96:11346-11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jasuja, R., J. Keyoung, G. P. Reid, D. R. Trentham, and S. Khan. 1999. Chemotactic responses of Escherichia coli to small jumps of photoreleased l-aspartate. Biophys. J. 76:1706-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung, A., T. Domratcheva, M. Tarutina, Q. Wu, W. H. Ko, R. L. Shoeman, M. Gomelsky, K. H. Gardner, and I. Schlichting. 2005. Structure of a bacterial BLUF photoreceptor: insights into blue light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 102:12350-12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan, S., and R. M. Macnab. 1980. The steady-state counterclockwise/clockwise ratio of bacterial flagellar motors is regulated by protonmotive force. J. Mol. Biol. 138:563-597. [DOI] [PubMed] [Google Scholar]
- 28.Khan, S., and D. R. Trentham. 2004. Biphasic excitation by leucine in Escherichia coli chemotaxis. J. Bacteriol. 186:588-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan, S., J. L. Spudich, J. A. McCray, and D. R. Trentham. 1995. Chemotactic signal integration in bacteria. Proc. Natl. Acad. Sci. USA 92:9757-9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan, S., S. Jain, G. P. Reid, and D. R. Trentham. 2004. The fast tumble signal in bacterial chemotaxis. Biophys. J. 86:4049-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan, S., F. Castellano, J. L. Spudich, J. A. McCray, R. S. Goody, G. P. Reid, and D. R. Trentham. 1993. Excitatory signaling in bacteria probed by caged chemoeffectors. Biophys. J. 65:2368-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, C., M. Jackson, R. Lux, and S. Khan. 2001. Determinants of chemotactic signal amplification in Escherichia coli. J. Mol. Biol. 307:119-135. [DOI] [PubMed] [Google Scholar]
- 33.Ludovici, C., R. Frohlich, K. Vogtt, B. Mamat, and M. Lubben. 2002. Caged O2. Reaction of cytochrome bo3 oxidase with photochemically released dioxygen from a cobalt peroxo complex. Eur. J. Biochem. 269:2630-2637. [DOI] [PubMed] [Google Scholar]
- 34.Lybarger, S. R., U. Nair, A. A. Lilly, G. L. Hazelbauer, and J. R. Maddock. 2005. Clustering requires modified methyl-accepting sites in low-abundance but not high-abundance chemoreceptors of Escherichia coli. Mol. Microbiol. 56:1078-1086. [DOI] [PubMed] [Google Scholar]
- 35.Macnab, R., and D. E. Koshland, Jr. 1974. Bacterial motility and chemotaxis: light-induced tumbling response and visualization of individual flagella. J. Mol. Biol. 85:399-406. [DOI] [PubMed] [Google Scholar]
- 36.Montrone, M., M. Eisenbach, D. Oesterhelt, and W. Marwan. 1998. Regulation of switching frequency and bias of the bacterial flagellar motor by CheY and fumarate. J. Bacteriol. 180:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkinson, J. S., P. Ames, and C. A. Studdert. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8:116-121. [DOI] [PubMed] [Google Scholar]
- 39.Pruss, B. M., J. W. Campbell, T. K. Van Dyk, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sourjik, V. 2004. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 12:569-576. [DOI] [PubMed] [Google Scholar]
- 42.Sourjik, V., and H. C. Berg. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740-751. [DOI] [PubMed] [Google Scholar]
- 43.Spikes, J. D., and R. Straight. 1967. Sensitized photochemical processes in biological systems. Annu. Rev. Phys. Chem. 18:409-436. [Google Scholar]
- 44.Studdert, C. A., and J. S. Parkinson. 2004. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. USA 101:2117-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swartz, T. E., S. B. Corchnoy, J. M. Christie, J. W. Lewis, I. Szundi, W. R. Briggs, and R. A. Bogomolni. 2001. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 276:36493-36500. [DOI] [PubMed] [Google Scholar]
- 46.Talora, C., L. Franchi, H. Linden, P. Ballario, and G. Macino. 1999. Role of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J. 18:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, B. L., and D. E. Koshland, Jr. 1975. Intrinsic and extrinsic light responses of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 123:557-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor, B. L., I. B. Zhulin, and M. S. Johnson. 1999. Aerotaxis and other energy-sensing behavior in bacteria. Annu. Rev. Microbiol. 53:103-128. [DOI] [PubMed] [Google Scholar]
- 49.Taylor, B. L., J. B. Miller, H. M. Warrick, and D. E. Koshland, Jr. 1979. Electron acceptor taxis and blue light effect on bacterial chemotaxis. J. Bacteriol. 140:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, B. L., A. Rebbapragada, and M. S. Johnson. 2001. The FAD-PAS domain as a sensor for behavioral responses in Escherichia coli. Antioxid. Redox Signal. 3:867-879. [DOI] [PubMed] [Google Scholar]
- 51.Yang, H., A. Sasarman, H. Inokuchi, and J. Adler. 1996. Non-iron porphyrins cause tumbling to blue light by an Escherichia coli mutant defective in hemG. Proc. Natl. Acad. Sci. USA 93:2459-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, H., H. Inokuchi, and J. Adler. 1995. Phototaxis away from blue light by an Escherichia coli mutant accumulating protoporphyrin IX. Proc. Natl. Acad. Sci. USA 92:7332-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, H. S., J. H. Saw, S. Hou, R. W. Larsen, K. J. Watts, M. S. Johnson, M. A. Zimmer, G. W. Ordal, B. L. Taylor, and M. Alam. 2002. Aerotactic responses in bacteria to photoreleased oxygen. FEMS Microbiol. Lett. 217:237-242. [DOI] [PubMed] [Google Scholar]