Abstract
When grown in divalent cation-limited medium, Pseudomonas aeruginosa becomes resistant to cationic antimicrobial peptides and polymyxin B. This resistance is regulated by the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems. To further characterize Mg2+ regulation in P. aeruginosa, microarray transcriptional profiling was conducted to compare wild-type P. aeruginosa grown under Mg2+-limited and Mg2+-replete conditions to isogenic phoP and pmrA mutants grown under Mg2+-limited conditions. Under Mg2+-limited conditions (0.02 mM Mg2+), approximately 3% of the P. aeruginosa genes were differentially expressed compared to the expression in bacteria grown under Mg2+-replete conditions (2 mM Mg2+). Only a modest subset of the Mg2+-regulated genes were regulated through either PhoP or PmrA. To determine which genes were directly regulated, a bioinformatic search for conserved binding motifs was combined with confirmatory reverse transcriptase PCR and gel shift promoter binding assays, and the results indicated that very few genes were directly regulated by these response regulators. It was found that in addition to the previously known oprH-phoP-phoQ operon and the pmrHFIJKLM-ugd operon, the PA0921 and PA1343 genes, encoding small basic proteins, were regulated by Mg2+ in a PhoP-dependent manner. The number of known PmrA-regulated genes was expanded to include the PA1559-PA1560, PA4782-PA4781, and feoAB operons, in addition to the previously known PA4773-PA4775-pmrAB and pmrHFIJKLM-ugd operons.
Pseudomonas aeruginosa is an important opportunistic pathogen that is capable of infecting a large number of hosts, including nematodes, insects, plants, animals, and especially humans. It is the third-leading cause of nosocomial infections and is also the leading cause of morbidity and mortality in cystic fibrosis (CF) patients (33). P. aeruginosa is also noted for its metabolic diversity, which allows it to colonize a large number of environmental habitats. The versatility of this organism is believed to be related to the large number of regulatory proteins found in its genome (469 of 5,570 open reading frames) (43).
The two-component response regulators constitute one of the larger families of regulatory proteins in P. aeruginosa (43). These systems typically contain a sensor protein that responds to some chemical or physical stimulus, which leads to phosphorylation of the sensor protein at a conserved histidine residue, thus altering the conformation of the sensor and promoting phosphotransfer to a cognate response regulator protein (9). The phosphorylated response regulator then recognizes and binds to a specific DNA sequence, leading to modulation of transcription from that promoter. Although this is often the mechanism, regulation may also occur through phosphatase activity of the sensor kinase with the response regulator (37) or through integration into the signaling cascade of multiple signals from other proteins (32). In P. aeruginosa, there are 64 response regulators and 63 histidine kinases, as well as 16 atypical kinases (36). The functions of the majority of these regulatory proteins have not been established yet.
In P. aeruginosa, two separate two-component regulatory systems, PmrA-PmrB (26) and PhoP-PhoQ (21), are known to respond to the presence of limiting concentrations of Mg2+ and to separately regulate certain operons. The PhoPQ system autoregulates the oprH-phoP-phoQ operon (21) under Mg2+-limiting growth conditions and is also involved in resistance to cationic antimicrobial peptides and polymyxin B and in virulence, as phoQ mutants exhibit increased resistance to cationic antimicrobial peptides and polymyxin B and have reduced virulence (20). Similarly, the PmrAB system regulates resistance to cationic antimicrobial peptides and polymyxin B in response to limiting concentrations of Mg2+. This regulatory redundancy occurs because both the PhoPQ and PmrAB systems separately contribute to regulation of the pmrHFIJKLM-ugd operon (PA3552 to PA3559) in response to limiting concentrations of Mg2+ (26). The PmrAB system also autoregulates the PA4773-PA4775-pmrAB-PA4778 operon under Mg2+-limiting conditions, and there is evidence that the first three genes in this operon, PA4773 to PA4775, also contribute to cationic antimicrobial peptide and polymyxin B resistance (26). Previous work has established that within the promoters controlled by PhoPQ- and PmrAB-containing operons there are related but distinct putative binding motifs that have been implicated in autoregulation (21, 26).
Because of the importance of Mg2+ regulation via the PhoP-PhoQ and PmrA-PmrB genes to virulence and cationic antimicrobial peptide resistance in P. aeruginosa, a transcriptional analysis of P. aeruginosa PAO1 under Mg2+-deficient conditions was performed. Mutants with mutations in PhoP and PmrA were also examined to determine the contributions of each of these regulatory systems to regulation by limiting concentrations of Mg2+. In addition to this transcriptional profiling, in silico searches were carried out to identify promoters that contain the previously identified PhoP and/or PmrA regulatory motifs. This analysis was conducted independent of the transcriptional profiling to allow a comparison of these different approaches for identifying and characterizing targets of the regulatory proteins. Using both techniques, several novel PhoP- and PmrA-regulated genes were identified and characterized phenotypically.
MATERIALS AND METHODS
Bacterial strains, primers, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. The sequences of DNA primers used in this study are available by request from us. All primers were synthesized by AlphaDNA (Montreal, QC, Canada). Cultures were routinely grown in Luria-Bertani broth or BM2-glucose minimal medium containing a low MgSO4 concentration (20 μM) or a high MgSO4 concentration (2 mM) (11). Antibiotics for selection were used at the following concentrations: tetracycline, 100 μg/ml; and gentamicin, 50 μg/ml. Routine genetic manipulations were carried out by using standard molecular biology procedures (23).
TABLE 1.
P. aeruginosa strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristics | Reference |
|---|---|---|
| H103 | Wild-type P. aeruginosa PAO1 | 21 |
| H851 | phoP::xylE-aacC1; Gmr | 21 |
| H974 | PA4773::luxCDABE derivative of H103; Tcr | 26 |
| H988 | pmrA::xylE-aacC1; Gmr | 26 |
| H993 | pmrH::xylE-aacC1; Gmr | This study |
| H1034 | feoA::luxCDABE derivative of H103 | 19a |
| pphoP-His6 | His6-phoP cloned into pET28a | This study |
| ppmrA-His6 | His6-pmrA cloned into pET28a | This study |
RNA extraction, cDNA synthesis, and hybridization to DNA microarrays.
For each condition microarray experiments were performed with five independent cultures. P. aeruginosa PAO1 was grown for 18 h in BM2-glucose medium supplemented with either 2 mM MgSO4 (high concentration) or 20 μM MgSO4 (low concentration) in acid-washed glassware. Cultures were diluted 1/100 into fresh media, and cells were harvested at the mid-log phase (optical density at 600 nm [OD600] for the high Mg2+ concentration, 0.5 to 0.6; OD600 for the low Mg2+ concentration, 0.3 to 0.4). RNA was isolated using a QIAGEN RNeasy midi RNA isolation kit according to the manufacturer's protocols (QIAGEN Inc., Canada). Contaminating genomic DNA was removed by treatment with a DNA-free kit (Ambion Inc., Austin, TX). RNA was stored at −80°C with 0.2 U/μl of SUPERase-In RNase inhibitor (Ambion Inc., Austin, TX). RNA quality was assessed by agarose gel electrophoresis and spectrophotometrically.
P. aeruginosa PAO1 microarray slides were provided by The Institute for Genomic Research (TIGR) Pathogenic Functional Genomics Resource Center (http://pfgrc.tigr.org/). Ten micrograms of total RNA was treated using a Microbe Express kit with the Pseudomonas module (Ambion) to remove rRNA. Two micrograms of cDNA was reverse transcribed using random hexamers (Invitrogen Canada Inc., Burlington, ON, Canada), deoxynucleoside triphosphates (Invitrogen), dUTP (Ambion), and Superscript II reverse transcriptase (Invitrogen) with a PTC-225 Peltier thermal cycler (MJ Research). cDNA labeling, purification, and analysis of the labeling reaction mixture were performed by using the TIGR “microbial RNA aminoallyl labeling” protocol (pfgrc.tigr.org/protocols/M007.pdf). Treated cDNA was labeled with cyanine-5 (GE Healthcare Canada), and control cDNA was labeled with cyanine-3 (GE Healthcare Canada). Labeled cDNA was hybridized by using the TIGR “hybridization of labeled DNA probes” protocol (pfgrc.tigr.org/protocols/M008.pdf). Briefly, 200 pmol of each cyanine-labeled cDNA was combined and hybridized to the array slides for 18 h in a solution containing 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate, and 0.6 μg/μl salmon sperm DNA at 42°C in chambers in which the humidity was maintained at 100%. Following hybridization, the slides were washed, dried, and scanned using a ScanArray Express scanner and software (Packard BioScience BioChip Technologies).
Analysis of DNA microarrays.
Images from microarray slides were quantified using the ImaGene 6.0 Standard Edition software (BioDiscovery, Inc., El Segundo, CA). Assessment of slide quality, normalization, detection of differential gene expression, and statistical analysis were carried out with ArrayPipe (version 1.7), which is web-based, semiautomated software specifically designed for processing of microarray data (http://koch.pathogenomics.ca/cgi-bin/pub/arraypipe.pl) (16), using genome annotation from the Pseudomonas genome database (51). The following processing steps were performed: (i) flagging of markers and control spots; (ii) subgrid-wise background correction, using the median of the lower 10% foreground intensity as the foreground intensity as an estimate for the background noise; (iii) data shifting, to rescue most of the negative spots; (iv) printTip LOESS normalization; (v) merging of replicate spots; (vi) two-sided one-sample Student's t test for the log2 ratios within each experiment; and (vii) averaging of biological replicates to obtain overall fold changes for each treatment group. Only genes that exhibited a change compared to the control of twofold or more and a P value of ≤0.05 were considered in this study.
Identification of PhoP and PmrA binding sites.
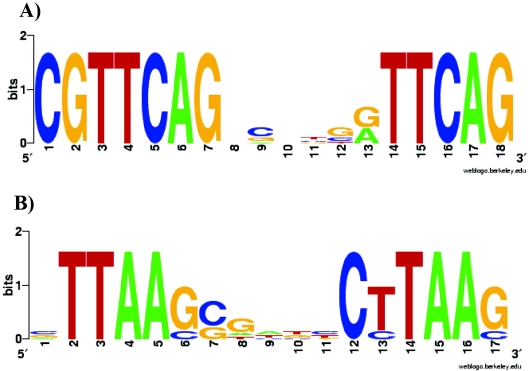
Sequence logos for the proposed PhoP binding sites upstream of the oprH-phoPQ and pmrH-ugd operons, as well as the proposed PmrA binding sites upstream of the PA4773-pmrB and pmrH-ugd operons, were first constructed using Weblogo (weblogo.berkeley.edu/logo.cgi) (5) (Fig. 1). Frequency matrices were then constructed using the Emboss program “Prophecy” (emboss.sourceforge.net/apps/prophecy.html). These matrices were used to parse the P. aeruginosa genome for additional matching binding sites, using a conservative cutoff score of 0.82 to 0.85. Any sites that were in a promoter between convergent genes were excluded from further analysis based on the likelihood that they did not represent real promoters. Regulation of the putative target genes was then examined by semiquantitative PCR (qPCR) for strains H103 (wild type), H851 (ΔphoP), and H988 (ΔpmrA) in response to a limiting concentration of Mg2+. Genes identified as genes that were regulated by PhoP and/or PmrA were then used to refine the binding motif and carry out a second iteration of frequency matrix construction and genomic searches for matching motifs.
FIG. 1.
Weblogos generated from the conserved sequences identified in the promoters of the PhoP- and PmrA-regulated genes. (A) PhoP weblogo. (B) PmrA weblogo.
His6-PhoP and His6-PmrA purification.
The constructs used to overexpress and purify His6-PmrA and His6-PhoP were created by PCR amplifying the phoP or pmrA genes and cloning them separately into pET28a (Invitrogen, Carlsbad, CA) as NdeI-BamHI fragments. The plasmids containing the His6-phoP or His6-pmrA genes were transformed into Escherichia coli BL21. Cells were grown to an OD600 of 0.5 before they were induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h. Cells were harvested and resuspended in sonication buffer containing 500 mM NaCl, 5 mM MgCl2, 50 mM sodium phosphate buffer (pH 7.8), and 10 mM imidazole. Cells were lysed by sonication on ice (three 1-min treatments). Cell debris and unbroken cells were removed by centrifugation at 7,500 × g. The supernatant was filtered through a 0.8-μm filter (Nalgene, Rochester, NY). The filtered supernatant was mixed with Ni2+-nitrilotriacetic acid resin (QIAGEN, Mississauga, ON, Canada) and gently shaken at 23°C for 1 h. The resin was washed sequentially with 5-ml portions of sonication buffer containing 30, 50, 100, and 200 mM imidazole. His6-phoP was eluted with sonication buffer containing 300 mM imidazole and 15% glycerol. Protein was collected on ice, aliquoted, and frozen in a dry ice-ethanol bath. Frozen aliquots were stored at −80°C.
qPCR assays.
Using RNeasy mini columns (QIAGEN, Mississauga, ON, Canada), total RNA was isolated from mid-log-phase P. aeruginosa cultures grown in BM2-glucose minimal medium with 20 μM Mg2+ or 2 mM Mg2+. RNA samples were treated with DNase I (Invitrogen, Carlsbad, CA) to remove contaminating genomic DNA. Four micrograms of total RNA was added to a reaction mixture containing each deoxynucleoside triphosphate at a concentration of 0.5 μM, 500 U/ml SuperaseIN (Ambion, Austin, TX), and 10 μM dithiothreitol in 1× reaction buffer and reverse transcribed for 1 h at 37°C and for 2 h at 42°C with 10,000 U/ml Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). The RNA was subsequently destroyed by addition of 170 mM NaOH and incubation at 65°C for 10 min. The reaction mixture was then neutralized by addition of HCl, and the cDNA was used as a template for PCR. The number of cycles used to amplify each gene of interest was chosen to ensure that the PCR was not saturated. All reactions were normalized to the rpsL gene encoding the 30S ribosomal protein S12.
DNA binding assays.
DNA binding assays were performed by using previously described methods (15). PCR-amplified promoter fragments of the entire intergenic region of the genes of interest were purified by excision from agarose gels using the Qiaquick column purification system (QIAGEN, Mississauga, ON, Canada). These fragments were labeled with digoxigenin (DIG) using a DIG gel shift kit (second generation) obtained from Roche Applied Science. The purified protein (His6-PhoP or His6-PmrA) was mixed with 40 pg of DIG-labeled probe in buffer consisting of 20 mM HEPES, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM dithiothreitol, 0.2% Tween 20, 30 mM KCl, and 0.5 mM MgCl2. For samples containing unlabeled probe, 100 ng of the probe was allowed to bind for 15 min at 20°C before addition of DIG-labeled probe and incubation for another 20 min. All other samples were incubated for 20 min at 20°C before electrophoresis. Following electrophoresis, the samples were blotted onto nylon membranes and exposed to film overnight at 23°C.
Growth curves.
Overnight cultures of P. aeruginosa PAO1 strains H103 (wild type) and H1034 (feoA::luxCDABE) were grown in HEPES-buffered minimal medium containing 20 mM glucose, 2 mM MgSO4, and 10 μM FeSO4. These cultures were washed in sterile saline (0.85% NaCl) and diluted to obtain an OD600 of 0.2 in saline. Five-microliter portions of these cultures were added to 200 μl of minimal medium containing a high concentration of MgSO4 (2 mM) or a low concentration of MgSO4 (20 μM) and 10 μM FeSO4 or FeCl3. Medium containing FeSO4 was also supplemented with 1 mM sodium ascorbate to maintain the iron in the ferrous form and with 300 μM desferrioxamine mesylate to chelate any contaminating ferric ions. Similarly, medium containing ferric iron was supplemented with 300 μM 2,2′-dipyridyl to chelate ferrous ions. The growth of the cultures at 37°C was monitored with a TECAN Spectrofluor Plus by determining the A620 every 20 min for 36 h. Growth experiments were carried out twice, and eight replicates were used in each experiment.
RESULTS
Microarray analysis of regulation by Mg2+ limitation, PhoP, and PmrA.
To determine the subsets of Mg2+-regulated genes that were regulated by PhoP and/or PmrA, microarray analyses were performed. For these studies we utilized quite stringent criteria, and the results presented below reflect only those genes that exhibited significantly altered expression (P values of <0.05 as determined by Student's t test) in five separate experiments. Growth of P. aeruginosa in BM2-glucose medium supplemented with either 20 μM Mg2+ (low concentration) or 2 mM Mg2+ (high concentration) led to a twofold or greater change in the expression of 158 genes (Table 2), which represented approximately 3% of the coding capacity of the genome.
TABLE 2.
Selected P. aeruginosa genes that are regulated by growth in the presence of a limiting concentration of Mg2+ in wild-type (H103), phoP::xylE (H851), and pmrA::xylE (H988) strains sorted by categorya
| PA no. | Gene designation | Comparison of growth
|
|||||
|---|---|---|---|---|---|---|---|
| H103 with low Mg2+ concn vs H103 with high Mg2+ concn
|
H851 with low Mg2+ concn vs H103 with high Mg2+ concn
|
H988 with low Mg2+ concn vs H103 with high Mg2+ concn
|
|||||
| Fold change | P value | Fold change | P value | Fold change | P value | ||
| Positively Mg2+ regulated | |||||||
| PA0762b | algU | 4.1 | 0.04 | 1.9 | <0.01 | 2.6 | <0.01 |
| PA0763b | mucA | 3.4 | <0.01 | 2.3 | 0.08 | 1.8 | 0.05 |
| PA0764b | mucB | 2.5 | <0.01 | 1.9 | 0.01 | 4.2 | 0.02 |
| PA1297 | 3.3 | <0.01 | 3.1 | <0.01 | 3.0 | <0.01 | |
| PA3552 | pmrH | 7.2 | <0.01 | 5.6 | <0.01 | 6.9 | <0.01 |
| PA3553 | pmrF | 12.5 | <0.01 | 5.3 | 0.05 | 7.6 | 0.05 |
| PA3554 | pmrI | 11.9 | <0.01 | 10.4 | <0.01 | 11.6 | <0.01 |
| PA3555 | pmrJ | 3.8 | <0.01 | 3.0 | <0.01 | 3.7 | <0.01 |
| PA3556 | pmrK, arnT | 10.6 | <0.01 | 9.6 | <0.01 | 7.8 | <0.01 |
| PA3557 | pmrL | 3.7 | <0.01 | 1.6 | 0.63 | −1.3 | 0.37 |
| PA3558 | pmrM | 3.6 | 0.01 | 2.0 | <0.01 | 1.8 | 0.01 |
| PA3559 | ugd | 8.0 | <0.01 | 5.3 | <0.01 | 3.9 | <0.01 |
| PA3920 | 5.4 | <0.01 | 8.6 | 0.01 | 6.2 | <0.01 | |
| Negatively Mg2+ regulated | |||||||
| PA2260 | kguE | −3.3 | 0.03 | −1.9 | <0.01 | −2.9 | <0.01 |
| PA2261 | kguK | −5.4 | <0.01 | −6.7 | <0.01 | −7.8 | <0.01 |
| PA2262 | kguT | −3.4 | 0.01 | −2.6 | <0.01 | −3.3 | <0.01 |
| PA2265 | gad | −2.8 | <0.01 | −2.2 | 0.04 | −2.1 | 0.01 |
| PA2266 | −2.7 | <0.01 | −2.3 | <0.01 | −2.6 | <0.01 | |
| PA2321 | −5.2 | 0.01 | −3.2 | <0.01 | −4.2 | <0.01 | |
| PA2322 | gntT | −16.9 | <0.01 | −16.4 | <0.01 | −9.5 | <0.01 |
| PA2323 | −11.1 | <0.01 | −9.2 | <0.01 | −10.7 | <0.01 | |
| PA4236 | katA | −2.5 | 0.05 | −2.7 | <0.01 | −3.1 | <0.01 |
| PA4463 | −6.6 | <0.01 | −4.7 | 0.01 | −4.6 | <0.01 | |
| PA4761 | dnaK | −2.9 | 0.02 | −2.8 | 0.03 | −1.8 | <0.01 |
| PA4835 | −2.2 | 0.01 | −2.4 | <0.01 | −2.4 | <0.01 | |
| PA4836 | −2.1 | 0.02 | −4.6 | <0.01 | −3.0 | <0.01 | |
| Positively Mg2+ and positively PhoP regulated | |||||||
| PA0265 | gabD | 5.2 | <0.01 | 1.4 | 0.06 | 4.0 | 0.01 |
| PA0266 | gabT | 5.1 | <0.01 | 1.2 | 0.09 | 4.0 | 0.05 |
| PA0921 | 2.2 | <0.01 | −1.1 | 0.57 | 2.6 | <0.01 | |
| PA1178 | oprH | 7.6 | <0.01 | −1.8 | 0.01 | 18.4 | 0.03 |
| PA1179b | phoP | 19.8 | <0.01 | −1.2 | 0.29 | 39.8 | <0.01 |
| PA1180b | phoQ | 5.9 | <0.01 | −1.2 | 0.09 | 8.0 | <0.01 |
| PA3522 | mexQ | 16.4 | <0.01 | 3.1 | <0.01 | 7.9 | <0.01 |
| PA3649 | 2.5 | <0.01 | −1.8 | 0.80 | 2.3 | <0.01 | |
| PA3885b | 2.7 | 0.05 | −1.5 | 0.05 | 1.4 | <0.01 | |
| PA4010 | 7.0 | <0.01 | −1.1 | 0.08 | 4.7 | <0.01 | |
| PA4011 | 2.0 | 0.01 | −1.1 | 0.47 | 2.3 | <0.01 | |
| PA4453 | 2.8 | <0.01 | −1.3 | 0.04 | 4.9 | <0.01 | |
| PA4454 | 2.2 | <0.01 | −1.3 | 0.07 | 3.1 | <0.01 | |
| PA4455 | 2.6 | <0.01 | −1.2 | 0.08 | 1.8 | <0.01 | |
| Negatively Mg2+ and negatively PhoP regulated | |||||||
| PA0836b | ack | −4.0 | <0.01 | −1.5 | 0.04 | −5.7 | <0.01 |
| PA1196b | −3.1 | <0.01 | −1.3 | 0.07 | −4.2 | <0.01 | |
| PA3309 | −3.2 | <0.01 | 1.6 | 0.14 | −8.6 | <0.01 | |
| PA4918 | −2.5 | 0.02 | −1.4 | 0.85 | −2.8 | <0.01 | |
| Not Mg2+ regulated and positively PhoP regulated | |||||||
| PA4366 | sodB | −1.3 | 0.26 | −5.2 | <0.01 | −1.1 | 0.49 |
| Positively Mg2+ regulated and positively PmrA regulated | |||||||
| PA0201 | 2.6 | 0.03 | 2.8 | <0.01 | −2.4 | 0.08 | |
| PA0202 | 2.2 | <0.01 | 1.1 | 0.12 | −1.2 | 0.26 | |
| PA0282 | cysT | 3.6 | 0.01 | 2.7 | <0.01 | −1.2 | 0.02 |
| PA0546 | metK | 2.7 | <0.01 | 1.8 | 0.06 | −1.0 | 0.60 |
| PA0913 | mgtE | 2.8 | 0.01 | 2.7 | <0.01 | −1.2 | 0.61 |
| PA1559 | 4.0 | 0.01 | 13.6 | <0.01 | 2.4 | <0.01 | |
| PA1560 | 3.2 | <0.01 | 8.0 | <0.01 | 2.9 | <0.01 | |
| PA2359b | 3.4 | 0.01 | 1.7 | <0.01 | 1.0 | 0.51 | |
| PA3446 | 2.3 | 0.02 | 2.8 | 0.01 | −1.6 | 0.06 | |
| PA4357 | 4.6 | 0.03 | 1.4 | 0.03 | −1.3 | 0.06 | |
| PA4358 | feoB | 3.3 | <0.01 | 5.4 | <0.01 | −1.7 | 0.03 |
| PA4359 | feoA | 4.0 | <0.01 | 2.2 | 0.10 | −2.3 | 0.01 |
| PA4773 | 4.1 | <0.01 | 3.2 | 0.04 | 1.0 | 0.45 | |
| PA4774 | 6.2 | 0.02 | 19.0 | <0.01 | 1.0 | 0.70 | |
| PA4775 | 1.0 | 0.22 | 1.1 | 0.08 | 1.0 | 0.25 | |
| PA4776b | pmrA | 3.3 | <0.01 | 4.5 | <0.01 | −1.3 | 0.05 |
| PA4777b | pmrB | 6.7 | <0.02 | 2.8 | <0.01 | −1.1 | 0.49 |
| PA4778b | 4.4 | <0.01 | 2.7 | <0.01 | 1.1 | 0.02 | |
| PA4781b | 2.2 | <0.01 | 2.0 | <0.01 | 1.1 | 0.67 | |
| PA4782 | 12.7 | <0.01 | 6.2 | <0.01 | −1.1 | 0.02 | |
| PA4822 | 7.7 | <0.01 | 6.9 | <0.01 | −1.9 | <0.01 | |
| PA4823 | 5.8 | <0.01 | 9.0 | <0.01 | −1.0 | 0.69 | |
| PA4824 | 8.1 | <0.01 | 13.3 | <0.01 | −1.1 | 0.02 | |
| PA4825 | mgtA | 10.0 | <0.01 | 30.5 | <0.01 | −2.4 | <0.01 |
| PA4826 | 5.2 | <0.01 | 3.6 | 0.02 | −1.4 | 0.03 | |
| Negatively Mg2+ regulated and positively PmrA regulated | |||||||
| PA0527b | dnr | −3.1 | 0.01 | −1.7 | 0.01 | −9.8 | <0.01 |
| Positively Mg2+ regulated and negatively PmrA regulated | |||||||
| PA2064 | pcoB | 3.2 | 0.01 | 2.0 | 0.04 | 4.4 | <0.01 |
| PA2065 | pcoA | 3.2 | 0.02 | 2.0 | 0.01 | 18.6 | <0.01 |
| PA3515 | 6.1 | <0.01 | 7.3 | 0.02 | 150.0 | <0.01 | |
| PA3516 | 3.6 | 0.01 | 1.3 | 0.16 | 5.9 | 0.02 | |
| PA3517 | 13.7 | <0.01 | 11.2 | <0.01 | 22.1 | <0.01 | |
| PA3518 | 8.0 | <0.01 | 7.2 | 0.01 | 55.5 | <0.01 | |
| Not Mg2+ regulated and negatively PmrA regulated | |||||||
| PA2274 | 1.2 | 0.18 | 1.5 | 0.21 | 5.4 | <0.01 | |
| PA4205 | mexG | 1.4 | 0.05 | 1.2 | 0.16 | 6.5 | <0.01 |
| PA4206 | mexH | 1.3 | 0.06 | 1.3 | 0.05 | 5.0 | <0.01 |
| PA4207 | mexI | 1.2 | 0.17 | 1.2 | 0.19 | 2.3 | <0.01 |
Only selected Mg2+ genes that were not affected by phoP::xylE or pmrA::xylE mutations are included. All results were standardized to expression of the wild-type strain in the fully repressed state (i.e., in the presence of 2 mM Mg2+).
Known or putative regulatory gene.
As expected, Mg2+ limitation induced the operons that encode PhoPQ and PmrAB, the two previously characterized (20, 26, 29) regulatory systems involved in resistance to cationic antimicrobial peptides and polymyxin B. The previously described pmrHFIJKLM-ugd operon, which is responsible for the addition of aminoarabinose to lipid A (29), was similarly upregulated (4- to 13-fold) with Mg2+ limitation, consistent with our previous observations (26).
Several genes involved in divalent cation uptake were upregulated under Mg2+ limitation conditions; these genes included PA0913 (mgtE) and the PA4826-PA4822 operon containing mgtA. The homologs of both mgtE and mgtA are involved in Mg2+ uptake in enteric organisms (24). In addition, the feoAB (PA4359-PA4357) operon, which has been proposed to be involved in ferrous iron uptake in other bacteria, was upregulated under Mg2+ limitation conditions (24), suggesting that divalent cation signaling has a potential role in iron uptake.
Interestingly, Mg2+-deficient growth conditions led to 2.5- to 4-fold upregulation of the algU-mucAB genes that encode the alternative sigma factor AlgU (AlgT) and its cognate anti-σ factor MucA (39), which combine to regulate alginate production, a common adaptation found in CF isolates of P. aeruginosa (25). This seems especially relevant since P. aeruginosa isolates from the lungs of CF patients, who suffer from chronic infection by mucoid Pseudomonas, have lipid A modifications that are equivalent to those of cells grown under Mg2+ limitation conditions (7).
Two groups of genes with similarity to genes encoding gluconate and 2-ketogluconate uptake systems were strongly downregulated when organisms were grown under Mg2+-limiting conditions. PA2260 to PA2263 (kguEKTD) are required for utilization of 2-ketogluconate in P. aeruginosa (44). Additionally, PA2321 (gntK, a homologue of the gene encoding gluconokinase), PA2322 (gntT, a homologue of the gene encoding high-affinity gluconate permease), and PA2323 (encoding a predicted glyceraldehyde phosphate dehydrogenase) were also downregulated 5- to 16-fold. While this system has not been well characterized in P. aeruginosa, mutants of the high-affinity gluconate uptake system in E. coli exhibit dramatically longer doubling times when they are grown aerobically on gluconate (8).
As expected based on previous studies, a number of known PhoP-PhoQ- and PmrA-PmrB-regulated genes were observed to be upregulated in response to a limiting concentration of Mg2+, including the oprH-phoPQ, pmrHFIJKLM-ugd, and PA4773-PA4775-pmrAB-PA4778 operons. To determine which Mg2+-regulated genes were independently regulated by each system, microarray analyses were performed with both phoP and pmrA mutants under Mg2+-limiting conditions. These analyses indicated that each system was responsible for the regulation of a subset of the total genes induced under Mg2+-limiting conditions in P. aeruginosa, indicating that other, as-yet-unidentified regulatory systems may also be involved in the response of P. aeruginosa to divalent cation limitation. Consistent with this concept, in addition to the previously described two-component regulatory systems PhoP-PhoQ and PmrA-PmrB, several other regulatory proteins were observed to have modulated expression under Mg2+-limiting conditions (Table 2). Significantly, the genes encoding regulatory proteins that exhibited Mg2+-regulated expression included the alginate regulator genes algU and mucA described above, dnr, which encodes a regulator that responds to nitrogen oxides (35), the RocR-type (4) regulators PA1196 and PA2359, and a MerR-type regulator, PA4778. Another regulated operon contains the PA4782 and PA4781 genes. PA4782 encodes a small protein with an unidentified function and is just upstream of PA4781, an uncharacterized response regulator gene. The PA4781-encoded protein is a hybrid protein that contains a typical two-component response regulator receiver domain and a conserved HD-GYP output domain that in some proteins has been proposed to have phosphohydrolase activity and may be involved in cyclic di-GMP signaling (2, 10, 40). These observations indicate that there may be a complex regulatory hierarchy that influences adaptation to growth with a limiting concentration of Mg2+.
In order to determine which Mg2+-regulated genes are controlled via the PhoP protein, transcriptional profiling of a phoP::xylE mutant of P. aeruginosa under Mg2+-limiting conditions was carried out, and the results were compared to the results for an isogenic wild-type strain grown under Mg2+-replete conditions. This analysis revealed that the PhoP protein regulates only a modest number of genes in P. aeruginosa. Furthermore, the majority of genes (14/19) that were PhoP regulated appeared to be subject to positive regulation in an Mg2+-dependent manner, confirming that PhoP acts primarily as a transcriptional activator. The genes regulated included the previously identified oprH-phoPQ operon; three genes of a four-gene operon, the PA4456-PA4453 operon; the gabD and gabT genes, which are involved in aminobutyrate/putrescine degradation; and PA3885, which encodes a hypothetical serine/threonine phosphatase. Also observed were several hypothetical genes, including PA0921 and the two-gene PA4010-PA4011 operon.
In addition to these positively Mg2+- and PhoP-regulated genes, we also found four genes that appeared to be negatively regulated by PhoP. These genes included two regulatory genes, one encoding acetate kinase and one (PA1196) encoding a RocR-type regulator, as well as two hypothetical genes, PA3309 and PA4918. Finally, the expression of the sodB gene encoding the Fe2+-dependent superoxide dismutase was reduced fivefold when the phoP gene was deleted. This was somewhat unusual since this gene was the only PhoP-regulated gene that was not also Mg2+ regulated.
Transcriptional analysis of a pmrA::xylE mutant identified 36 genes with significantly altered expression. As observed for the phoP::xylE mutant, the majority (25/36) of these genes were positively regulated by both Mg2+ limitation and PmrA. These genes included a large number of genes involved in metal transport, such as the feoAB-PA4357 operon involved in ferrous iron uptake and the mgtE and PA4826-mgtA-PA4822 operons involved in Mg2+ transport. In addition to these transport genes, several regulatory proteins were observed, including the proteins encoded by the previously identified PA4773-PA4775-pmrAB operon (26), a RocR-type regulatory protein encoded by PA2659, and the protein encoded by PA4781, a hypothetical two-component response regulator with a CheY-like receiver domain and an HD-GYP motif-containing output domain. PA4778, encoding a MerR-like regulator, was also upregulated by, and may even be cotranscribed with, the pmrAB-containing operon. In addition, we observed that the dnr gene, which is involved in sensing nitrogen oxides, was also dysregulated in the pmrA::xylE mutant.
One interesting observation was the strong derepression of the PA3515-PA3518 operon observed in the pmrA::xylE mutant strain. The function of this operon is unknown, but the genes are similar to genes involved in aliphatic compound catabolism. These genes were also relatively mildly (4- to 16-fold) upregulated under Mg2+-limiting conditions. A similar pattern was observed for the pcoAB operon, which is involved in resistance to copper (17). Similar observations were made for the PA2274 and mexGHI genes, and loss of pmrA led to increased expression of these genes under Mg2+-limiting conditions. These results suggest that PmrA has a role in the control of these genes that is more complex than simply causing increased transcription.
Identification of conserved PhoP and PmrA binding motifs in the promoters of Mg2+-regulated genes.
It seems possible that the analysis of microarray data could have revealed indirectly regulated genes or even missed important directly regulated genes since stringent criteria were used for identification of alterations in gene expression. Therefore, we used an alternative bioinformatic approach to discover genes in the PhoPQ and PmrAB regulons. Putative DNA binding sites for the regulatory proteins PhoP and PmrA were identified in previous studies (21, 26). These sequences were used to generate frequency matrices with the Emboss program Prophecy. These matrices were then used to search the P. aeruginosa genome sequence for sequences with high levels of similarity to the PmrA or PhoP matrix. This motif searching was carried out independent of the microarray analysis in order to validate both approaches for identifying Mg2+-regulated genes. As new sequences were found and confirmed by qPCR, they were incorporated into the matrix to obtain a larger list of putatively regulated genes. A number of PmrA-like and PhoP-like sequences were identified, suggesting that these genes may be regulated by either of these systems. The promoters identified by this analysis are shown in Table 3 (PhoP) and Table 4 (PmrA). Some operons had single sites, while others had two sites; the pmrH-ugd promoter region was unique because it had two PhoP binding sites and two PmrA binding sites.
TABLE 3.
PhoP-like promoters identified by consensus sequence searching, patterns of regulation detected by qPCR, and results of gel shift assays
| Sequence or operon | Sequencea | Genome coordinates | Mg2+ regulatedb | PhoP regulatedb | Gel shift |
|---|---|---|---|---|---|
| Consensus sequence | CGTTCAGNNNNNRTTCAG | ||||
| oprH-phoP-phoQ | CGTTCAGCCCGGGTTCAG | 1276906-1276923 | Yes | Yes | Yesd |
| CGTTCAGGGGCGGTTCAG | 1276928-1276945 | ||||
| pmrH-ugd | CGTTCAGTCTTCATTCAG | 3979746-3979763 | Yes | Yes | Yes |
| CGTTCAGGCAGCATTCAG | 3979789-3979806 | ||||
| PA1343 | CGTTCAGAAATTGTTCAG | 1457051-1457068 | Yes | Yes | Yes |
| CGTTCAGGCCCGATTCAG | 1457073-1457109 | ||||
| PA0921 | CGTTCAGCGATGGTTCAG | 1006780-1006797 | Yes | Yes | Yes |
| PA4457-PA4461 | CGTTCAGCTTGGATTCAG | 4989024-4989041 | No | NTc | NT |
| PA1851 | CGTCCAGGCCTGTTTCAG | 2010955-2010972 | No | NT | NT |
| PA3925 | CGTTCAGACCCTATCCAG | 4400087-4400104 | No | NT | NT |
| PA0918 | CATTCAGGCTGGCTTCAG | 1002608-1002591 | No | NT | NT |
| PA2775-PA2774 | CCTTCACGGATGATTCAG | 3133288-3133271 | No | NT | NT |
| spuABCD | CGTTGAGGCCGTTTTCAG | 334565-334582 | No | NT | NT |
Nucleotides that differ from the nucleotides in the consensus sequence are indicated by boldface type. Abbreviations: N = A, C, G, or T; R = A or G.
Determined by reverse transcription-qPCR.
NT, not tested (no Mg2+ regulation could be demonstrated, and no regulation was observed in array experiments).
Promoter regions were tested for binding to PhoP and PmrA. Only the pmrH-ugd promoter fragment bound to both. The other promoters bound only to PhoP and not to PmrA.
TABLE 4.
PmrA-like promoters identified by consensus sequence searching
| Sequence or operon | Sequencea | Genome coordinates | Mg2+ regulatedb | PmrA regulatedb | Gel shift |
|---|---|---|---|---|---|
| Consensus sequence | NTTAASNNNNNCTTAAS | ||||
| pmrH-ugd | GTTAAGCGAGTCTTAAC | 3979769-3979785 | Yes | Yes | Yesc |
| ATTAATGAACCCTTAAA | 3979614-3979630 | ||||
| PA4773-pmrAB | GTTAAGCACCTCTTAAG | 5361502-5361518 | Yes | Yes | Yes |
| feoAB-PA4357 | CTTAAGCGAGCCTTAAG | 4887593-4887577 | Yes | Yes | Yes |
| PA1559-PA1560 | ATTAACGGTTCCTTAAG | 1697095-1697111 | Yes | Yes | Yes |
| PA4782-PA4781 | CTTAAGCGATCCCTAAG | 5370750-5370734 | Yes | Yes | NDe |
| PA0327 | TTTAAGCTTGGCTTCAG | 367370-367386 | No | NTd | NT |
| algD-8-44-KEGXLIGFA | CTTAAGGTTTGCTTAAG | 3962241-3962257 | No | NT | NT |
| PA0545 | GTTAAGGGACAATTAAG | 602272-602288 | No | NT | NT |
| PA0053 | TTTAAGCATGTCGCAAG | 69049-69065 | No | NT | NT |
| aprA (PA1249) | TTTAAGTGCAGCTTAAT | 1355477-1355493 | No | NT | NT |
| PA2050-PA2051 | TTTAAGTGCAGCTTAAT | 1355477-1355493 | No | NT | NT |
| PA4500 | TTTCAGCATGACTTAAT | 5036896-5036912 | No | NT | NT |
| PA2505-PA2506 | ATTAAGTGCGAGTTAAG | 2824124-2824108 | No | NT | NT |
| PA3868 | ATTAAGGCCTGCTCAAC | 4331298-4331314 | No | NT | NT |
| PA4498-PA4499 | TTTAAGCTCGACTTAAA | 5036126-5036110 | No | NT | NT |
| PA5106 | TTTACGCACCGCTTAAT | 5749388-5749404 | No | NT | NT |
Nucleotides that differ from the nucleotides in the consensus sequence are indicated by boldface type. Abbreviations: N = A, C, G, or T; S = C or G.
Determined by reverse transcription-qPCR.
Promoter regions were tested for binding to PhoP and PmrA. Only the pmrH-ugd promoter fragment bound to both. The other promoters bound only to PmrA and not to PhoP.
NT, not tested (no Mg2+ regulation could be demonstrated, and no regulation was observed in array experiments).
ND, not determined.
The transcript levels of the candidate genes in wild-type, phoP::xylE, and pmrA::xylE strains grown in minimal media were examined under high-Mg2+ conditions (2 mM Mg2+) and low-Mg2+ conditions (20 μM Mg2+). Analysis of the qPCR results (Fig. 2) confirmed that there was PmrA-dependent Mg2+ regulation of the feoAB (PA4359-PA4358), PA4782, and PA1559-PA1560 operons, as well as the previously identified pmrH-ugd (PA3552-PA3559) and PA4773-pmrAB operons. PhoP-dependent Mg2+ regulation was observed for PA0921 and PA1343, as well as for the previously identified oprH-phoPQ and pmrH-ugd operons. We did not observe any Mg2+ regulation for PA1851, PA3925, PA0918, PA2775, or PA0297 (spuA). Similarly, we examined but did not observe Mg2+ regulation for PA0327, PA0545, PA0053, PA4500, PA2505, PA2506, PA3868, PA4498, PA4499, and PA5106. These results suggest either that these genes are not regulated by Mg2+ and PhoP/PmrA at all or that their expression is codependent on the presence of a second regulatory signal (e.g., release from repression) that was not present under the conditions used.
FIG. 2.
Selected reverse transcription-qPCR results. Primers were designed to amplify a 100- to 150-bp fragment in the 5′ region of each gene indicated. RNA was isolated from strains H103 (wild type [wt]), H851 (phoP::xylE), and H988 (pmrA::xylE) under Mg2+-limiting (−) and Mg2+-replete (+) conditions.
Gel mobility shift assays.
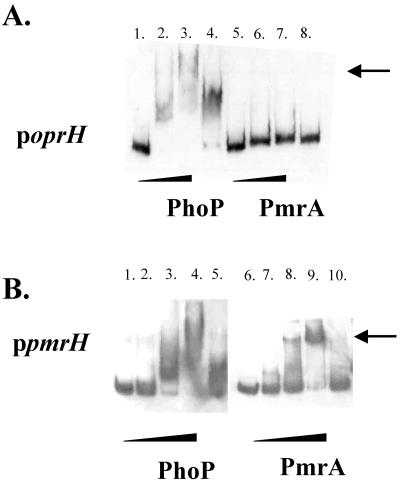
To obtain further evidence that PhoP was directly involved in the PhoP regulation of the PA0921, PA1343, oprH-phoPQ, and pmrH-ugd operons, gel mobility shift assays were performed using the entire PCR-amplified upstream intergenic region of these operons. When recombinant PhoP was incubated with the promoters upstream of the oprH, pmrH (Fig. 3), PA0921, and PA1343 genes (Table 3), marked retardation of the mobility of these fragments was observed. The mobility shift was eliminated by preincubating the PhoP protein with excess unlabeled probe. Similarly, mobility shift analysis of the putative PmrA-regulated promoters revealed that the promoters upstream of the pmrH (Fig. 3), PA4773, PA1559, and feoA (Table 4) genes were all bound by the PmrA protein, and this binding was specifically inhibited by an excess of unlabeled probe. Most promoters regulated by PmrA did not interact with His6-PhoP, nor did most PhoP-regulated genes interact with His6-PmrA; the only exception was the region upstream of the pmrH-ugd operon, which contains both PhoP and PmrA binding motifs (Fig. 3) and is known to be regulated by both regulatory systems (26). These results indicate that the PhoP and PmrA systems of P. aeruginosa regulate separate gene sets (with a single overlapping target operon) and that these gene sets independently respond to the same Mg2+ limitation signal.
FIG. 3.
Demonstration of binding of PhoP and PmrA to selected promoter regions: examples of results of gel shift assays for oprH and pmrH. (A) The entire intergenic region between napE (PA1177) and oprH (PA1178) was amplified by PCR and end labeled with DIG. The preparation was incubated with different concentrations of purified His6-PhoP or His6-PmrA as described in Materials and Methods. All lanes contained 40 pg of labeled probe. Lane 1, no protein; lane 2, 2.5 μg His6-PhoP; lane 3, 5 μg His6-PhoP; lane 4, 5 μg His6-PhoP preincubated with 100 ng unlabeled probe; lane 5, no protein; lane 6, 2.5 μg His6-PmrA; lane 7, 5 μg His6-PmrA; lane 8, 5 μg His6-PmrA preincubated with 100 ng unlabeled probe. (B) The entire intergenic region between algA (PA3551) and pmrH (PA3552) was amplified by PCR and end-labeled with DIG. The preparation was incubated with different concentrations of purified His6-PhoP or His6-PmrA as described in Materials and Methods. All lanes contained 40 pg of labeled probe. Lane 1, no protein; lane 2, 1.25 μg His6-PhoP; lane 3, 2.5 μg His6-PhoP; lane 4, 5 μg His6-PhoP; lane 5, 5 μg His6-PhoP preincubated with 100 ng unlabeled probe; lane 6, no protein; lane 7, 1.25 μg His6-PmrA; lane 8, 2.5 μg His6-PmrA; lane 9, 5 μg His6-PmrA; lane 10, 5 μg His6-PmrA preincubated with 100 ng unlabeled probe.
Limiting concentration of Mg2+ reduces growth on glucose and gluconate.
Due to the strong downregulation of two operons that are probably involved in metabolism of 2-ketogluconate and gluconate, the ability of P. aeruginosa to grow on glucose (which in Pseudomonas is oxidized to glucose in the periplasm prior to transport), gluconate, and succinate was examined under Mg2+-replete and Mg2+-deficient conditions. As shown in Fig. 4, in all cases the final density of cells was less when cells were grown in the presence of 20 μM Mg2+, but a limiting concentration of Mg2+ also led to a substantially longer doubling time when cells were grown on either glucose (doubling time with high Mg2+ concentration, 47 min; doubling time with low Mg2+ concentration, 70 min) or gluconate (doubling time with high Mg2+ concentration, 45 min; doubling time with low Mg2+ concentration, 64 min), but not when they were grown on succinate (doubling time with high Mg2+ concentration, 44 min; doubling time with low Mg2+ concentration, 46 min).
FIG. 4.
Effect of Mg2+ limitation on growth using carbon sources metabolized via the Entner-Douderhoff pathway. The growth of H103 (wild type) was monitored under Mg2+-replete conditions (high Mg or high) and Mg2+-limiting conditions (low Mg or low) in BM2 minimal medium with different carbon sources. The carbon sources used included glucose, gluconate, and succinate.
Mutants with mutations in P. aeruginosa feoB are defective for growth with Fe2+ as an iron source.
To investigate the potential effect of Mg2+ regulation of P. aeruginosa FeoAB, growth studies were performed in media containing either Fe2+ or Fe3+ as the sole source of iron, and a polar knockout mutant with a mutation in the feoA gene was compared with the isogenic wild-type strain (Fig. 5). These studies clearly demonstrated that there was no change in the growth of the feoA mutant when Fe3+ was the sole iron source, but there was a severe growth defect when Fe2+ was provided as the iron source, a result that was consistent with analogous observations made for Helicobacter pylori (50). This defect was apparent under both Mg2+-limiting and Mg2+-replete conditions, but the phenotype was more pronounced when Mg2+ was limiting, as under these conditions growth of the feoA mutant was completely eliminated (Fig. 5).
FIG. 5.
Involvement of feoAB in growth on ferrous iron. The growth of H103 (wild type) and the growth of H1034 (feoA::luxCDABE) were measured under Mg2+-limiting and Mg2+-replete conditions with only Fe2+ or Fe3+ available as an iron source. (A) Mg2+-replete conditions. (B) Mg2+-limiting conditions. Symbols: •, H103 plus Fe3+; ○, H103 plus Fe2+; ▪, H1034 plus Fe3+; □, H1034 plus Fe2+.
DISCUSSION
Here we describe a comprehensive examination of the response of P. aeruginosa to growth under Mg2+-limiting conditions. Although a large number of genes (158 genes) were found to be regulated by growth in the presence of a limiting concentration of Mg2+, much smaller numbers of genes were found to be influenced by mutations in the genes encoding two-component response regulators, phoP (19 genes) and pmrA (36 genes). This contrasts with the situation in Salmonella, in which the PhoP protein regulates, either directly or indirectly, 214 genes located in 189 operons (28), while PmrA regulates at least 47 genes (46). The limited number of proteins that are directly regulated by these regulators in P. aeruginosa, combined with the substantial number of regulatory proteins found to be differentially expressed in the presence of a limiting concentration of Mg2+, indicates that there may be other, as-yet-unidentified regulators that contribute to the total Mg2+ stimulon, and the limited number may also reflect the greater complexity of gene regulation in P. aeruginosa (43).
A separate bioinformatic approach was used to search for genes that may be regulated by either the PhoP response regulator or the PmrA response regulator in silico. This study was based on a previous observation that there are conserved binding sites in genes regulated by PhoP or PmrA (21, 26). Using these sequences as a starting point, a bioinformatic search for promoters containing these putative PhoP and/or PmrA binding sites was conducted. Examination of the sequence logos generated (Fig. 2B and 2C) indicated that there was a higher level of plasticity in the PmrA binding site than in the PhoP binding site. It was also clear that the PmrA and PhoP consensus sequences are quite strongly related to one another. Prokaryotic transcription factor binding sites are often direct or inverted repeats, and consistent with this, the PhoP consensus sequence was approximately two GTTCAG half-sites separated by five nucleotides, while the PmrA consensus sequence was approximately two CTTAAG half-sites separated by five nucleotides. The PhoP consensus sequence of Pseudomonas is quite different from that of Salmonella ([G/T]GTTTA[A/T][G/T]GTTTA[A/T]) (53), while the consensus sequence of PmrA is almost identical ([C/T]YTTAA[G/T]-N5-[C/T]YTTAA[G/T]) (1). This difference in the PhoP binding site may reflect the higher G+C content of P. aeruginosa than of E. coli and Salmonella.
Using qPCR and/or microarray analysis, only a subset of the putative PhoP- and PmrA-regulated genes (Table 2) were also observed to be regulated by growth in the presence of a limiting concentration of Mg2+. There was an overlap between the promoters that were predicted to contain PhoP and/or PmrA binding sites and the genes that were regulated by a limiting concentration of Mg2+ in microarray analyses. However, microarray analyses revealed a much larger repertoire of P. aeruginosa open reading frames that were Mg2+ regulated than was predicted by the bioinformatic analyses of PmrA and PhoP binding sites. Conversely, there was substantial overlap between the PhoP and PmrA promoters identified bioinformatically and confirmed by qPCR (Tables 3 and 4) and the PhoP and PmrA promoters shown by microarray analyses to be regulated in the phoP and pmrA mutants, respectively (Table 2).
In contrast to the situation in Salmonella enterica serovar Typhimurium, the microarray analysis of P. aeruginosa revealed no overlap between the PhoP and PmrA regulons (Table 2). This was not expected since previous work involving reporter fusions indicated that both systems are involved in regulation of the pmrH-ugd (PA3552-PA3559) operon (26), a result consistent with the observation that the pmrH-ugd promoter region contained binding sites for both transcription factors and the observation that these binding sites were likely to be functional based on gel shift assays (Fig. 5). However, qPCR and microarray experiments (Fig. 2) indicated that there was not a major decrease in the level of transcript from the pmrH-ugd operon in either the phoP or the pmrA mutant. This indicates that the two activators, PhoP and PmrA, can support activation of this operon to similar extents, so eliminating either one could permit altered expression to be observed only by a higher-resolution method, like analysis of transcriptional fusions. Thus, these data illustrate the utility of combining microarray and bioinformatic analyses for determining the contributions of PhoP and PmrA to the total Mg2+ stimulon of P. aeruginosa. It is important, however, that the pmrH-ugd operon was the only operon for which both PhoP and PmrA binding sites were predicted.
A limiting concentration of Mg2+ leads to increased transcription of the genes encoding several divalent cation transporter proteins, including MgtA and MgtE, and a putative copper-transporting ATPase; PA3920; pcoAB, which is involved in resistance to copper; and the feoAB operon encoding a ferrous iron transport system. Interestingly, almost all of the Mg2+-regulated divalent cation transporters required a functional copy of pmrA for normal transcription (Table 2). This contrasts with the situation in S. enterica serovar Typhimurium, in which the Mg2+ transporter MgtA (22, 47) is regulated by PhoPQ (53). However, no PhoP or PmrA binding motif was identified in the mgtA promoter, indicating that the Mg2+ regulation of mgtA may be indirectly regulated via PmrA.
MgtE from Bacillus firmus OF4 was first described as a multicopy suppressor that was able to complement the deficient growth on Mg2+ of a Salmonella strain with mutations in the corA, mgtA, and mgtB genes (41). MgtE is not similar to other types of metal transporters and has limited distribution among the bacterial species that have been examined (49). In P. aeruginosa, microarray analysis demonstrated that mgtE was induced by a limiting concentration of Mg2+ and that mgtE expression was reduced when pmrA was deleted, indicating that induction is dependent on this system.
This study also produced some evidence of regulatory processes that are more complicated than those described above. Notably, four genes, PA2274 and the mexGHI operon, were shown to be dysregulated in the pmrA::xylE mutant but were not regulated by Mg2+ (Table 2). Similarly, the sodB gene, encoding an Fe2+-dependent superoxide dismutase, is dysregulated in the phoP::xylE mutant but is not regulated by Mg2+ (Table 2). This suggests that phoP and/or pmrA mutants have altered physiology, even under Mg2+-replete conditions. It is also worth noting that the PA2274 and mexGHI-opmD genes were identified as the only dysregulated genes in a study examining the role of SoxR in resistance to paraquat in P. aeruginosa (30) and that sodB has been identified as a gene that is crucially important to P. aeruginosa during normal aerobic metabolism and in resistance to paraquat (14).
Since we demonstrated here that PmrA and a limiting concentration of Mg2+ regulated the feoAB operon, we investigated the role of feoAB in promoting growth in response to limiting iron and limiting Mg2+ conditions. As shown in Fig. 4, strains having a mutation in feoB were defective for growth when Fe2+ iron was the only iron source provided. Furthermore, the defect became more prominent when Mg2+ was also limiting. FeoAB is a well-conserved system that is involved in the transport of ferrous iron (24), and feoB mutants of E. coli (3, 19) and H. pylori (50) have reduced virulence and/or colonization activity.
It is not yet clear whether P. aeruginosa utilizes Fe2+ or Fe3+ as its primary iron source during infection. There is clear evidence for the presence of Fe3+-chelating siderophores in the sputum of CF patients, indicating that iron is limiting in CF lungs (12). Also, mutants with mutations in Fe3+ siderophore, especially ferri-pyoverdine, uptake have somewhat reduced virulence in a number of model systems (27, 45). However, pyoverdine-deficient mutants have been found in CF lungs (6), indicating that other iron uptake systems may be important. Recent results have demonstrated that the thickened and dehydrated mucus in CF lungs has a reduced oxygen tension that approaches complete hypoxia (∼5 mm Hg), even though elsewhere in CF lungs the partial O2 pressure is apparently normal (∼200 mm Hg) (52). Consistent with this, P. aeruginosa grows anaerobically in CF lungs. The decreased oxygen tension would tend to stabilize iron in the Fe2+ state. Furthermore, the lungs of CF patients have concentrations of ferritins that are 70-fold higher than the normal concentrations (34, 42). Indeed, CF patients have 20-fold-higher proportions of H-type ferritins, which are important for detoxifying Fe2+ ions by oxidizing them to Fe3+ before they are sequestered within the ferritin shell (13). These results collectively indicate that P. aeruginosa growing in the CF lung mucus layer may utilize Fe2+ to satisfy its iron requirements. The observation that the lungs of CF patients contain P. aeruginosa isolates with lipid A modifications equivalent to those of cells grown under Mg2+-limiting conditions (7) may thus reflect greater activation of feoAB in the CF lungs.
One of the most prominent transcriptional changes in response to Mg2+ limitation that was independent of PhoP and PmrA was the downregulation of two operons that seem likely to be involved in gluconate and 2-ketogluconate metabolism. Both of these carbon sources, as well as glucose, are metabolized via the Entner-Douderhoff pathway. To enter this pathway, glucose is first taken up across the outer membrane and then oxidized to gluconate in the periplasm via a membrane-bound dehydrogenase (38). The gluconate is then taken up via a gluconate permease (PA2322) (18) or is oxidized further to 2-ketogluconate by a gluconate dehydrogenase (PA2265) (44). The 2-ketogluconate produced is likely taken up via a permease protein, encoded by PA2262, which is in the same operon as the gene encoding gluconate dehydrogenase. Once inside the cell, glucose, gluconate, and 2-ketogluconate are all routed through the intermediate 6-phosphogluconate before they are degraded to pyruvate and glyceraldehyde-3-phosphate (48). As expected from the regulatory patterns observed, growth on glucose or gluconate as a carbon source was substantially slower under Mg2+-limiting conditions than under Mg2+-replete conditions. In contrast, there was virtually no difference in the doubling times on succinate, which is independently transported and enters directly into the tricarboxylic acid cycle.
It is not clear why P. aeruginosa exhibits this behavior unless growth of P. aeruginosa on substrates other than glucose represents a favorable adaptation to in vivo growth conditions. Consistent with this possible explanation and our observations, it was recently demonstrated that gluconate permease (PA2322) and the probable glyceraldehyde-3-phosphate dehydrogenase (PA2323) were downregulated in P. aeruginosa growing in CF sputum (31). This phenotype requires further study to elucidate a more detailed mechanism for the regulatory patterns observed; however, this observation and other observations described above are consistent with a role for the Mg2+ limitation regulon in adaptation of Pseudomonas to the CF lung.
In summary, this work expanded the number of known targets of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems. Our results demonstrate that these regulatory systems each regulate, either directly or indirectly, only a modest number of genes. The contribution of each system is also relatively small compared to the contribution of the entire Mg2+ stimulon of P. aeruginosa.
Acknowledgments
This work was supported by a grant from the Canadian Cystic Fibrosis Foundation to R.E.W.H. and by a grant from the US Cystic Fibrosis Foundation to R.E.W.H. and F.S.L.B. to support maintenance and development of the Pseudomonas.com web page. J.B.M. was a recipient of a CCFF studentship, and S.L. was a recipient of a CCFF postdoctoral fellowship. F.S.L.B. was a Michael Smith Foundation for Health Research Scholar, while R.E.W.H. holds a Canada Research Chair in Microbiology.
REFERENCES
- 1.Aguirre, A., S. Lejona, E. G. Vescovi, and F. C. Soncini. 2000. Phosphorylated PmrA interacts with the promoter region of ugd in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:3874-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469-472. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, E., I. Bergevin, D. Malo, P. Gros, and M. F. Cellier. 2002. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6032-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calogero, S., R. Gardan, P. Glaser, J. Schweizer, G. Rapoport, and M. Debarbouille. 1994. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J. Bacteriol. 176:1234-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Vos, D., M. De Chial, C. Cochez, S. Jansen, B. Tummler, J. M. Meyer, and P. Cornelis. 2001. Study of pyoverdine type and production by Pseudomonas aeruginosa isolated from cystic fibrosis patients: prevalence of type II pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch. Microbiol. 175:384-388. [DOI] [PubMed] [Google Scholar]
- 7.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 8.Faik, P., and H. L. Kornberg. 1973. Isolation and properties of E. coli mutants affected in gluconate uptake. FEBS Lett. 32:260-264. [DOI] [PubMed] [Google Scholar]
- 9.Foussard, M., S. Cabantous, J. Pedelacq, V. Guillet, S. Tranier, L. Mourey, C. Birck, and J. Samama. 2001. The molecular puzzle of two-component signaling cascades. Microbes Infect. 3:417-424. [DOI] [PubMed] [Google Scholar]
- 10.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 11.Gilleland, H. E., Jr., J. D. Stinnett, and R. G. Eagon. 1974. Ultrastructural and chemical alteration of the cell envelope of Pseudomonas aeruginosa, associated with resistance to ethylenediaminetetraacetate resulting from growth in a Mg2+-deficient medium. J. Bacteriol. 117:302-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas, B., J. Kraut, J. Marks, S. C. Zanker, and D. Castignetti. 1991. Siderophore presence in sputa of cystic fibrosis patients. Infect. Immun. 59:3997-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison, P. M., and P. Arosio. 1996. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta 1275:161-203. [DOI] [PubMed] [Google Scholar]
- 14.Hassett, D. J., H. P. Schweizer, and D. E. Ohman. 1995. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J. Bacteriol. 177:6330-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haydel, S. E., W. H. Benjamin, Jr., N. E. Dunlap, and J. E. Clark-Curtiss. 2002. Expression, autoregulation, and DNA binding properties of the Mycobacterium tuberculosis TrcR response regulator. J. Bacteriol. 184:2192-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hokamp, K., F. M. Roche, M. Acab, M. E. Rousseau, B. Kuo, D. Goode, D. Aeschliman, J. Bryan, L. A. Babiuk, R. E. Hancock, and F. S. Brinkman. 2004. ArrayPipe: a flexible processing pipeline for microarray data. Nucleic Acids Res. 32:W457-W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huffman, D. L., J. Huyett, F. W. Outten, P. E. Doan, L. A. Finney, B. M. Hoffman, and T. V. O'Halloran. 2002. Spectroscopy of Cu(II)-PcoC and the multicopper oxidase function of PcoA, two essential components of Escherichia coli pco copper resistance operon. Biochemistry 41:10046-10055. [DOI] [PubMed] [Google Scholar]
- 18.Hunt, J. C., and P. V. Phibbs, Jr. 1983. Regulation of alternate peripheral pathways of glucose catabolism during aerobic and anaerobic growth of Pseudomonas aeruginosa. J. Bacteriol. 154:793-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Lewenza, S., R. K. Falsafi, G. Winsor, W. J. Gooderham, J. B. McPhee, F. S. Brinkman, and R. E. W. Hancock. 2005. Construction of a mini-Tn5- luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macfarlane, E. L., A. Kwasnicka, and R. E. W. Hancock. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146:2543-2554. [DOI] [PubMed] [Google Scholar]
- 21.Macfarlane, E. L., A. Kwasnicka, M. M. Ochs, and R. E. W. Hancock. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305-316. [DOI] [PubMed] [Google Scholar]
- 22.Maguire, M. E. 1992. MgtA and MgtB: prokaryotic P-type ATPases that mediate Mg2+ influx. J. Bioenerg. Biomembr. 24:319-328. [DOI] [PubMed] [Google Scholar]
- 23.Maniatis, T., E. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Plainview, N.Y.
- 24.Marlovits, T. C., W. Haase, C. Herrmann, S. G. Aller, and V. M. Unger. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. USA 99:16243-16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, D. W., B. W. Holloway, and V. Deretic. 1993. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J. Bacteriol. 175:1153-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McPhee, J. B., S. Lewenza, and R. E. Hancock. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205-217. [DOI] [PubMed] [Google Scholar]
- 27.Meyer, J. M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monsieurs, P., S. De Keersmaecker, W. W. Navarre, M. W. Bader, F. De Smet, M. McClelland, F. C. Fang, B. De Moor, J. Vanderleyden, and K. Marchal. 2005. Comparison of the PhoPQ regulon in Escherichia coli and Salmonella typhimurium. J. Mol. Evol. 60:462-474. [DOI] [PubMed] [Google Scholar]
- 29.Moskowitz, S. M., R. K. Ernst, and S. I. Miller. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186:575-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palma, M., J. Zurita, J. A. Ferreras, S. Worgall, D. H. Larone, L. Shi, F. Campagne, and L. E. Quadri. 2005. Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infect. Immun. 73:2958-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer, K. L., L. M. Mashburn, P. K. Singh, and M. Whiteley. 2005. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 187:5267-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perego, M. 1998. Kinase-phosphatase competition regulates Bacillus subtilis development. Trends Microbiol. 6:366-370. [DOI] [PubMed] [Google Scholar]
- 33.Rajan, S., and L. Saiman. 2002. Pulmonary infections in patients with cystic fibrosis. Semin. Respir. Infect. 17:47-56. [DOI] [PubMed] [Google Scholar]
- 34.Reid, D. W., N. J. Withers, L. Francis, J. W. Wilson, and T. C. Kotsimbos. 2002. Iron deficiency in cystic fibrosis: relationship to lung disease severity and chronic Pseudomonas aeruginosa infection. Chest 121:48-54. [DOI] [PubMed] [Google Scholar]
- 35.Rinaldo, S., G. Giardina, M. Brunori, and F. Cutruzzola. 2005. N-oxide sensing in Pseudomonas aeruginosa: expression and preliminary characterization of DNR, an FNR-CRP type transcriptional regulator. Biochem. Soc. Trans. 33:184-186. [DOI] [PubMed] [Google Scholar]
- 36.Rodrigue, A., Y. Quentin, A. Lazdunski, V. Mejean, and M. Foglino. 2000. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 8:498-504. [DOI] [PubMed] [Google Scholar]
- 37.Sanowar, S., and H. Le Moual. 2005. Functional reconstitution of the Salmonella typhimurium PhoQ histidine kinase sensor in proteoliposomes. Biochem. J. 390:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schleissner, C., A. Reglero, and J. M. Luengo. 1997. Catabolism of d-glucose by Pseudomonas putida U occurs via extracellular transformation into d-gluconic acid and induction of a specific gluconate transport system. Microbiology 143:1595-1603. [DOI] [PubMed] [Google Scholar]
- 39.Schurr, M. J., H. Yu, J. M. Martinez-Salazar, J. C. Boucher, and V. Deretic. 1996. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 178:4997-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slater, H., A. Alvarez-Morales, C. E. Barber, M. J. Daniels, and J. M. Dow. 2000. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38:986-1003. [DOI] [PubMed] [Google Scholar]
- 41.Smith, R. L., L. J. Thompson, and M. E. Maguire. 1995. Cloning and characterization of MgtE, a putative new class of Mg2+ transporter from Bacillus firmus OF4. J. Bacteriol. 177:1233-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stites, S. W., M. W. Plautz, K. Bailey, A. R. O'Brien-Ladner, and L. J. Wesselius. 1999. Increased concentrations of iron and isoferritins in the lower respiratory tract of patients with stable cystic fibrosis. Am. J. Respir. Crit. Care Med. 160:796-801. [DOI] [PubMed] [Google Scholar]
- 43.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 44.Swanson, B. L., P. Hager, P. Phibbs, Jr., U. Ochsner, M. L. Vasil, and A. N. Hamood. 2000. Characterization of the 2-ketogluconate utilization operon in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 37:561-573. [DOI] [PubMed] [Google Scholar]
- 45.Takase, H., H. Nitanai, K. Hoshino, and T. Otani. 2000. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 68:1834-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamayo, R., S. S. Ryan, A. J. McCoy, and J. S. Gunn. 2002. Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6770-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tao, T., M. D. Snavely, S. G. Farr, and M. E. Maguire. 1995. Magnesium transport in Salmonella typhimurium: mgtA encodes a P-type ATPase and is regulated by Mg2+ in a manner similar to that of the mgtB P-type ATPase. J. Bacteriol. 177:2654-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Temple, L. M., A. E. Sage, H. P. Schweizer, and P. V. Phibbs, Jr. 1998. Carbohydrate catabolism in Pseudomonas aeruginosa, p. 35-72. In T. M. Montie (ed.), Pseudomonas, vol. 10. Plenum Press, New York, N.Y. [Google Scholar]
- 49.Townsend, D. E., A. J. Esenwine, J. George III, D. Bross, M. E. Maguire, and R. L. Smith. 1995. Cloning of the mgtE Mg2+ transporter from Providencia stuartii and the distribution of mgtE in gram-negative and gram-positive bacteria. J. Bacteriol. 177:5350-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velayudhan, J., N. J. Hughes, A. A. McColm, J. Bagshaw, C. L. Clayton, S. C. Andrews, and D. J. Kelly. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274-286. [DOI] [PubMed] [Google Scholar]
- 51.Winsor, G. L., R. Lo, S. J. Sui, K. S. Ung, S. Huang, D. Cheng, W. K. Ching, R. E. Hancock, and F. S. Brinkman. 2005. Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 33:D338-D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto, K., H. Ogasawara, N. Fujita, R. Utsumi, and A. Ishihama. 2002. Novel mode of transcription regulation of divergently overlapping promoters by PhoP, the regulator of two-component system sensing external magnesium availability. Mol. Microbiol. 45:423-438. [DOI] [PubMed] [Google Scholar]