Abstract
The Bacillus anthracis chromosome encodes four unique, putative superoxide dismutase (sod) genes. During exponential growth and sporulation, sodA1, sodA2, and sodC are transcribed constitutively throughout the growth cycle as individual genes. In contrast, the transcription of sod15 occurs mainly during late exponential and sporulation phases as part of a four-gene operon that may be involved in spore formation. Vegetative cell and spore lysates of wild-type Sterne and superoxide dismutase deletion (Δsod) mutants show detectable SOD activity for SODA1 and SODA2, and protein analysis suggests that these two proteins form active homodimers and heterodimers. A comparison of the growth of parental versus Δsod mutants under various chemical oxidative stresses indicates that ΔsodA1 mutants are particularly sensitive to endogenously produced superoxide, whereas ΔsodA2, Δsod15, and ΔsodC mutants remain as resistant to this stress as the parental strain. In addition, in mouse survival assays, Δsod15 and ΔsodA1 were responsible for less end-point death, but the level of decreased virulence does not fall within a statistically significant range. Collectively, these data show that sodA1 acts as a major protectant from intracellular superoxide stress, that sod15 is transcribed as part of an operon that may play a role in cell morphology, and that sodA2 and sodC may have minor roles that are not apparent in the conditions tested here.
The plasmid-encoded virulence factors (toxin and capsule) of the endospore-forming bacterium Bacillus anthracis, the causative agent of the disease anthrax, have been studied extensively (11, 46). However, the functions of the approximately 5,500 chromosomally encoded genes in this pathogen's biology and disease-causing capability are now beginning to be explored (8). The genome of B. anthracis has the potential for a high level of redundancy; for example, there are four phospholipases, five catalases, four superoxide dismutases, etc. By utilizing multiple genes in a modular fashion, bacteria are able to adjust quickly to various environmental stresses and insults, such as heat, changes in osmolarity, nutrient and metabolite deprivation, and highly oxidative conditions (54). This adaptability is of particular importance to pathogens, since they face multiple stresses within a host. B. anthracis responds to inhospitable conditions in the soil by forming dormant, metabolically inert endospores. The endospore is the form of the bacterium that can enter a mammalian host via various routes (respiratory, cutaneous, or gastrointestinal) and cause the disease anthrax. Once inside the host, the spores germinate and outgrow, effectively transforming into a replicative, metabolically active vegetative form. The pathogenesis of this microorganism is particularly complex since it is marked by two unique forms of the bacterium, a transition from one form to the other, and a spatial and temporal shift from one locale (the lung, skin, or gastrointestinal tract) to another (regional lymph nodes and the circulatory system) (10, 20).
B. anthracis is a facultatively aerobic organism and so, like all aerobes, must protect itself from toxic forms of oxygen that are produced during normal metabolism. Various antioxidant enzymes (superoxide dismutases, catalases, and peroxidases) and radical-neutralizing metabolites are the main mechanisms by which oxygen-utilizing organisms protect themselves from oxidative damage (3, 55). Pathogens must protect themselves from additional oxidative insults within a host environment, such as the oxidative burst of professional phagocytic cells and the varying oxidative environments within cellular and extracellular compartments. Although the phagocytic oxidative burst is known to be an important bactericidal weapon of the immune system (24, 42), the exact mechanism by which it works is still unclear and a subject of debate (49, 50). In addition, the role that self-generated reactive metabolites play in prokaryotic cellular regulation is also now beginning to be addressed (22).
Superoxide dismutase (SOD) proteins were discovered and characterized by McCord and Fridovich in the 1960s (36). There are two main classes of SOD proteins differentiated by their metal specificity: Mn or Fe versus Cu-Zn. It was at first thought that only eukaryotic species utilized Cu-Zn SODs, but it has since been shown that Cu-Zn SODs are quite ubiquitous in the prokaryotic world (30). SOD enzymes are highly conserved and exist in almost all aerobic organisms studied and even in many strict anaerobes (6). All SODs perform one chemical reaction: the dismutation of superoxide anion (O2·−), the first reduction product of molecular oxygen, to hydrogen peroxide (H2O2) and molecular oxygen. By catalyzing this reaction, SODs act as scavengers of O2·−, which can cause direct cellular damage or lead to the formation of other more reactive species such as hydroxyl radical or peroxynitrite (26). Both classes of SODs have been identified in many bacterial species. In some, such as Salmonella enterica serovar Typhimurium, SODs have been implicated in the pathogen's ability to cause disease (15, 16, 59). The case of Mycobacterium tuberculosis is more complex, with conflicting data about which SOD, the Cu-Zn or the Fe form, is more important to the disease-causing ability of this important pathogen (5, 12, 14, 41). SODs have also been implicated as being important for virulence in Shigella flexneri (18) and Streptococcus agalactiae (44).
Previously, a proteomic study of the B. anthracis endospore showed that two of the four genomically encoded sod's of B. anthracis, SODA1 and SOD15, are resident proteins of the endospore (33), the infectious form of the microorganism. Because of this and because B. anthracis encodes both classes of SODs, we used a genetic approach to determine whether either of the resident spore SODs (SOD15 and SODA1) or the nonresident SODC and SODA2 proteins are important for the survival of B. anthracis during normal growth, growth under oxidative stress, and growth during infection of mice.
MATERIALS AND METHODS
Plasmid and bacterial strain construction.
Strains and plasmids used in the present study are listed in Table 1. The creation of Δsod mutants was performed as follows. Oligonucleotide primers (Invitrogen) for PCR amplification of SOD genes (all primer sequences available upon request) were designed by using the Bacillus anthracis Ames strain genomic sequence from the TIGR Comprehensive Microbial Resource and Manatee Server. All initial cloning strategies were facilitated with the use of the Escherichia coli strains listed in Table 1. The strategy was as follows. The SOD genomic regions were PCR amplified with either Sigma KlenTaq or Invitrogen Platinum Taq high-fidelity systems using B. anthracis Sterne strain 34F2 genomic DNA with a 5′ BamHI restriction site and a 3′ KpnI restriction site. These fragments were ligated into the Promega pGEM-T Easy vector and selected for by lacZ selection on LB-ampicillin plates. “Inside-out PCR” was then performed (all primers are available upon request) creating XhoI and XbaI restriction sites such that 292 to 354 bp were omitted from the wild-type sequence. A kanamycin resistance cassette PCR amplified from vector pDG783 (21) with XhoI and XbaI restriction sites was ligated into these fragments and transformed for passage in E. coli DH5α or XL1-Blue. Standard minipreps (QIAGEN) were used to isolate plasmid, and the entire construct was PCR amplified and digested for insertion into recombination vector pKSV7 (52) by standard techniques utilizing Invitrogen T4 DNA ligase. pKSV7 constructs were passaged through dam+ dcm+ and dam dcm strains listed in Table 1. Transformation of pKSV7::Δsod plasmids into Bacillus anthracis Sterne 34F2 was done according to methods described previously (45) with the following modification: cells were allowed to recover after electroporation for 1 to 4 h in brain heart infusion (BHI) broth at 30°C with no shaking. Allelic exchange was performed as follows. After electroporation and recovery, cells were plated on BHI-kanamycin plates and grown at 30°C overnight to 2 days. Colonies were picked, grown in BHI-kanamycin broth at the nonpermissive temperature of 39.5°C, shaken at 300 rpm, and back-diluted 1:1,000 five to seven times to facilitate plasmid integration. These cultures were then shifted to the permissive temperature of 30°C in BHI broth with no antibiotic to facilitate allelic exchange. These cultures were back-diluted 1:1,000 every 12 h eight to eleven times. The cultures were then shifted to the nonpermissive temperature of 39.5°C to facilitate clearance of any cytoplasmic plasmid and back-diluted 1:1,000 every 12 h eight to eleven times. Cultures were streaked onto BHI plates and patch plated to screen for loss of chloramphenicol resistance and gain of kanamycin resistance. Candidate transformants were verified by Southern blotting and/or multiple PCRs. All strains were screened by PCR for the presence of the pXO1 plasmid.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant phenotype | Source or reference |
|---|---|---|
| Strains | ||
| Bacillus anthracis | ||
| Sterne 34F2 | pXO1+, pXO2- | Sterne (1937) |
| KDC1 | 34F2 Δsod15::Kmr | This study |
| KDC2 | 34F2 ΔsodC::Kmr | This study |
| KDC3 | 34F2 ΔsodA1::Kmr | This study |
| KDC4 | 34F2 ΔsodA2::Kmr | This study |
| KDCom3 | 34F2 ΔsodA1::Kmr, pBJ258(sodA1::Ermr) | This study |
| KDCom4 | 34F2 ΔsodA2::Kmr, pBJ258(sodA2::Ermr) | This study |
| Escherichia coli | ||
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 | Stratagene |
| supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | ||
| One Shot TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ[ρ](ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| CGSC 6478 | GM272 (dam-3, dcm-6) | Palmer and Marinus (1994) |
| INV110 | F′ (traΔ 36 proAB lacIqlacZΔ[ρ]M15) rpsL (Strr) thr leu endA thi-1 lacY galK galT ara tonA tsx dam dcm supE44 Δ[ρ](lac-proAB) Δ(mcrC-mrr)102::Tn10(Tetr) | Invitrogen |
| Plasmids | ||
| pGEM-T Easy | Ampr | Promega |
| pKSV7 | pUCori pE194ori(ts)aprcmr | 52 |
| pDG783 | pSB118::Kmr | 21 |
| pBJ258 | Ermr | Brian Janes |
Strains created for this study were made utilizing the TIGR Comprehensive Microbial Resources Genome sequence for the virulent Bacillus anthracis Ames strain since the Sterne 34F2 genome sequence was not available at the time this study was started. Therefore, BA numbers referenced in this study are from the Ames sequence; however, the sequences of all four sod genes are 100% identical in Sterne 34F2 and Ames. Ampr, ampicillin resistance; Kmr, kanamycin resistance; Ermr, erythromycin resistance; Tetr, tetracycline resistance.
All strains were sporulated as follows. Briefly, overnight BHI broth cultures (<12 h old) were diluted 1:20 into modified G medium and shaken at 30°C at 300 rpm for 3 days. The cultures were pelleted by centrifugation at 3,000 rpm for 30 min and washed five times in 45 ml of sterile Milli-Q water. The cultures were heat treated at 65°C for 30 min to kill any remaining vegetative bacilli and then pelleted by centrifugation once more and transferred into 1.5-ml volume screw-cap tubes in 1-ml volumes of sterile Milli-Q water. The cultures were then pelleted by centrifugation and washed two to three times with sterile Milli-Q water and stored at room temperature. The purity of the spore preparations was confirmed by phase-contrast microscopy, and concentrations were determined by serial dilution.
The antibiotic concentrations were as follows: kanamycin (30 to 50 μg/ml), chloramphenicol (30 μg/ml [E. coli] and 7 to 10 μg/ml [B. anthracis]), ampicillin (100 μg/ml), and erythromycin (300 to 400 μg/ml [E. coli] and 5 μg/ml [B. anthracis]).
B. anthracis growth under oxidative and nonoxidative conditions.
Cultures of B. anthracis Sterne strain (34F2) and Δsod mutant strains (34F2 parental strain) were grown overnight in LB broth or LB plus selective antibiotic for less than 12 h. In the morning, cultures were diluted into fresh LB broth to an optical density at 600 nm (OD600) of 0.1 to 0.2 and allowed to recover for 1.0 to 1.5 h at 37°C with shaking at 300 rpm. These cultures were then diluted and adjusted to an OD600 of 0.01 in fresh LB or LB plus selective antibiotic in a final volume of 50 or 70 ml and then shaken at 250 rpm at 37°C. OD600 was measured at 1-h intervals. For growth curves testing sensitivity to oxidative stress, redox cycling reagents were added when cultures reached exponential growth (OD600 = 0.4 to 0.6), usually at time point 2.5 h. A flask with water added instead of paraquat served as a growth control. Paraquat at 194 mM (Ultra Scientific PST-740) was added to a final concentration of 300 or 800 μM. Growth curves were determined three times on separate days utilizing three unique spore stocks of each strain.
Disk diffusion assays of tolerance to redox cycling compounds.
Cultures of B. anthracis Sterne strain (34F2) and Δsod mutant strains (34F2 parental strain) were started from a colony grown on BHI agar plates in BHI medium or BHI medium plus selective antibiotic. Cells were grown at 37°C shaking at 300 rpm to an OD600 of 0.4 to 0.5. Plate assays were performed by adding 200 μl of mid-log-phase cultures to 2 ml of 0.7% sterile soft agar. Then, 3 ml of the agar suspensions was spread onto BHI plates. Sterile paper disks (6 mm in diameter) were permeated with 10 μl of either 5% paraquat (Ultra Scientific PST-740 or Sigma methyl viologen M-2254), 0.08 M menadione (Sigma M5625) dissolved in ethanol, or 66 mM plumbagin (Sigma P7262) dissolved in ethanol or distilled water (negative control). Disks were placed on plates and incubated overnight at 37°C. Zones of inhibition were measured in millimeters. For each experiment, 10 disks (2 disks per plate) were used.
Change in OD600 germination assay.
Spores of B. anthracis Sterne (34F2) or Δsod mutant strains (34F2 parental strain) were placed in a microcuvette (Bio-Rad 22309955) to an OD600 0.3. in 1 ml of germination buffer (100 mM phosphate-buffered saline [PBS] plus 100 mM l-alanine). The change in the OD600 was measured every 60 s for 20 min using the kinetic read function on a Beckman DU530 spectrophotometer. The percent fall in the OD was calculated as follows: {[OD600(time zero) − OD600(time 20 min)]/OD600(time zero)} × 100. As shown previously, a decrease in the OD of ∼60% correlates to loss of heat resistance in >99% of cultures (17). Experiments were performed with at least three different spore preparations.
Nondenaturing polyacrylamide gel electrophoresis (PAGE) assay of SOD activity.
B. anthracis Sterne strain (34F2) and Δsod mutants (34F2 parental strain) were grown in modified G medium to late exponential phase (OD600 of ∼0.9). Portions (6 ml) of cultures or 200 μl of purified spore preps were pelleted at 4°C, washed twice in cold PBS, and resuspended in 1 ml of 0.05 M potassium phosphate buffer (pH 8.0) plus 25 mM benzamidine. Suspensions were transferred to 2.0-ml screw-cap tubes filled halfway with acid-washed glass beads (diameter, 150 to 200 μm; Sigma G-1145). Cells were disrupted mechanically via bead beating with a Biospec mini-bead beater on high speed 10 times for 1 min with at least a 5-min rest on ice between pulses. Tubes were spun at 4°C for 10 min at 1,000 rpm. Supernatants were transferred to fresh tubes and spun at 4°C for 10 min at 1,000 rpm. Then, 2 μl of DNase I, RNase-free (Roche), was added, followed by incubation on ice for 1 h and spinning at 5,000 rpm for 10 min at 4°C. Supernatants were transferred to a new tube and centrifuged at 14,000 rpm for 30 to 40 min. Supernatants were quantified for soluble protein by using the Bio-Rad protein assay. A total of 25 μl (ca. 30 to 80 μg of protein) was run on an Invitrogen 12% pre-cast Tris-glycine gel in Invitrogen nondenaturing Tris-glycine buffer. After electrophoresis, the gel was soaked in 0.05 M potassium phosphate buffer (pH 8.0) plus 50 mM EDTA plus 2 ml of 20 mg of nitroblue tetrazolium (NBT; Sigma N-5514)/ml and 6 ml of 0.5 mM riboflavin for 20 min. Gels were developed for 15 min under a miniature UV lamp.
Cell growth conditions and RNA isolation.
For quantitative reverse transcription-PCR (RT-PCR) of sod expression during sporulation, overnight BHI broth cultures of B. anthracis Sterne strain (34F2) were diluted 1:1,000 in the morning into nutrient-limiting (sporulation) modified G medium. Cells were harvested at an OD600 of 0.4 to 0.5, 0.7, 0.8, 0.9, and 1.1. Portions (5 to 7 ml) of cultures were collected and pelleted by centrifugation at 4°C. For end-point RT-PCR to determine transcriptional products of the sod15 operon, B. anthracis Sterne (34F2) and the Δsod15 strain were grown similarly but harvested at an OD600 of 0.9 to 1.0. RNA isolation was performed by using the Ambion RiboPure-bacteria kit according to the manufacturer's instructions with the following modifications: cell disruption with zirconia beads was done for 15 min, 400 μl of RNAwiz reagent was used, BCP phase separation reagent was used in place of chloroform, and 50 μl of RNase/DNase-free distilled water was added during extraction. The QIAGEN RNEasy mini-kit with a DNase digestion step was used according to the manufacturer's RNA cleanup protocol. RNA was quantitated via the A260/A280 ratio on a Beckman DU530 spectrophotometer. Then, 1 μg of RNA was run on a denaturing formaldehyde gel to verify the RNA integrity. These procedures were carried out in three separate experiments utilizing unique cultures each time.
SYBR green quantitative RT-PCR.
For quantitative RT-PCR, RNA was collected as described above, and cDNA was made from RNA samples utilizing Invitrogen random primers and Invitrogen SuperScript II reverse transcriptase. Briefly, random primers were used at 300 ng/μl with 1 μg of RNA and incubated overnight at 42°C. All samples were also run with no reverse transcriptase. A control PCR was performed on all cDNA samples to verify no genomic DNA contamination. Then, 1.5 μl of each cDNA reaction was used in 20-μl PCRs using an Applied Biosystems SYBR green master mix. Each sample was run twice in a full reaction: once with no cDNA and once with cDNA made with no RT. Reactions were run in a 384-well plate at the University of Michigan Comprehensive Cancer Center cDNA core facility on an ABI Prism 7900 HT SDS with the SDS Software version 2.0 sequence detection system using an annealing temperature of 56.4°C and an extension at 72°C for 1 min with 35 cycles. All primers were sequence specific for each gene analyzed, with PCR products of between 171 and 191 bp. The mean of the CT (threshold cycle) values for the duplicate runs and triplicate biological samples were used.
Endpoint RT-PCR.
A total of 750 ng of RNA collected from B. anthracis Sterne 34F2 grown as explained above was used to perform endpoint RT-PCR using Invitrogen one-step RT-PCR with Platinum Taq according to the manufacturer's instructions. Briefly, RT was performed at 50°C for 30 min. PCR was performed with 0.25 pg of operon/gene-specific primers (the sequences are available upon request) for 35 or 37 cycles with an elongation temperature of 70°C and an extension time of 1 min and 10 s. Next, 5 μl of endpoint PCR product was run on 0.7% agarose gels and visualized by ethidium bromide staining. Negative controls omitting reverse transcriptase and positive controls with B. anthracis Sterne (34F2) genomic DNA were done with each experiment. Operon/gene-specific primers were designed to result in 0.6- to 1.0-kb products.
Transmission electron microscopy.
B. anthracis spores or vegetative bacilli were fixed in 2.5% glutaraldehyde in 0.1 M Sorensen buffer (pH 7.4) overnight at 4°C. After several buffer rinses, the spores were postfixed in 1% osmium tetroxide in the same buffer. They were then rinsed in double-distilled water to remove phosphate and then stained en bloc with aqueous 3% uranyl acetate for 1 h. They were dehydrated in ascending concentrations of ethanol, treated with propylene oxide, and embedded in Spurr's epoxy resin over the course of 7 days. Ultrathin sections, 70 nm in thickness, were poststained with uranyl acetate and lead citrate. Sections were then examined by using a Philips CM100 electron microscope at 60 or 80 kV. Images were recorded digitally using a Hamamatsu ORCA-HR digital camera system operated using AMT software (Advanced Microscopy Techniques Corp., Danvers, MA).
Determination of primary protein sequence distances.
The DNASTAR Lasergene v6 program MegAlign was used for determining the protein sequence identities and divergence. Multiple alignments were performed by using the CLUSTAL W method set on a “slow-accurate” parameter with a gap penalty of 10.00. MegAlign calculates the divergence between sequences by comparing sequence pairs in relation to a reconstructed phylogeny that is generated by the program. It should be noted that the percent identity is a direct comparison, but divergence is calculated by comparing sequence pairs to a phylogeny reconstructed by the program used and so is not an inverse of percent identity (i.e., this number can be >100).
Infection of DBA/2 mice with B. anthracis Sterne and Δsod spores.
Intratracheal infection of mice. Briefly, mice were anesthetized with ketamine-xylazine, and a small incision was made through the skin above the trachea. A 30-μl inoculum containing approximately 100, 1,000, 10,000, or 100,000 spores suspended in water was delivered to the lung with a 30-gauge needle plus a 1-ml syringe. Spores were quantitated pre- and postinfection via serial dilutions on BHI plates. Postmortem necropsies were performed, and B. anthracis strains were isolated on BHI or BHI-kanamycin plates from lung and/or spleen homogenates of all terminal animals and on one surviving animal of each group at the end of the experiments. Mice were monitored three times a day for morbidity and mortality for 10 days. The 50% lethal dose (LD50) was calculated by the method of Reed and Muench (48). For each dose of spores, the total number of mice utilized was as follows: (i) 102 spores, n = 4 for all strains; (ii) 103 spores, n = 9 mice for all strains except ΔsodC, where n = 4; and (iii) 104 spores, n = 9 for Sterne, Δsod15, and ΔsodA2, n = 4 for ΔsodC, and n = 18 for ΔsodA1. Information regarding the log-rank test may be found online (http://bioinf.wehi.edu.au/software/russell/logrank/).
RESULTS
Bioinformatics of the four B. anthracis SOD genes.
The B. anthracis genome encodes four distinct proteins with conserved SOD domains (Fig. 1A), one of likely Cu-Zn specificity (BA5139 [sodC]), two that are putatively manganese (BA4499 [sodA1] and BA5696 [sodA2]), and one of unknown metal specificity (BA1489 [sod15]) (47). As of January 2006, nine different strains of B. anthracis are contained in the TIGR Comprehensive Microbial Resource (www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl), including virulent strains and the attenuated Sterne strain. Although the location of the four sod genes displayed in Fig. 1A was derived from the virulent Ames Ancestor strain, the sod's of the toxin-producing, unencapsulated Sterne strain (utilized in the present study) are 100% identical at the nucleotide level and are located on the chromosome in the same regions. The four sod's are located quite far from each other, with two on the leading strand (sod15 and sodA2) and two on the lagging strand (sodA1 and sodC) and no apparent linkages between them. The nomenclature for sod15 was coined for the present study to underscore its unknown metal specificity and to differentiate it from the other paralogous sod's A1 and A2.
FIG. 1.
Genomic location of B. anthracis SOD genes (sod) and SOD activity assay. (A) Distribution of the four putative SOD (sod) open reading frames on the B. anthracis Ames genome (BA1489, BA5139, BA4499, and BA5696, respectively) and the sizes of the intergenic spaces. (B and C) SOD activity on native PAGE-nitroblue tetrazolium gels from vegetative cell lysates of Sterne and Δsod mutants (B) and vegetative cell lysates of complementation strains for sodA1 and sodA2 (C).
The Mn-containing SODA of B. subtilis has been characterized (7, 27), as have the Cu-Zn SODs of Salmonella serovar Typhimurium and M. tuberculosis (9, 15, 53). Multiple protein alignments were constructed to ascertain the level of primary amino acid sequence conservation that exists between the two classes of these proteins (see Materials and Methods) comparing the following primary amino acid sequences: (i) the three B. anthracis Mn-SODs (SODA1, SODA2, and SOD15) with B. subtilis SODA and (ii) B. anthracis SODC with the Cu-Zn SODs of the two bacterial pathogens Mycobacterium tuberculosis and Salmonella enterica serovar Typhimurium.
Of the three putatively Mn-containing SODs of B. anthracis, SODA1 is the one most closely related to B. subtilis SODA with 76.7% identity, while SODA2 maintains a lower identity at 55.4%. Because SODA of B. subtilis is the only active SOD seen during all stages of growth and this enzyme is essential for resistance to superoxide in B. subtilis (7), this suggested to us that either SODA1 or SODA2, or both, would most likely play the normal role of an antioxidant protectant during aerobic growth or under oxidative stress. SOD15 has only a 36.5% identity with the B. subtilis ortholog and is almost completely divergent from the other two B. anthracis Mn-SOD paralogs (>100% divergent from SODA1 and SODA2). This degree of divergence is mainly due to an additional N-terminal 135 amino acids encoded in the B. anthracis sod15 gene. The predicted molecular mass of SOD15 is approximately 36 kDa, which is substantially larger than the average size of most SODs (∼23 kDa). SOD15 has identical homologs only in the other pathogenic Bacillus species, B. cereus and B. thuringiensis, suggesting that this particular SOD may have evolved a unique function in these species.
Cu-Zn SODs represent an entirely different class of enzyme from the Mn- or Fe-chelating class of proteins and are present in many bacteria, often periplasmically located in gram-negative species (30, 37, 53). B. anthracis encodes one putative Cu-Zn SOD (SODC), and this gene is conserved in the pathogenic B. cereus group. We compared the primary sequence of B. anthracis SODC to the Cu-Zn SOD of two pathogens, M. tuberculosis and S. enterica serovar Typhimurium, that are capable of surviving intracellularly, as has been shown for B. anthracis (51). The role of SODC in M. tuberculosis is unclear, but it has been shown to possibly play a role in intracellular survival (12, 41), and the B. anthracis SODC shares a very low identity at with it at only 21.2%. In S. enterica serovar Typhimurium, all strains encode a Cu-Zn enzyme called SODCII, but only highly virulent Salmonella species contain an additional phage-encoded Cu-Zn SOD, named SODCI (15). In our CLUSTAL W analysis, the two serovar Typhimurium enzymes share ∼58% identity with each other, but neither one is particularly similar to the SODC of B. anthracis, which only shares 25% identity with each. Oddly, the Cu-Zn SODs of prokaryotes tend to be very divergent, often lacking obvious metal ligands entirely (1), which is contrary to the high level of conservation in eukaryotic Cu-Zn SODs. The high level of divergence between the B. anthracis SODC and the enzymes of serovar Typhimurium and M. tuberculosis suggested to us that this putative SOD might play a novel, perhaps specialized, role in the biology of B. anthracis or, conversely, might be a relic with no overt physiological role at all.
Since the B. anthracis SODs differ considerably from one another and from those in other bacterial species, we hypothesized that these enzymes might play unique roles during different phases of growth and in different growth environments. In addition, proteomic analysis of the B. anthracis spore revealed that SOD15 and SODA1 are resident members of the spore, the infectious form of the microbe (33), further suggesting that these SODs may be important during pathogenesis.
Superoxide scavenging by B. anthracis SODs from cell lysates.
In order to reveal which SOD(s) might be the main superoxide scavenging enzyme of B. anthracis and to determine whether any of the four proteins is important for the establishment of the disease anthrax, single-deletion mutants in each of the four sod genes were generated. Single-deletion strains were created via homologous recombination involving the removal of approximately 300 nucleotides from each sod open reading frame and the insertion of a kanamycin resistance cassette (sod::Kmr mutants, hereafter referred to as Δsod mutants). Mutants were then assayed for defects in normal growth, for growth under various oxidative stresses, and for the ability to generate disease in susceptible mice.
Since the enzymatic function of SODs is to scavenge superoxide, we set out to determine whether each of the four B. anthracis SODs is present in active form in spore and vegetative cell lysates. To determine which form(s) of the proteins are present in whole bacterial cells, we performed a nondenaturing PAGE-SOD activity assay with cell lysates prepared by mechanical disruption of B. anthracis cells (Fig. 1B and C). Three distinct bands of SOD activity can be detected in the wild-type Sterne strain, as well as in the two single deletion mutants Δsod15 and ΔsodC for both vegetative cells (Fig. 1B) and spores (not shown). However, both the ΔsodA1 and the ΔsodA2 mutant display only one band of SOD activity each; the highest band in the former and the lowest band in the latter (Fig. 1B). Complementing ΔsodA1 and ΔsodA2 in trans with a plasmid-borne copy of the wild-type gene leads to a restoration of the two missing bands for each strain (Fig. 1C). This not only implies that the bands of activity are due to SODA1 and SODA2 homodimers but also strongly suggests that SODA1 and SODA2 form an active heterodimer. Although the uppermost band of activity, which is most likely SODA2, in Fig. 1B for Sterne cells appears to display a qualitatively stronger band of SOD activity, it should be mentioned that this assay is only semiquantitative and that from gel to gel the intensities of each band varied with no apparent pattern. In addition, we believe that the SODA2 bands of activity that we detected from spore lysates are, indeed, from proteins resident in the spore and not from vegetative detritus in the spore preparation, since it seems unlikely that a sufficient amount of nonspore enzyme would be abundant enough to show a clear band of activity.
Iron- and manganese-containing SODs have very similar tertiary structures, and it is sometimes possible to distinguish the two from their primary structures (39, 40). However, anomalies exist, and it is not always possible to distinguish these types of enzymes from specific metal-chelating residues alone (28). Recently, the crystal structures of B. anthracis SODA1 and SODA2 were determined (4). In that study, the recombinant proteins were chelated with Mn, the structures determined were homodimers, and the same nondenaturing gel assay was used to confirm that the homodimers of each protein were able to scavenge superoxide. However, the possibility that SODA1 and/or SODA2 might be cambialistic (i.e., able to chelate and utilize both Mn and Fe) has not yet been tested. We performed the NBT gel assay under several conditions, but additional bands of SOD activity corresponding to SOD15 and SODC from cell lysates were not found. At this point, it is unclear whether SOD15 and SODC proteins provide SOD activity at all, whether they simply are not abundant enough in the cell to show a signal in this assay, or whether they had been degraded or inactivated during the gathering of lysate.
Expression of B. anthracis sod's during exponential growth and entry into sporulation.
A lack of SOD activity for SOD15 and SODC in the previous assay led us to elucidate the general transcriptional pattern of the four sod genes to determine their general expression levels. A previous microarray study (33) suggested that sod15 was differentially expressed upon entry into late exponential phase. Therefore, we used SYBR green quantitative RT-PCR to verify that that each of the four genes is actively transcribed (Fig. 2 and Table 2). The cells were grown in modified G medium, a specialized medium that promotes the formation of a high number of spores upon entry into stationary phase. sodA1 consistently showed the highest abundance of mRNA compared to the internal control translation elongation factor g (fusA) and was transcribed constitutively with a slight decrease in transcript abundance at OD600 higher than 0.9. A constitutive pattern of expression was also seen for sodC and sodA2, but to different levels. Whereas sodC showed almost equal transcript abundance with the internal control fusA, sodA2 had a consistently lower expression than both sodA1 and sodC. Only sod15 revealed a differential pattern of expression in this growth curve (Fig. 2 and Table 2) where the transcript was ∼200-fold less abundant than fusA at OD600 0.5 with a steadily increasing amount of mRNA coinciding with an increasing OD600. These data indicate that all four sod's are expressed during the growth cycle, albeit to different levels, and suggest a specialized role for sod15 after entry into stationary phase and/or during the formation of endospores.
FIG. 2.
SYBR green quantitative RT-PCR analysis of B. anthracis sod expression during exponential growth and sporulation. Negative threshold cycles (CT values) listed in Table 2 are plotted to show the pattern of changes in expression levels with increasing OD600, where an OD600 of 0.5 represents early to mid-log phase, an OD600 of 0.7 to 0.8 represents late log phase, and an OD600 of 0.9 to 1.1 represents entry into sporulation phase, where phase-bright spores are visible in the culture. Because lower CT values are indicative of a higher level of transcript, negative values were used to reflect increases or decreases in mRNA level over time, accordingly.
TABLE 2.
SYBR green quantitative RT-PCR of sod genes of B. anthracis during exponential growth and entry into sporulationa
| OD600 | fusAb (mean CT) |
sod15
|
sodC
|
sodA1
|
sodA2
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Mean CT | FD | Mean CT | FD | Mean CT | FD | Mean CT | FD | ||
| 0.5 | 17.7 | 25.6 | −200.0 | 21.6 | −15 | 15.9 | +3.5 | 23.0 | −39.4 |
| 0.7 | 20.6 | 25.7 | −34 | 20.2 | +1.3 | 16.5 | +17.1 | 22.5 | −3.7 |
| 0.8 | 21.3 | 20.7 | +1.5 | 21.4 | −1.0 | 17.1 | +18.4 | 23.0 | −3.2 |
| 0.9 | 19.5 | 18.5 | +2 | 21.0 | −1.5 | 18.4 | +2.1 | 24.5 | −32.0 |
| 1.1 | 19.6 | 17.7 | +3.7 | 20.6 | −2.0 | 17.8 | +3.5 | 22.7 | −8.5 |
SYBR green quantitative RT-PCR is interpreted via CT values, which are the values at which the PCR reaction crosses a set threshold into exponential phase; therefore, a lower CT value indicates a more abundant transcript. The fold difference (FD) is as compared to the reference gene “elongation factor g” and was calculated as: CT higher − CT lower = x. The lower-value CT transcript is approximately 2x times more abundant than the higher CT transcript.
The translation elongation factor G gene fusA (BA0107 or BAS0107) was chosen as a reference since previous microarray data (33) indicate that, during growth and sporulation, transcript levels of this gene are never more than 1× higher or lower than the pooled RNA reference.
sod15 is part of an operon during late exponential phase, and Δsod15 mutant spores show only slight ultrastructural differences relative to parental spores.
Because sod15 appeared to be differentially expressed during the late exponential and stationary (sporulation) phases, we looked more closely at the expression of this gene and at the ultrastructure of Δsod15 mutant spores. A role for SODs in spore formation has been suggested for B. subtilis, where sodA-deficient strains form aberrant endospore coats (23). Other genes linked to a similar endospore phenotype in B. subtilis include a three-gene operon composed of dacB (a d-alanyl-d-alanine carboxypeptidase/penicillin-binding protein) and spmA and spmB (spore maturation proteins), where these genes appear to be necessary for the formation of heat-resistant spores with a normal ultrastructure (43). In B. subtilis, sodA and the dacB operon are about 161 kb apart from each other. In B. anthracis, one of the four putative sods, sod15, is located only 116 bp upstream of the dacB operon homologs (BA1490-1492; referred to here as dacBBa, spmA, and spm). This gene order that includes sod15 is conserved only within the pathogenic B. cereus group. Neither the intergenic distance (116 bp) between sod15 and dacBBa, spmA, and spm (∼50, 66, and 66% amino acid identity with B. subtilis dacB, spmA, and spmB, respectively) nor their putative protein functions overtly imply operon-linkage. However, a new operon-prediction algorithm developed by N. H. Bergman and Z. S. Qin (unpublished data) suggested that these four genes might be transcribed as one unit. To determine whether B. anthracis sod15, dacBBa, spmA, and spm are transcribed as a single mRNA unit, we performed endpoint RT-PCR from RNA collected from wild-type Sterne and Δsod15 cells grown to both a low and a high OD600 (0.4 and 1.0, respectively) using various primer sets designed to overlap the four genes (Fig. 3A and B). In both wild-type Sterne cells and Δsod15 mutants at an OD600 of 0.4, no transcript overlapping sod15 and DacBBa was detected, but transcript within the sod15 gene in Sterne was seen, suggesting that transcription of these genes during early log phase is separate. The tightly linked three-gene operon of dacBBa, spmA, and spm is seen as a contiguous unit at a low OD600. Interestingly, at an OD600 of 1.0, a transcript overlapping all four genes was readily detected in both Sterne and Δsod15. Predictably, no transcript from within the sod15 gene was detected in the Δsod15 mutant. The presence of transcripts overlapping sod15 and dacBBa in both strains but only during late log phase implies the presence of multiple promoters. This indicates that, at least during late exponential phase and entry into stationary phase, the products of these four genes may serve a cooperative function and also suggests that they may be part of a stress-induced regulon.
FIG. 3.
sod15 is part of a four-gene operon at high OD600, and Δsod15 spores are similar to parental Sterne spores as seen by transmission electron microscopy. Endpoint RT-PCR shows that sod15 is transcribed as part of a contiguous transcript at high OD600. (A and B) Primers (arrows) and their predicted gene-overlap products (lines) are indicated above and below the open reading frames of the sod15 (BA1489) and Δsod15 genomic regions: d-alanyl-d-carboxypeptidase (DacBBa [BA1490]), spore maturation protein A (spmA [BA1491]), and spore maturation protein (spm [BA1492]). Gels showing contiguous products at low (0.4) and high (1.0) OD600s from the indicated primer pairs. Two lanes on each side of ladder in panel A are biological replicates. Non-reverse-transcriptase-containing negative controls revealed no genomic DNA contamination (not shown). (C) Transmission electron microscopy of Sterne (34F2) and Δsod15 whole spores at ×64,000 magnification (left two panels) and details of spore coat structures at ×245,000 magnification (right two panels). cx, cortex; sc, spore coat; ex, exosporium; om, outer membrane; im, inner membrane.
To determine whether the removal of sod15 affected endospore ultrastructure, we performed transmission electron microscopy on Sterne (34F2) and Δsod15 endospores (preparations were also made for ΔsodA1 and ΔsodC mutants [not shown]). Figure 3C shows micrographs of endospores of both strains at ×64,000 (left two panels) and ×245,000 (right two panels) magnifications. The lower magnification shows no overt differences in the ultrastructure of the spores, and the average size of the spores (900 nm to 1 μm) did not differ between the two strains. An intact exosporium and thick cortex was observed in both strains. At a higher magnification, the spore coat and cortex can be seen in more detail. The spore coat of B. anthracis differs markedly from that of B. subtilis in thickness, and the protein composition of the endospore coats differs from species to species (31). Sterne spores consistently showed a very clear, double-layered protein coat with a visible outer membrane located between the spore coat and the cortex. Spore coats of the Δsod15 mutant typically had a more diffuse, multilayered appearance (Fig. 3C) but by no means revealed the striking phenotype of B. subtilis sodA mutants (23). Although it remains formally possible that this appearance is an artifact of thin sectioning, this form of the spore coat was not detected in several preparations of the parental Sterne strain. The Δsod15 strain sporulates in modified G medium as efficiently as Sterne, and the spores show only a slight increase in sensitivity to wet heat at 70°C (not shown). In addition, Δsod15 germinates identically to the parental Sterne strain as measured by the change in OD600 with the addition of the germinant l-alanine (100 mM in PBS) with an average drop in OD600 of ca. 53% over the course of 21 min for both strains. The modest changes in the spore structure of the B. anthracis Δsod15 mutant differ radically from the dramatic phenotypic changes seen in B. subtilis by removal of either its sodA or the dacB operon. We conclude that in the late exponential and stationary phases, there is transcriptional linkage between B. anthracis sod15 and three downstream genes that have been implicated in spore formation in B. subtilis. The obvious controlled expression of sod15 and its operon linkage strongly suggest that this protein has evolved a novel role in spore formation, but it appears to be less critical than B. subtilis sodA. In addition, although a contiguous mRNA unit was detected for the dacBBa operon in the Δsod15 mutant at both low and high OD600 values, it cannot be ruled out that a polar effect has occurred in the more downstream genes, either due to frameshift or to changes in the dacBBa promoter. The possible functional relationship between the genes in the sod15 transcriptional unit remains unclear and is currently under investigation.
SODA1 is responsible for protection from intracellular superoxide.
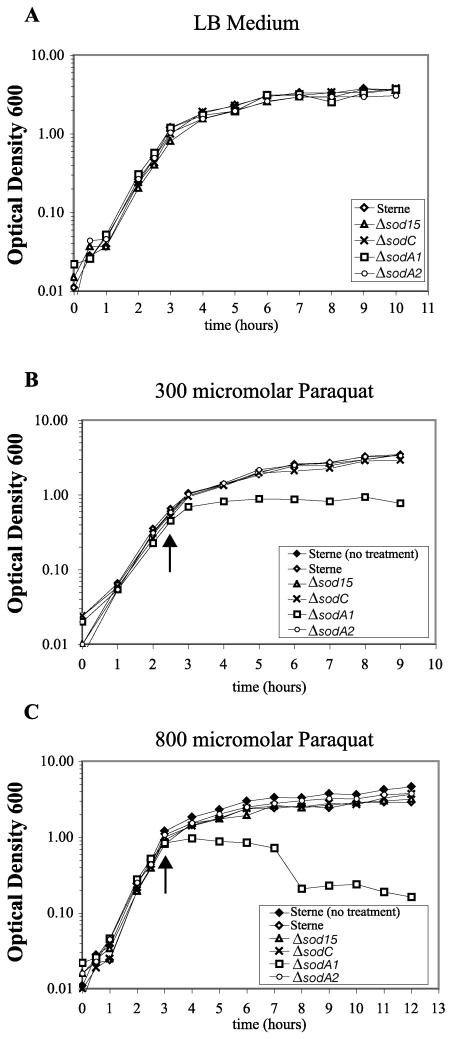
The typical role of SODs in cells is to scavenge superoxide as a first defense in preventing the oxidative damage of biomolecules. Superoxide is a byproduct of electron transport during aerobic growth (26), and although in itself is not a particularly reactive oxygen radical (13), its presence can lead to the release of free iron in the cell from proteins that contain [4Fe-4S] and [2Fe-2S] clusters, which can then, in the presence of H2O2, lead to the formation of the highly reactive hydroxyl radical via the Fenton reaction (25, 29, 57). We tested each single Δsod mutant for sensitivity to compounds that cause the generation of intracellular superoxide radical. In broth growth assays, all Δsod mutants were indistinguishable from wild-type Sterne in rich medium (LB) at 37°C (Fig. 4A). With the addition of the redox cycling compound paraquat added at mid-exponential phase to final concentrations of 300 and 800 μM, only the ΔsodA1 mutant showed an obvious growth defect (Fig. 4A and B). At 300 μM, ΔsodA1 leveled off in growth after the addition of paraquat, with all other strains attaining stationary phase equivalently (Fig. 4B). At the higher paraquat concentration of 800 μM, the ΔsodA1 strain was highly sensitive, leveling off in growth for several hours and then showing a decrease in OD600 (Fig. 4C). At this concentration, all other strains, including Sterne, grew to only a slightly lower OD600 in the stationary phase than Sterne cells with no treatment. We conclude that in broth growth, under high superoxide stress, the presence of SODA1 is necessary to allow for robust growth.
FIG. 4.
Growth of Sterne and Δsod strains in liquid broth under paraquat-induced superoxide stress. Arrows indicate the time at which paraquat was added to indicated concentration. (A) Growth of four Δsod mutants is equivalent to the parental Sterne strain in liquid Luria-Bertani (LB) broth at 37°C. (B) Growth of Δsod mutants in the presence of a low concentration of paraquat (300 μM) suggests that the lack of only one SOD protein is compensated for by the remaining paralogs; at a higher concentration (800 μM) (C), SODA1 is essential for survival into exponential phase, and a drop in OD600 suggests a lysis response. (The growth curves shown are representative of three independent experiments performed from three separate spore stocks all with the same trend.)
Disk diffusion plate assays were performed to test the sensitivity to the redox cycling compounds paraquat (5%) and menadione (80 mM) at a higher oxygen tension (cell to air interface). These assays were performed with both vegetative bacilli and purified spore preparations to differentiate the sensitivity of actively growing cells from the ability of dormant endospores to germinate and outgrow in an oxidatively stressful environment. Only the vegetative cells and spores of ΔsodA1 are significantly more sensitive to paraquat and menadione than the parental Sterne strain and the other three Δsod strains, showing significantly larger zones of inhibition (Fig. 5A to D). In addition, spores of ΔsodA1 display an increased sensitivity to these compounds relative to replicating bacilli, suggesting that the amount of SODA1 present within the endospore is not sufficient for normal outgrowth under high oxidative stress. The ΔsodA1 complemented strain [ΔsodA1(A1)] showed significantly smaller zones of inhibition compared to strains carrying an empty vector [ΔsodA1(pBJ258) and Sterne(pBJ258)] (Fig. 5E), although the zones of the complemented strain were not entirely reduced to the size of those for Sterne, suggesting that transcription from a low-copy plasmid provides partial complementation.
FIG. 5.
Disk diffusion assays of the sensitivity of Δsod mutants to the superoxide generating compounds paraquat and menadione. Photos are of plates from experiments listed in Table 3. Zones of inhibition of vegetative cells and spores from oxidative stress induced by 5% paraquat (A, B, and E) or 80 mM menadione (C and D). n = 10 disks per experiment.
As mentioned above, B. subtilis sodA mutants display a striking spore morphology phenotype (23), and this particular SOD has been shown to be the main protective enzyme in B. subtilis from paraquat-induced oxidative stress (7). Of the four B. anthracis SODs, SODA1 shows the closest amino acid sequence homology (∼77%) to the B. subtilis enzyme and also protects B. anthracis from intracellular superoxide stress. However, transmission electron microscopy of ΔsodA1 mutants shows neither a change in average spore size nor any obvious morphological defect (not shown).
The results of bacterial growth under oxidatively stressful conditions both in broth and on plates confirm that, of the four SODs of B. anthracis, SODA1 protects against endogenously generated superoxide and therefore is the predominant protective enzyme during aerobic growth. It should also be noted that colonies of the ΔsodA1 mutant on plates are generally smaller than those of parental Sterne and the other three Δsod mutants, suggesting that at the agar-air interface, where oxygen tension is higher than in broth, SODA1 is needed for optimal growth.
Intratracheal infections of mice with Δsod mutants.
The degree to which chromosomally encoded factors contribute to the establishment of disease in B. anthracis is only now beginning to be addressed. To determine whether one of the four B. anthracis SODs is important for bacterial survival within a host, we performed survival studies on DBA/2 mice, a strain that is sensitive to infection with Sterne spores. The intratracheal route of infection was chosen to most closely mimic an inhalational route of spore entry. Table 4 indicates the LD50 determined for each strain in the present study, as well as final percent survivals and a mean times to death at an infectious dose of 104 spores. Experiments were also performed with 102, 103, and, for Sterne and the ΔsodA1 mutant, 105 spores (not shown) to determine the LD50s by the method of Reed and Muench (48). After 10 days, the percent survival values for mice infected with 104 spores of Sterne, ΔsodC, and ΔsodA2 strains were similar: 22, 25, and 33%, respectively (Table 4). However, mice infected with an equivalent dose of Δsod15 and ΔsodA1 showed higher rates of survival after 10 days, with 44 and 56% surviving, respectively. Log-rank tests performed on the stated data indicate P values of 0.4 for Δsod15 and 0.2 for ΔsodA1. These P values do not fall under the typical cutoff for statistical significance (P ≤ 0.05) so, although the trends are very consistent, attenuation is only suggested. Interestingly, dissemination to the spleen was inconsistent for all strains; some of the dead and living mice had detectable bacterial loads in the spleen, and some did not. The lungs, however, all maintained a load of bacteria, both at the time of death and even in mice that had survived 10 days. Considering the complexity of modeling an inhalational route of infection via intratracheal delivery of spores and the lung environment in general, it does appear that the SOD15 and SODA1 may contribute slightly to bacterial fitness in a lung infection but are not essential for the microorganism to establish disease. Whether the Δsod15 and ΔsodA1 mutants are slightly more sensitive to bacterial killing or are unable to grow efficiently at later time points is uncertain and is still under investigation.
TABLE 4.
LD50 and mean-time-to-death data for intratracheal infections of DBA/2 mice with 104 spores of B. anthracis Sterne and Δsod mutants
| Strain | LD50 (48) | % Survival | Mean days to death |
|---|---|---|---|
| Sterne (34F2) | ∼4.5 × 104 | 22 | 2.4 |
| Δsod15 mutant | ∼8.0 × 104 | 44 | 3.6 |
| ΔsodC mutant | ∼5.0 × 104 | 25 | 3.0 |
| ΔsodA1 mutant | ∼1.0 × 105 | 56 | 2.4 |
| ΔsodA2 mutant | ∼5.0 × 104 | 33 | 2.3 |
DISCUSSION
Since the discovery of SOD in the 1960s (36), this important antioxidant enzyme has been characterized in numerous species, both eukaryotic and prokaryotic (2, 19, 38). The roles that SODs play in the disease-causing ability of pathogenic microorganisms are diverse, underlining the adaptability that prokaryotes have evolved to optimize growth in varied physical environments (see reference 34 for an excellent review). Two of the four encoded putative SODs of B. anthracis, SOD15 and SODA1, were found to be resident proteins of the endospore, the infectious form of this microorganism (33). In this proteomic analysis, SOD15 was identified in the soluble, membrane, and exosporium fractions, whereas SODA1 was eluted in the soluble and exosporium fractions. It should be noted, however, that the exosporium fractions in the present study most likely also contained some proteins from the spore coat; therefore, the exact location of these two enzymes in the spore is still not entirely clear. In addition, B. anthracis carries two other putative sod genes on the genome (sodA2 and sodC). The quantity and diversity of the encoded SODs prompted us to create single-deletion mutants in each of the four B. anthracis sod genes to determine whether these proteins work cooperatively as antioxidants or whether they might have unique functions in different growth environments.
Only sod15 shows a transcriptional profile that suggests some form of adaptive regulation, where mRNA abundance is low during exponential growth but increases dramatically during the late exponential and sporulation phases. However, although the other sod genes displayed constitutive expression under our experimental conditions, they may, indeed, be differentially regulated under conditions different from those tested here such as, for example, under alternative stresses or during infection. The transcriptional profile of sod15 suggests that it may be a member of a stress-induced regulon, since entry into sporulation phase is initiated by signals such as DNA damage and nutrient depletion (56). Its transcription at high OD600 as part of a four-gene operon that includes homologs to B. subtilis genes that are important for the formation of normal endospores (dacB operon) (43) suggests that sod15 may have a minor role in the formation of spores. B. subtilis cells lacking sodA or the dacB operon have strong spore structure defects (23, 43). Electron microscopy of B. anthracis Δsod15 mutant spores reveals only minor, sporadic differences in spore ultrastructure. Also, Δsod15 spores do not have a germination defect as measured by a drop in the OD600 in 100 mM l-alanine compared to the parental Sterne strain. This differs from the radical phenotypes of B. subtilis dacB and sodA mutants. Henriques et al. (23) suggest that the role SOD could play in B. subtilis spore formation is indirectly catalytic, changing the oxidative microenvironment during coat assembly and facilitating dityrosine linkages of coat proteins. However, the only biochemical precedent describing a similar phenomenon is in eukaryotes (32, 58) and has not yet been substantiated in prokaryotes. Further in vitro biochemical analysis with recombinant proteins may aid in explaining why sod15 is part of an operon putatively involved in cell morphology. Since Δsod15 strain causes slightly less mortality in intratracheal infections of mice, it is possible that spore integrity is compromised in a way that is not visible at the ultrastructure level.
The ΔsodA1 strain consistently revealed a major phenotype in terms of sensitivity to endogenous superoxide stress. During exponential growth and entry into the stationary or sporulation phase in sporulation medium, the transcriptional levels of sodA1 are consistently the highest of the four sod genes, whereas sodA2 shows the lowest abundance of transcript, further supporting a leading role for the sodA1 paralog. Growth under intracellular superoxide stress induced in vitro, both in broth (300 and 800 μM paraquat) and on agar plates (paraquat and menadione), shows that SODA1 is an extremely important enzyme for protection from endogenously generated superoxide stress. Also, spores of this strain are even more sensitive to a strongly oxidative environment than replicating bacilli, suggesting that the amount of SOD in the spore is important for efficient outgrowth under high oxidative stress. Of the four B. anthracis SODs, SODA1 shares the highest amino acid identity with B. subtilis SODA, but differences in spore ultrastructure were not apparent in the ΔsodA1 mutant strain by transmission electron microscopy, again underlining differences in the role of SODs in these two species. In mouse intratracheal infections, the ΔsodA1 strain caused less mortality than the parental Sterne strain, although not within a statistically significant range. A recent study of the ability of neutrophils to kill B. anthracis (35) suggests that, at least in these phagocytes, killing is independent of the oxidative burst, since cells treated with the NADPH oxidase inhibitor DPI were just as efficient at killing as those without treatment, suggesting that B. anthracis SODs may not be strongly protective from the host oxidative burst. Because ΔsodA1 mutants struggle in the presence of intracellular superoxide, sodA1 most likely serves B. anthracis by protecting it from metabolically generated oxidative stress during rapid growth during infection and in vitro growth.
The crystal structures of SODA1 and SODA2 were recently determined as homodimers with chelated Mn as the catalytic metal (4). Our nondenaturing PAGE-nitroblue tetrazolium SOD activity assays of Δsod mutants and complemented strains strongly suggest that SODA1 and SODA2 form both homodimers and heterodimers. The possibility that one or both of these enzymes might be able to utilize Fe as a catalytic metal has not been tested. Since the ΔsodA2 mutant is not sensitive to superoxide stress and is able to cause the same amount of mortality in mouse intratracheal infections as parental Sterne, it is unclear how important this gene or the A1/A2 heterodimer form of the enzyme is for normal growth. A possible level of redundancy that might exist between the four putative SODs may not be apparent in the phenotypes of single mutants but could be apparent in multiple knockouts.
In conclusion, our initial survey of the four B. anthracis SODs shows sodA1 to be the prototypical, cellular antioxidant enzyme needed for growth under endogenously generated superoxide stress. sod15, on the other hand, may or may not actually function as a SOD in vivo, but its presence in the endospore, its conservation in the B. cereus group, and its unusual transcriptional pattern as part of an operon putatively involved in cell morphology make this an intriguing target for future biochemical characterization. The role of sodA2, which has been functionally isolated from both endospores and vegetative cells and which most likely forms an active heterodimer with sodA1, may serve a minor role in cellular antioxidant protection that is not apparent in single deletion strains. The removal of sodC did not evoke an obvious phenotype in any of the assays performed here, even though this gene is actively transcribed in wild-type cells. B. anthracis encodes multiple antioxidant enzyme systems, such as catalases and peroxidases, underlining the complex defense strategies that this important pathogen has evolved to cope with variably generated oxidative stresses.
TABLE 3.
Disk diffusion assay of sensitivity to superoxide-generating compounds (with 6-mm paper disks)a
| Compound | Zone of inhibition (diam) in mm ± SD
|
||||
|---|---|---|---|---|---|
| Sterne | Δsod15 mutant | ΔsodC mutant | ΔsodA1 mutant | ΔsodA2 mutant | |
| Spores | |||||
| 5% Methyl viologen | 31.1 ± 0.7 | 31.6 ± 0.5 | 30.8 ± 0.4 | 42.6 ± 0.8* | 31.0 ± 1.1 |
| 80 mM menadione | 31.4 ± 0.7 | 30.6 ± 0.5 | 30.4 ± 0.5 | 49.1 ± 1.0* | 31.6 ± 0.7 |
| Bacilli | |||||
| 5% Methyl viologen | 27.1 ± 0.6 | 27.5 ± 0.7 | 26.9 ± 0.6 | 38.3 ± 1.6* | 27.4 ± 0.5 |
| 80 mM menadione | 24.4 ± 0.5 | 24.0 ± 0.5 | 24.8 ± 0.4 | 32.6 ± 1.1* | 26.1 ± 0.6 |
| Sterne + pBJ258 | ΔsodA1 + pBJ258 | ΔsodA1(A1) + sodA1 | ΔsodA2 + pBJ258 | ΔsodA2(A2) + sodA2 | |
| Bacilli (complementation strains) | |||||
| 5% Methyl viologen | 24.4 ± 1.0 | 56.7 ± 1.6 | 34.1 ± 1.0** | 26.1 ± 1.0 | 24.9 ± 1.2 |
✽, P ≤ 0.001 (different from wild type; two-tailed t test); ✽✽, P ≤ 0.001 (different from wild-type, original strain, and original strain with plasmid alone; two-tailed t test). n = 10 disks per compound (two disks per BHI plate). All H2O controls: zones = 0 mm.
Acknowledgments
Special thanks go to Brian Janes, Dotty Sorenson (University of Michigan Microscopy and Imaging Lab), Joe Washburn (University of Michigan cDNA Core), and Rod McDonald for all manner of technical assistance.
Funding for this project was provided in part by the NIH-funded Cellular Biotechnology Training Program at the University of Michigan. This study was also supported in part by HHS contract N266200400059C/N01-AI-40059, by NIH grant AI08649, and by the Great Lakes and the Southeast Regional Centers of Excellence for Biodefense and Emerging Infections.
REFERENCES
- 1.Banci, L., I. Bertini, V. Calderone, F. Cramaro, R. Del Conte, A. Fantoni, S. Mangani, A. Quattrone, and M. S. Viezzoli. 2005. A prokaryotic superoxide dismutase paralog lacking two Cu ligands: from largely unstructured in solution to ordered in the crystal. Proc. Natl. Acad. Sci. USA 102:7541-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battistoni, A. 2003. Role of prokaryotic Cu,Zn superoxide dismutase in pathogenesis. Biochem. Soc. Trans. 31:1326-1329. [DOI] [PubMed] [Google Scholar]
- 3.Beyer, W., J. Imlay, and I. Fridovich. 1991. Superoxide dismutases. Prog. Nucleic Acids Res. Mol. Biol. 40:221-253. [DOI] [PubMed] [Google Scholar]
- 4.Boucher, I. W. K. A., V. M. Levdikov, E. V. Blagova, M. J. Fogg, J. A. Brannigan, K. S. Wilson, and A. J. Wilkinson. 2005. Structures of two superoxide dismutases from Bacillus anthracis reveal a novel active center. Acta Crystallogr. Sect. F Struct. Biol. Crystal. Commun. 2005:621-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunstein, M., B. J. Espinosa, J. Chan, J. T. Belisle, and W. R. Jacobs, Jr. 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48:453-464. [DOI] [PubMed] [Google Scholar]
- 6.Brioukhanov, A. L., and A. I. Netrusov. 2004. Catalase and superoxide dismutase: distribution, properties, and physiological role in cells of strict anaerobes. Biochemistry 69:949-962. [DOI] [PubMed] [Google Scholar]
- 7.Casillas-Martinez, L., and P. Setlow. 1997. Alkyl hydroperoxide reductase, catalase, MrgA, and superoxide dismutase are not involved in resistance of Bacillus subtilis spores to heat or oxidizing agents. J. Bacteriol. 179:7420-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cendrowski, S., W. MacArthur, and P. Hanna. 2004. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol. Microbiol. 51:407-417. [DOI] [PubMed] [Google Scholar]
- 9.De Groote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon, T. C., M. Meselson, J. Guillemin, and P. C. Hanna. 1999. Anthrax. N. Engl. J. Med. 341:815-826. [DOI] [PubMed] [Google Scholar]
- 11.Drysdale, M., S. Heninger, J. Hutt, Y. Chen, C. R. Lyons, and T. M. Koehler. 2005. Capsule synthesis by Bacillus anthracis is required for dissemination in murine inhalation anthrax. EMBO J. 24:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dussurget, O., G. Stewart, O. Neyrolles, P. Pescher, D. Young, and G. Marchal. 2001. Role of Mycobacterium tuberculosis copper-zinc superoxide dismutase. Infect. Immun. 69:529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberhardt, M. K. 2001. Reactive oxygen metabolites: chemistry and medical consequences. CRC Press, Boca Raton, Fla.
- 14.Edwards, K. M., M. H. Cynamon, R. K. Voladri, C. C. Hager, M. S. DeStefano, K. T. Tham, D. L. Lakey, M. R. Bochan, and D. S. Kernodle. 2001. Iron-cofactored superoxide dismutase inhibits host responses to Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 164:2213-2219. [DOI] [PubMed] [Google Scholar]
- 15.Fang, F. C., M. A. DeGroote, J. W. Foster, A. J. Baumler, U. Ochsner, T. Testerman, S. Bearson, J. C. Giard, Y. Xu, G. Campbell, and T. Laessig. 1999. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc. Natl. Acad. Sci. USA 96:7502-7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrant, J. L., A. Sansone, J. R. Canvin, M. J. Pallen, P. R. Langford, T. S. Wallis, G. Dougan, and J. S. Kroll. 1997. Bacterial copper- and zinc-cofactored superoxide dismutase contributes to the pathogenesis of systemic salmonellosis. Mol. Microbiol. 25:785-796. [DOI] [PubMed] [Google Scholar]
- 17.Fisher, N., and P. Hanna. 2005. Characterization of Bacillus anthracis germinant receptors in vitro. J. Bacteriol. 187:8055-8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franzon, V. L., J. Arondel, and P. J. Sansonetti. 1990. Contribution of superoxide dismutase and catalase activities to Shigella flexneri pathogenesis. Infect. Immun. 58:529-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frealle, E., C. Noel, E. Viscogliosi, D. Camus, E. Dei-Cas, and L. Delhaes. 2005. Manganese superoxide dismutase in pathogenic fungi: an issue with pathophysiological and phylogenetic involvements. FEMS Immunol. Med. Microbiol. 45:411-422. [DOI] [PubMed] [Google Scholar]
- 20.Guarner, J., J. A. Jernigan, W. J. Shieh, K. Tatti, L. M. Flannagan, D. S. Stephens, T. Popovic, D. A. Ashford, B. A. Perkins, and S. R. Zaki. 2003. Pathology and pathogenesis of bioterrorism-related inhalational anthrax. Am. J. Pathol. 163:701-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 22.Gusarov, I., and E. Nudler. 2005. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc. Natl. Acad. Sci. USA 102:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henriques, A. O., L. R. Melsen, and C. P. Moran, Jr. 1998. Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J. Bacteriol. 180:2285-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heyworth, P. G., A. R. Cross, and J. T. Curnutte. 2003. Chronic granulomatous disease. Curr. Opin. Immunol. 15:578-584. [DOI] [PubMed] [Google Scholar]
- 25.Imlay, J. 2006. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59:1073-1082. [DOI] [PubMed] [Google Scholar]
- 26.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 27.Inaoka, T., Y. Matsumura, and T. Tsuchido. 1998. Molecular cloning and nucleotide sequence of the superoxide dismutase gene and characterization of its product from Bacillus subtilis. J. Bacteriol. 180:3697-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson, S. M., and J. B. Cooper. 1998. An analysis of structural similarity in the iron and manganese superoxide dismutases based on known structures and sequences. Biometals 11:159-173. [DOI] [PubMed] [Google Scholar]
- 29.Keyer, K., and J. A. Imlay. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. USA 93:13635-13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kroll, J. S., P. R. Langford, K. E. Wilks, and A. D. Keil. 1995. Bacterial [Cu,Zn]-superoxide dismutase: phylogenetically distinct from the eukaryotic enzyme, and not so rare after all! Microbiology 141(Pt. 9):2271-2279. [DOI] [PubMed] [Google Scholar]
- 31.Lai, E. M., N. D. Phadke, M. T. Kachman, R. Giorno, S. Vazquez, J. A. Vazquez, J. R. Maddock, and A. Driks. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 185:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larios, J. M., R. Budhiraja, B. L. Fanburg, and V. J. Thannickal. 2001. Oxidative protein cross-linking reactions involving l-tyrosine in transforming growth factor-β1-stimulated fibroblasts. J. Biol. Chem. 276:17437-17441. [DOI] [PubMed] [Google Scholar]
- 33.Liu, H., N. H. Bergman, B. Thomason, S. Shallom, A. Hazen, J. Crossno, D. A. Rasko, J. Ravel, T. D. Read, S. N. Peterson, J. Yates III, and P. C. Hanna. 2004. Formation and composition of the Bacillus anthracis endospore. J. Bacteriol. 186:164-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch, M., and H. Kuramitsu. 2000. Expression and role of superoxide dismutases (SOD) in pathogenic bacteria. Microbes Infect. 2:1245-1255. [DOI] [PubMed] [Google Scholar]
- 35.Mayer-Scholl, A., R. Hurwitz, V. Brinkmann, M. Schmid, P. Jungblut, Y. Weinrauch, and A. Zychlinsky. 2005. Human neutrophils kill Bacillus anthracis. PLoS Pathog. 1:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCord, J. M., and I. Fridovich. 1969. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244:6049-6055. [PubMed] [Google Scholar]
- 37.Munoz-Montesino, C., E. Andrews, R. Rivers, A. Gonzalez-Smith, G. Moraga-Cid, H. Folch, S. Cespedes, and A. A. Onate. 2004. Intraspleen delivery of a DNA vaccine coding for superoxide dismutase (SOD) of Brucella abortus induces SOD-specific CD4+ and CD8+ T cells. Infect. Immun. 72:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nozik-Grayck, E., H. B. Suliman, and C. A. Piantadosi. 2005. Extracellular superoxide dismutase. Int. J. Biochem. Cell. Biol. 37:2466-2471. [DOI] [PubMed] [Google Scholar]
- 39.Parker, M. W., and C. C. Blake. 1988. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 229:377-382. [DOI] [PubMed] [Google Scholar]
- 40.Parker, M. W., C. C. Blake, D. Barra, F. Bossa, M. E. Schinina, W. H. Bannister, and J. V. Bannister. 1987. Structural identity between the iron- and manganese-containing superoxide dismutases. Protein Eng. 1:393-397. [DOI] [PubMed] [Google Scholar]
- 41.Piddington, D. L., F. C. Fang, T. Laessig, A. M. Cooper, I. M. Orme, and N. A. Buchmeier. 2001. Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect. Immun. 69:4980-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollock, J. D., D. A. Williams, M. A. Gifford, L. L. Li, X. Du, J. Fisherman, S. H. Orkin, C. M. Doerschuk, and M. C. Dinauer. 1995. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat. Genet. 9:202-209. [DOI] [PubMed] [Google Scholar]
- 43.Popham, D. L., B. Illades-Aguiar, and P. Setlow. 1995. The Bacillus subtilis dacB gene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration. J. Bacteriol. 177:4721-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poyart, C., E. Pellegrini, O. Gaillot, C. Boumaila, M. Baptista, and P. Trieu-Cuot. 2001. Contribution of Mn-cofactored superoxide dismutase (SodA) to the virulence of Streptococcus agalactiae. Infect. Immun. 69:5098-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinn, C. P., and B. N. Dancer. 1990. Transformation of vegetative cells of Bacillus anthracis with plasmid DNA. J. Gen. Microbiol. 136:1211-1215. [DOI] [PubMed] [Google Scholar]
- 46.Rainey, G. J., D. J. Wigelsworth, P. L. Ryan, H. M. Scobie, R. J. Collier, and J. A. Young. 2005. Receptor-specific requirements for anthrax toxin delivery into cells. Proc. Natl. Acad. Sci. USA 102:13278-13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 48.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 49.Reeves, E. P., H. Lu, H. L. Jacobs, C. G. Messina, S. Bolsover, G. Gabella, E. O. Potma, A. Warley, J. Roes, and A. W. Segal. 2002. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 416:291-297. [DOI] [PubMed] [Google Scholar]
- 50.Reeves, E. P., M. Nagl, J. Godovac-Zimmermann, and A. W. Segal. 2003. Reassessment of the microbicidal activity of reactive oxygen species and hypochlorous acid with reference to the phagocytic vacuole of the neutrophil granulocyte. J. Med. Microbiol. 52:643-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruthel, G., W. J. Ribot, S. Bavari, and T. A. Hoover. 2004. Time-lapse confocal imaging of development of Bacillus anthracis in macrophages. J. Infect. Dis. 189:1313-1316. [DOI] [PubMed] [Google Scholar]
- 52.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 53.Spagnolo, L., I. Toro, M. D'Orazio, P. O'Neill, J. Z. Pedersen, O. Carugo, G. Rotilio, A. Battistoni, and K. Djinovic-Carugo. 2004. Unique features of the sodC-encoded superoxide dismutase from Mycobacterium tuberculosis, a fully functional copper-containing enzyme lacking zinc in the active site. J. Biol. Chem. 279:33447-33455. [DOI] [PubMed] [Google Scholar]
- 54.Storz, G., and R. Hengge-Aronis. 2000. Bacterial stress responses. ASM Press, Washington, D.C.
- 55.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 56.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 57.Sutton, V. R., A. Stubna, T. Patschkowski, E. Munck, H. Beinert, and P. J. Kiley. 2004. Superoxide destroys the [2Fe-2S]2+ cluster of FNR from Escherichia coli. Biochemistry 43:791-798. [DOI] [PubMed] [Google Scholar]
- 58.Thannickal, V. J. 2003. The paradox of reactive oxygen species: injury, signaling, or both? Am. J. Physiol. Lung Cell Mol. Physiol. 284:L24-L25. [DOI] [PubMed] [Google Scholar]
- 59.Tsolis, R. M., A. J. Baumler, and F. Heffron. 1995. Role of Salmonella typhimurium Mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect. Immun. 63:1739-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]