Abstract
6S RNA is a highly abundant small RNA that regulates transcription through direct interaction with RNA polymerase. Here we show that 6S RNA directly inhibits transcription of pspF, which subsequently leads to inhibition of pspABCDE and pspG expression. Cells without 6S RNA are able to survive at elevated pH better than wild-type cells due to loss of 6S RNA-regulation of pspF. This 6S RNA-dependent phenotype is eliminated in pspF-null cells, indicating that 6S RNA effects are conferred through PspF. Similar growth phenotypes are seen when PspF levels are increased in a 6S RNA-independent manner, signifying that changes to pspF expression are sufficient. Changes in survival at elevated pH most likely result from altered expression of pspABCDE and/or pspG, both of which require PspF for transcription and are indirectly regulated by 6S RNA. 6S RNA provides another layer of regulation in response to high pH during stationary phase. We propose that the normal role of 6S RNA at elevated pH is to limit the extent of the psp response under conditions of nutrient deprivation, perhaps facilitating appropriate allocation of diminishing resources.
6S RNA is a noncoding small RNA conserved in many bacteria (4, 30, 35). 6S RNA function remained a mystery for several decades until the discovery that this RNA makes specific interactions with RNA polymerase (RNAP) (32). Bacterial RNAP is a multisubunit enzyme consisting of a catalytically active core enzyme (α2, β, β′, ω) that requires an additional specificity subunit (σ) to form the holoenzyme (Eσ) required for transcription initiation. 6S RNA forms stable, specific complexes with the housekeeping holoenzyme of RNAP (i.e., Eσ70 in Escherichia coli and EσA in Bacillus subtilis) but does not interact with other holoenzymes, the core enzyme, or free σ subunits (30). E. coli 6S RNA makes direct contact with the σ70 subunit within this 6S RNA-Eσ70 complex (32), suggesting that RNA binding specificity may be mediated through the σ70 subunit in a manner analogous to that of DNA binding specificity. The 6S RNA is primarily double-stranded, with a large single-stranded region in the center. This highly conserved secondary structure is essential for the ability of 6S RNA to form stable complexes with Eσ70 (30). These data have suggested models in which 6S RNA functions as a direct competitor for promoter DNA interactions with RNAP (4, 30, 31, 32).
6S RNA interactions with Eσ70 result in altered transcription at specific promoters, either directly at several σ70-dependent promoters or indirectly at several σS-dependent promoters (29). Observed transcriptional changes are greatest during late-stationary phase (≥18 h of growth), a time when 6S RNA levels are maximal. Intriguingly, not all σ70-dependent promoters are sensitive to 6S RNA, even though most Eσ70 is complexed with 6S RNA during this time frame (29, 32). Several promoters directly regulated by 6S RNA contain an extended −10 element, which is defined by a conserved TGN sequence immediately upstream of the −10 sequence element. Promoters with extended −10 elements make additional, specific interactions with Eσ70 that facilitate relatively high levels of transcription initiation, often in the absence of conserved −35 sequence elements (3, 22, 25). These features of extended −10 promoters, together or independently, may make them especially susceptible to 6S RNA inhibition.
Cells lacking 6S RNA are at a disadvantage for competitive survival when cocultured with wild-type cells; the timing of such growth defects in late-stationary phase (≥18 h of growth) coincides with the timing of maximal 6S RNA levels and 6S RNA-dependent transcriptional regulation (29). When not in competitive growth, 6S RNA-deficient cells exhibit a decreased ability to persist during long-term nutrient deprivation in a time frame of weeks (29). Although it is presumed that changes in growth and cell survival are a consequence of 6S RNA-mediated changes in transcription, no specific gene products responsible for these observed growth defects had been identified. Here we describe the first example of a direct connection between 6S RNA regulation of transcription of one gene (pspF) and a specific growth phenotype (altered survival at high pH).
Stationary-phase cells are well adapted to survive a variety of environmental stresses, even with suboptimal nutrients (10, 13). Environmental conditions can be determined by exogenous forces in natural habitats, such as low pH in the stomach or high pH upon exposure to pancreatic secretions. Alternatively, environmental conditions can be a consequence of changing metabolic activities associated with shifting nutrient pools in liquid culture, such as acidification of a medium with growth on acetogenic carbon sources or alkalization due to ammonia release when amino acids are catabolized (18, 20, 36). Regulators must respond to complex signals and changing conditions to allow multiple levels of control of the many gene products needed to facilitate survival.
We have uncovered a role for 6S RNA in modulating the ability of E. coli to respond to elevated pH during stationary phase. 6S RNA directly regulates transcription of pspF, which encodes a transcriptional activator named for its role in phage shock protein response (reviewed in references 7 and 23). PspF specifically activates σ54-dependent transcription of two target operons (pspABCDE and pspG) (12, 14, 17). Although the precise details of how the psp response leads to protection against elevated pH and other stresses are not entirely clear, it has been suggested that PspA plays a role in maintenance of proton motive force (15). A complex network of interactions between PspA and other Psp members (PspF, PspB, and PspC) is thought to regulate PspA activity (1, 9, 19). We propose that 6S RNA regulation of pspF places an upper limit on the extent of the psp response when nutrients are limiting in stationary phase.
MATERIALS AND METHODS
Strains.
Cells were grown in unbuffered LB-Lennox medium at 37°C unless otherwise indicated. LB pH 7.0 was buffered with 100 mM morpholinepropanesulfonic acid (MOPS), and LB pH 9.3 was buffered with 100 mM CAPS [3-(cyclohexylamino)-1-propanesulfonic acid]. Chloramphenicol (Cm; 25 μg/ml) was used in plates or liquid culture when cells with plasmid pKK* or derivatives were grown for <24 h. For longer-term cultures, fresh Cm was added (final concentration, 12.5 μg/ml) after 18 h of growth and at 24-h intervals after that. Prior to addition of new Cm, cultures were plated on LB and LB-plus-Cm plates to check the proportion of cells with plasmids. Cultures with <95% retention of plasmid (as measured by the ratio of CFU on LB-plus-Cm plates to CFU on LB plates) were discarded. See Tables 1 and 2 for strains and oligonucleotides used here, respectively.
TABLE 1.
Bacterial strains
| Strain name | Genotype | Reference |
|---|---|---|
| ZK126 | W3110 ΔlacU169 tna-2 | 6 |
| KW437 | ZK126 ssrS1 | This work |
| KW72 | Wild-type E. coli K-12 laboratory strain | 32 |
| GS075 | KW72 ssrS1 (Ampr) | 32 |
| KW435 | KW72 ssrS2 (Tetr) | This work |
| KW438 | KW72 ssrS3 (no drug marker) | This work |
| KW448 | KW72 ΔpspF | This work |
| KW449 | KW72 ssrS1 ΔpspF | This work |
| RLG3499 | MG1655 pyrE+lacI lacZ (VH1000) | 11 |
| KW348 | RLG3499 ssrS1 | This work |
| ZK1142 | W3110 ΔlacU169 tna-2 (Nalr) | 38 |
| ZK1143 | W3110 ΔlacU169 tna-2 (Strr) | 38 |
| KW371 | ZK1143 ssrS1 (Strr Ampr) | 29 |
| KW439 | RLG3499 λpspF-lacZ (−151 +33 relative to P1) | This work |
| KW440 | KW439 ssrS1 | This work |
| KW441 | RLG3499 λpspA (−127 +15)-lacZ | This work |
| KW442 | KW441 ssrS1 | This work |
| KW443 | KW441 ΔpspF | This work |
| KW444 | KW441 ssrS1 ΔpspF | This work |
| KW445 | RLG3499 λpspG(−96 +13)-lacZ | This work |
| KW446 | KW445 ssrS1 | This work |
| KW447 | KW445 ΔpspF | This work |
| KW450 | KW445 ssrS1 ΔpspF | This work |
| KW376 | RLG3499 λgalP2 (−89 +50) (−37C-T)-lacZ | 29 |
| KW377 | KW376 ssrS1 | 29 |
| RLG6641 | RLG3499 λi21 λPR(−40 +20)-lacZ | 2 |
| KW325 | RLG6641 ssrS1 | 29 |
TABLE 2.
Oligonucleotide sequences
| Construct | Oligonucleotide sequence (5′-3′) |
|---|---|
| ssrS2 | CTTTTCGGTTTACTGTGGTAGAGTAACCGTGAAGACAAA-GAAGTTCCTATCAACAGGGTCATTATATTTCG |
| CTGGTAGTTGCGTCATGGCTGGTGCCAGATAAGAAG-GAAGTTCCTATACTCGACATCTTGGTTACCG | |
| ΔpspF | GCGAATTACACTAACAAGTGGCGAATTTCATC-GAAGTTCCTATCAACAGGGTCATTATATTTCG |
| CCGCATCCGGCAAGTTGTATTGCTCAACTTCGG-GAAGTTCCTATACTCGACATCTTGGTTACCG | |
| ΔpspABC | CGCAGAAATCAAGAGGACAACATT-GAAGTTCCTATCAACAGGGTCATTATATTTCG |
| GGCCTGTTGCCAGCGAGTATTCATAACTTTCC-GAAGTTCCTATACTCGACATCTTGGTTACCG | |
| pspF-lacZ | GGGAATTCCGCGAGAAAAAATACCATAATG |
| GGAAGCTTCCGATGAAATTCGCCACTTGTTAG | |
| pspA-lacZ | GGGAATTCCGATGATGAAATTCGCCACTTCTTAG |
| GGAAGCTTCCCAGGATTGTCCTGCTGAACTG | |
| pspG-lacZ | GCGAATTCAGAGTAGTCAAATTCACCACGCCC |
| GGCAAGCTTGAAATATGAAATCTCTTGC | |
| pKK-pspF | GGCGAATTCGGATGGCAGAATACAAAGATAATTTA |
| GGCAAGCTTGGCTAAATCTGGTGCTTTTTCAAC | |
| pKK-6S(M5)* | GGGCCAGTCCCCTGAGCCCACGAATGTGGCGCTCCGCGGTTGG |
| CCAACCGCGGAGCGCCACATTCGTGGGCTCAGGGGACTGGCCC |
Three 6S RNA-null alleles were used. ssrS1 contains an ampicillin resistance gene inserted within the 6S RNA coding sequence (16), and no detectable 6S RNA is expressed in this strain background (16, 32). ssrS2 is a precise replacement of the 6S RNA coding sequence with a tetracycline resistance gene flanked by flippase (FLP) recombination target (FRT) sites, and ssrS3 was generated from ssrS2 by removal of the tetracycline resistance gene by FLP recombinase, which results in replacement of the 6S RNA coding sequence by a FRT site. ssrS2, ΔpspF, and ΔpspABC alleles were generated by homologous chromosomal recombination. PCR products were generated to allow precise replacement of the gene of interest with a tetracycline resistance gene flanked by FRT sites (see oligonucleotide sequences in Table 2 for junction sequences). PCR products were transformed into DY330 according to the method of Yu et al. (37). P1 transductions were carried out as described elsewhere (26) to move the ssrS1, ssrS2, ΔpspF, and ΔpspABC alleles into various strain backgrounds. ssrS3 was generated from ssrS2 according to the method of Datsenko and Wanner (8).
For chromosomal reporter constructs (pspF-lacZ, pspA-lacZ, pspG-lacZ), promoter regions were amplified by PCR from genomic DNA (see Table 2 for oligonucleotides) and cloned into pMSB1, followed by generation of λ-phage lysogens using “system II” as described elsewhere (24).
Plasmids.
All vectors with a pKK designation are derivatives of pKK-177-3; an asterisk designates the version carrying chloramphenicol resistance. The pKK vector control (pKK*) and pKK-6S* have been described previously (32). pKK-6S(M5)* was generated from pKK-6S* using QuikChange (Strategene). The 6S(M5) RNA has a deletion of 14 nucleotides in the central single-stranded region of the RNA; this mutant RNA has an altered secondary structure and does not interact with RNA polymerase or exhibit any 6S RNA activity (30). pKK-pspF was generated by PCR amplification of the pspF coding sequence and insertion into the EcoRI and HindIII sites of pKK-177-3. pKK-pspF* was generated from pKK-pspF by insertion of a Cm resistance gene into the PvuII site (see reference 32). pKK-177-3 and all derivatives here contain the inducible Ptac promoter to express cloned genes; however, uninduced levels from this promoter are quite high and all experiments here were done using constitutive expression of pspF and 6S RNAs from pKK-177-3 plasmid derivatives. 6S RNA expressed from pKK-6S* is at ∼2-fold higher levels than endogenously expressed 6S RNA (32).
Cell growth/survival assays.
Cell densities in growth experiments were determined by plating as previously described (29). Cultures were grown from a single colony and at the times indicated were serially diluted into M9 salts and plated onto LB plates. For cells containing a plasmid or for competition assays, cells were plated on LB with appropriate antibiotics. Plates from dilutions that gave 100 to 250 colonies per plate were used to minimize statistical variation due to small sample sizes. These numbers allow accuracy down to at least twofold changes. Typically, three to five culture replicates were done per experiment, and all growth experiments were repeated at least three times.
Competition assays were carried out as previously described (29); briefly, equal numbers of each cell type were mixed in a single culture, and cell densities of each cell type over time were determined by plating.
Note that the magnitude of changes in cell survival we observe for pspF-null and pspABC-null cells (see Fig. 4) (data not shown) and previous reports describing 1,000-fold-decreased survival of pspA-disrupted cells at high pH (33) most likely result from strain background effects, as also noted in reference 33.
FIG. 4.
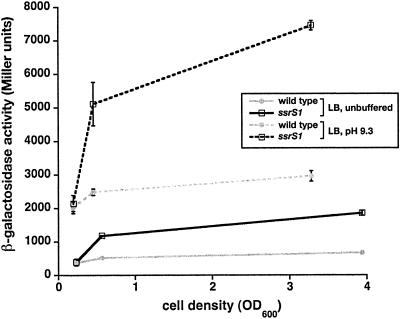
Induction of pspA transcription at high pH is not dependent on 6S RNA. The β-galactosidase activity of pspA-lacZ was measured in Miller units. (A) Wild-type (KW441) or ssrS1 (KW442) cells containing pspA-lacZ were grown in unbuffered LB (solid lines) or LB pH 9.3 (dotted lines) at 30°C to the cell densities indicated (OD600). Data shown are from one representative experiment including three duplicates per time point. Error bars, standard deviations from the averages.
β-Galactosidase assays.
β-Galactosidase activity assays were done at 30°C as previously described. Activity is expressed in Miller units (optical density at 420 nm [OD420] per min per OD600 unit) (21). For time courses, cells were grown overnight from a single colony in LB (buffered or unbuffered as indicated) at 30°C, diluted 1:100, grown to the indicated OD600, and lysed with sodium dodecyl sulfate-chloroform, and β-galactosidase activity was measured. Stationary-phase measurements were done on cells grown for 18 h at 30°C after dilution. At least three independent cultures per strain were assayed for each experiment, and experiments were repeated at least three times.
These assays were performed on cells grown at 30°C because we have found that promoter fusions with high expression in exponential phase had significant carryover of β-galactosidase into stationary phase when grown at 37°C. Carryover effects appear to be minimized by growth at 30°C, presumably due to the slower growth rate at 30°C and potential changes in the stability of β-galactosidase. Promoter fusions that do not have strong exponential-phase expression show similar 6S RNA-dependent regulation at 30°C and 37°C. In addition, primer extension analysis on mRNAs from several promoter fusions isolated from cells grown at 37°C and 30°C confirmed that β-galactosidase activities measured on cells grown at 30°C were representative of actual mRNA level differences between wild-type and ssrS1 cells (29).
Zone-of-inhibition assays.
A 200-μl volume of an overnight culture was plated in 3 ml top agar onto LB plates. Disks (5.5 mm) of filter paper containing 10 μl of 10 M NaOH were placed on the plates, and cells were allowed to grow overnight at 37°C. The diameter of the cell-free zone, including the disks, was measured.
RESULTS
Cells survive better at high pH in the absence of 6S RNA.
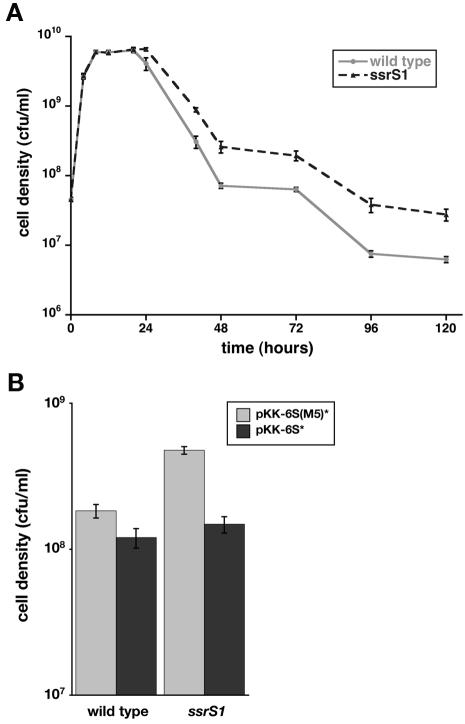
Cells without 6S RNA are at a disadvantage in competitive growth after 2 days in coculture with wild-type cells in LB medium (29). Growth in unbuffered LB results in increasing pH of the medium over time: the pH is >8 by 24 h of growth, increases to ∼8.8 by 6 days, and remains just under 9 over extended growth periods (data not shown). These observations led us to examine whether decreased survival of 6S RNA-deficient cells in coculture is due to sensitivity to pH changes. However, we found that 6S RNA-deficient cells were able to survive better than wild-type cells when grown at high pH independently (i.e., not in coculture). Specifically, 6S RNA-deficient cells (KW437) and wild-type cells (ZK126) cultured in LB pH 9.3 at 37°C grew similarly through log phase, but by 48 h of growth, 6S RNA-deficient cells were at ∼3- to 5-fold-higher density (Fig. 1A). The precise timing of onset of the growth phenotype varied some from day to day and in different strain backgrounds (KW72, ZK126, and RLG3499) but was consistently within 20 to 40 h, and cell density differences were ≥3-fold by 48 h (data not shown) (see Fig. 1B, 5, and 6). Different 6S RNA-null alleles (ssrS1, ssrS2, and ssrS3) behaved indistinguishably in all assays reported here; data shown are in ssrS1 backgrounds.
FIG. 1.
6S RNA-deficient cells survive at elevated pH better than wild-type cells. (A) Wild-type (ZK126) (gray line) or ssrS1 (KW437) (black dashed line) cells were inoculated into LB pH 9.3 medium and grown at 37°C. At the times indicated, cell densities were determined by plating. The data shown are from one representative experiment containing five duplicate cultures per strain. (B) Wild-type (KW72) or ssrS1 (GS075) cells containing pKK-6S(M5)* (shaded bars) or pKK-6S* (solid bars) were inoculated into LB pH 9.3 medium and grown at 37°C for 48 h. Cell densities were measured by plating. The data shown are averages from two representative experiments with two duplicates for each experiment. Similar results were obtained in at least four independent experiments. Error bars, standard deviations from the averages. At several time points in panel A, the error bars are smaller than the symbols and therefore are not visible.
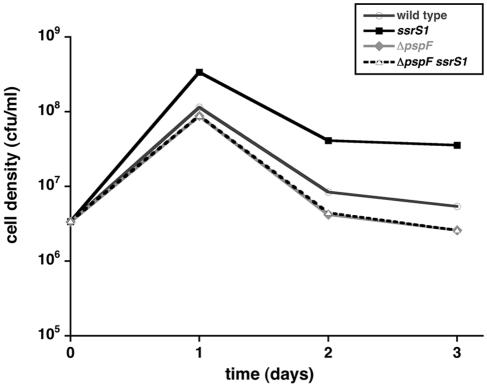
FIG. 5.
6S RNA effects on cell survival at high pH require pspF. Wild-type (KW72), ssrS1 (GS075), ΔpspF (KW448), or ΔpspFssrS1 (KW449) cells were inoculated into LB pH 9.3 medium and grown at 37°C. At the times indicated, cell densities were determined by plating. Data shown are from one representative experiment including three duplicates per time point. Standard deviations from averages were all <6% and are too small to be visible here. Similar results were obtained from five independent experiments.
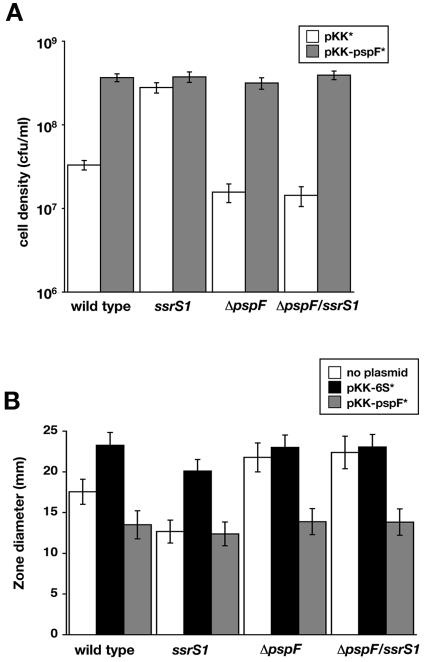
FIG. 6.
Increased pspF expression is sufficient to mediate increased cell survival at high pH. (A) Wild-type (KW72), ssrS1 (GS075), ΔpspF (KW448), or ΔpspFssrS1 (KW449) cells containing pKK* (open bars) or pKK-pspF* (shaded bars) were inoculated into LB pH 9.3 medium and grown at 37°C for 48 h. Cell densities were measured by plating. Data shown are averages of two experiments with two duplicates each. (B) Wild-type (KW72), ssrS1 (GS075), ΔpspF (KW448), or ΔpspFssrS1 (KW449) cells containing either no plasmid (open bars), pKK-6S* (solid bars), or pKK-pspF* (shaded bars) were tested for sensitivity to 10 M NaOH using the zone-of-inhibition assay. Measurements indicate the diameter of the cell-free zone surrounding the disk. Data shown are from one representative experiment with three duplicate cultures. Similar results were obtained from seven independent experiments. Error bars, standard deviations from the averages.
Growth in LB at pH 9.1 resulted in approximately two- to threefold-increased cell densities of ssrS1 (GS075) compared to wild-type (KW72) cells after 48 h, but growth in LB pH 8.9 showed minimal differences between the two strains even after several days (data not shown). Neither cell type was able to grow in LB pH 9.5 (data not shown), and growth in unbuffered LB is indistinguishable between wild-type and 6S RNA-deficient cells in these time frames (29). Growth in LB pH 9.3 at 30°C showed similar patterns of growth of wild-type compared to ssrS1 cells, although the timing of cell density changes was different. Wild-type and ssrS1 cells grew indistinguishably for at least 24 h, and by 48 h of growth the wild-type cell densities had decreased about ∼4-fold compared to those of ssrS1 cells (data not shown).
To test that the increased survival of ssrS1 cells in LB pH 9.3 was due to loss of 6S RNA, we examined cell growth after introduction of a plasmid expressing wild-type 6S RNA (pKK-6S*) or an inactive mutant [pKK-6S(M5)*]. Expression of 6S(M5) RNA in ssrS1 cells (GS075) did not alter cell growth; these cells achieved cell densities similar to those of ssrS1 cells without plasmid. In contrast, introduction of pKK-6S* into ssrS1 cells resulted in a reduced survival phenotype similar to that observed for wild-type cells (Fig. 1B).
Previous coculture experiments were done in unbuffered LB, and the time of the loss of ssrS1 cells was similar to the time at which the medium pH would be increasing (29). Since ssrS1 cell survival is favored at elevated pH in independent culture, we tested if the decreased competitive fitness of ssrS1 cells could be exaggerated under conditions that maintained a neutral pH. However, ssrS1 cells (KW371) exhibited a similar level of decreased survival in coculture with wild-type cells (ZK1142) in LB pH 7.0, LB pH 9.3, and unbuffered LB (data not shown). Wild-type cells also outcompete 6S RNA-deficient cells in minimal glucose medium, which becomes acidified in stationary phase (29).
6S RNA down-regulates transcription of pspF.
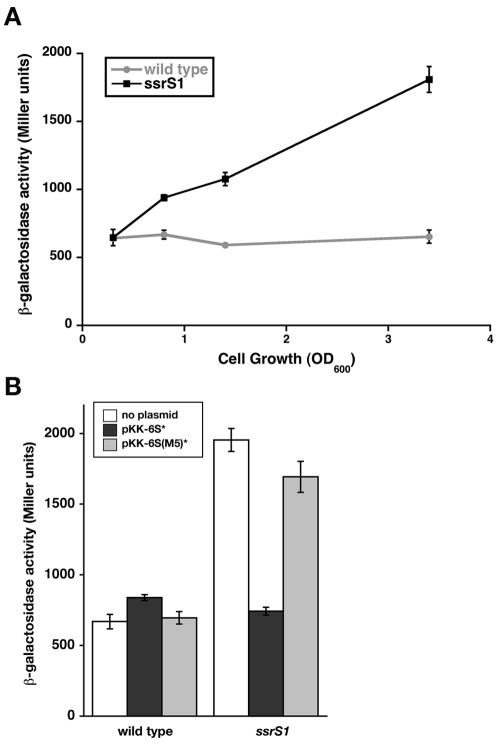
The cell growth/survival phenotypes of 6S RNA-deficient cells most likely result from 6S RNA-regulated changes in transcription. To identify potential promoter targets for 6S RNA regulation, we searched for genes important for cell growth or survival at high pH during long-term stationary phase. pspABC deletion results in decreased survival of cells at elevated pH relative to the wild type after 24 h of growth (33). The pspA operon is transcribed by Eσ54 (34); thus, it most likely is not directly regulated by 6S RNA (see below). However, transcription of this operon requires the transcriptional activator PspF (14), which might be a target for 6S RNA regulation. A reporter gene with the pspF promoter region fused to lacZ (pspF-lacZ) was examined in an ssrS1 compared to a wild-type background (Fig. 2; see also Fig. 3A). Expression of pspF-lacZ in wild-type cells (KW439) remains relatively constant over time, consistent with previous reports (14). However, expression of pspF-lacZ in 6S RNA-deficient cells (KW440) increased upon entry into stationary phase and reached levels 3- to 3.5-fold higher than those in wild-type cells after 18 h of growth in unbuffered LB (OD600 = 3.5).
FIG. 2.
pspF transcription is inhibited by 6S RNA. The β-galactosidase activity of pspF-lacZ was measured in Miller units. (A) Wild-type (KW439) (gray line) or ssrS1 (KW440) (black line) cells containing pspF-lacZ were grown in unbuffered LB at 30°C to the cell densities indicated (OD600). Data shown are from one representative experiment including three duplicates per time point. Similar results were obtained from three independent experiments. (B) Wild-type (KW439) or ssrS1 (KW440) cells containing pspF-lacZ and either no plasmid (white bars), pKK-6S* (solid bars), or pKK-6S(M5)* (shaded bars) were grown in unbuffered LB at 30°C for 18 h. Data shown are from one representative experiment with three duplicates. Similar results were seen in three independent experiments. Error bars, standard deviations from the averages.
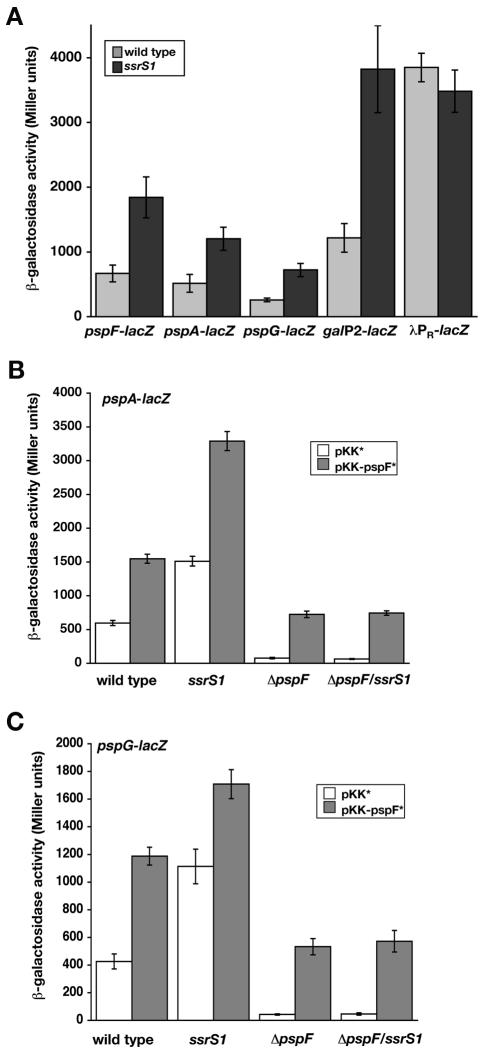
FIG. 3.
Transcription from pspA-lacZ and pspG-lacZ is inhibited in ssrS1 cells through changes in PspF levels. β-Galactosidase activities of indicated promoter-lacZ fusions were measured in Miller units. (A) Expression from pspF-lacZ, pspA-lacZ, pspG-lacZ, galP2-lacZ, and λpR-lacZ was examined in wild-type (shaded bars) or ssrS1 (solid bars) strain backgrounds when strains were grown at 30°C for 18 h. Data shown are averages from at least two experiments. (B) Wild-type (KW441), ssrS1 (KW442), ΔpspF (KW443), or ΔpspFssrS1 (KW444) cells containing pspA-lacZ and either the control plasmid pKK* (open bars) or pKK-pspF* (shaded bars) were grown in unbuffered LB at 30°C for 18 h. (C) Wild-type (KW445), ssrS1 (KW446), ΔpspF (KW447), or ΔpspFssrS1 (KW450) cells containing pspG-lacZ and either the control plasmid pKK* (open bars) or pKK-pspF* (shaded bars) were grown in unbuffered LB at 30°C for 18 h. Data in panels B and C are averages of two representative experiments with three duplicates. Error bars, standard deviations from the averages.
pspF-lacZ expression in the ssrS1 strain background (KW440) was restored to wild-type levels by expression of 6S RNA from a plasmid (pKK-6S*) but not by expression of the inactive 6S(M5) RNA (Fig. 2B), indicating that loss of 6S RNA in the ssrS1 background is responsible for changes to pspF transcription. Therefore, transcription of pspF is normally down-regulated by 6S RNA during stationary phase.
The pH of the growth medium did not alter expression of the pspF-lacZ reporter; similar levels of activity for both strain backgrounds (KW439 and KW440) were observed when cells were grown in unbuffered LB compared to LB pH 9.3 (data not shown). 6S RNA levels in wild-type cells (KW72) are likewise unchanged by growth medium pH; 6S RNA accumulates similarly through growth and reaches the highest levels in late-stationary phase (>18 h of growth) when cells are grown in LB pH 7.0, LB pH 9.3, or unbuffered LB (data not shown) (32).
6S RNA regulation of pspF transcription leads to regulation of pspABCDE and pspG.
To assess whether the changes in transcription of pspF lead to changes in PspF protein activity in the cell, we next examined transcription of the two promoters that require PspF for activation: pspA and pspG (12, 14, 17). Expression from pspA-lacZ and pspG-lacZ reporters was increased 2.5- to 3-fold during stationary phase in ssrS1 (KW442, KW446) compared to wild-type (KW441, KW445) strains grown in LB (Fig. 3A). This level of inhibition is similar to that for identified 6S RNA-sensitive promoters such as galP2-lacZ (Fig. 3A) (29). Other promoters, including λpR-lacZ, are not inhibited by 6S RNA (Fig. 3A) (29).
6S RNA-dependent changes in expression of pspA-lacZ and pspG-lacZ most likely result from altered PspF levels. As expected, expression of pspA-lacZ and pspG-lacZ was dependent on PspF; background expression levels were seen in pspF-null strains (KW443, KW447) (see Fig. 3B and C). Next, we expressed PspF in a 6S RNA-independent manner by introducing a plasmid constitutively expressing pspF (pKK-pspF*) compared to the vector control (pKK*) (Fig. 3B and C). Expression of PspF from the plasmid is able to restore the activity of pspA-lacZ and pspG-lacZ in the ΔpspF strain background (KW443 and KW450), indicating that active protein is expressed. The level of pspA-lacZ expression is slightly higher from plasmid-expressed PspF (see ΔpspF/pKK-pspF*) compared to normal PspF levels (see wild-type/pKK*), suggesting that at least wild-type levels of PspF are being made from the plasmid. Expression of pspA-lacZ and pspG-lacZ in the ΔpspF compared to the ΔpspF ssrS1 background is indistinguishable when PspF is expressed from the plasmid, pKK-pspF*. These data confirm that pspABCDE and pspG are not regulated directly by 6S RNA.
pspA-lacZ and pspG-lacZ activities increase further when PspF levels are overexpressed by introducing pKK-pspF* in the wild-type background (KW441, KW445), which also expresses endogenous pspF. Interestingly, plasmid-expressed PspF in the ssrS1 background (KW442, KW446) resulted in higher expression of pspA-lacZ and pspG-lacZ than either alone; therefore, these two mechanisms for increasing PspF activity act independently. Together, these results demonstrate that increasing PspF, either by introduction of the plasmid or by loss of 6S RNA, results in increased transcription of pspABCDE and pspG.
Expression from pspA-lacZ increased ∼4- to 5-fold when wild-type cells (KW441) were grown in LB pH 9.3 compared to unbuffered LB in late-exponential and stationary phase (Fig. 4). Expression of pspA-lacZ also was induced ∼4- to 5-fold in ssrS1 cells (KW442) grown in LB pH 9.3 compared to unbuffered LB. In contrast, expression from pspG-lacZ was not altered by the pH of the medium (data not shown).
6S RNA regulation of pspF is responsible for altered survival at high pH.
Next, we set out to determine if the increased survival of 6S RNA-null cells was directly due to altered PspF levels. The survival of pspF-null cells (KW448) was slightly decreased at elevated pH after 2 days relative to that of wild-type cells (KW72) (Fig. 5). In a ΔpspF ssrS1 double mutant (KW449), cells likewise showed decreased survival at high pH, indicating that the ability of 6S RNA to increase survival is lost in the absence of pspF.
To confirm that altered sensitivity in ssrS1 cells was due to changes in PspF, survival at elevated pH was tested when PspF levels were increased by expression from a plasmid (pKK-pspF*) compared to the vector control (pKK*) (Fig. 6A). In the wild-type strain background (KW72), increased PspF levels from pKK-pspF* led to increased cell survival at 48 h of growth in LB pH 9.3, similar to the survival in ssrS1 cells (GS075) with the vector control (pKK*) (Fig. 6A; see also Fig. 5). Survival in both ΔpspF (KW448) and ΔpspF ssrS1 (KW449) strain backgrounds were increased when pKK-pspF* was present compared to these cells containing the vector control (pKK*). Interestingly, the combination of ssrS1 and pKK-pspF* does not lead to an additive effect on survival. The maximal cell density is similar (∼5 × 108 cells/ml at 48 h) for all cells with increased pspF expression, whether from a lack of 6S RNA or from pKK-pspF*.
To further confirm growth phenotypes in strains containing plasmids, we assayed pH sensitivity using a zone-of-inhibition assay (Fig. 6B). The diameter of cell-free zones surrounding a disk containing 10 M NaOH was used as a measure of sensitivity to increased pH. In agreement with growth in culture, wild-type cells (KW72) exhibit a larger zone of clearing than ssrS1 cells (GS075), indicating that wild-type cells are more sensitive to increased pH. Similar trends in sensitivity were observed with 5 M NaOH and with 5 M or 10 M KOH (data not shown). ΔpspF cells (KW448) are slightly more sensitive than wild-type cells (KW72), and the ΔpspF ssrS1 double mutant (KW449) behaves similarly to the ΔpspF single mutant, in agreement with the 48-h culture growth assay. Increasing 6S RNA levels by expression from pKK-6S* leads to decreased survival in either the wild-type (KW71) or the ssrS1 (GS075) background, while increasing 6S RNA levels in the ΔpspF (KW448) or ΔpspF ssrS1 (KW449) background has little effect on survival. As seen above, these data strongly suggest that 6S RNA effects on cell survival at elevated pH are mediated primarily through the 6S RNA-dependent changes in pspF expression.
To further test whether an increase in PspF was sufficient to obtain the level of survival in 6S RNA-null strains in the zone-of-inhibition assay, PspF was overexpressed from the plasmid (pKK-pspF*) in wild-type cells. Indeed, increasing PspF levels from pKK-pspF* leads to increased survival in the wild-type (KW72), ΔpspF (KW448), and ΔpspF ssrS1 (KW449) backgrounds; the level of survival in all cases where pKK-pspF* is present is similar to that seen for ssrS1 cells (GS075) alone. Again, increasing PspF levels from the plasmid in the ssrS1 background did not result in further increased cell resistance to pH.
DISCUSSION
We have found an additional growth phenotype associated with the loss of 6S RNA activity: increased survival at elevated pH during late-stationary phase. We have identified a direct 6S RNA-regulated gene target, pspF, which mediates the 6S RNA changes in cell survival. Transcription of pspF is normally down-regulated by 6S RNA such that in the absence of 6S RNA, pspF transcription is increased. The increased transcription of pspF results in increased PspF protein activity, as measured by PspF-dependent activation of pspA and pspG transcription. Changes in 6S RNA levels do not mediate altered cell survival at high pH in the absence of pspF, indicating that pspF is required for these changes in survival. Increasing pspF expression in a 6S RNA-independent manner by introduction of pspF on a plasmid (pKK-pspF*) also leads to increased survival at high pH, indicating that increased PspF activity is sufficient to mediate the altered survival at high pH. Altogether, these data provide the first connection between a direct 6S RNA-dependent change in transcription of a single gene (i.e., pspF) and a specific growth phenotype associated with loss of 6S RNA (i.e., increased survival at high pH).
6S RNA roles in survival during stationary phase.
We have shown that wild-type cells are less fit for survival at elevated pH than cells without 6S RNA, which might lead to the question of why cells maintain their 6S RNA. In fact, conserved 6S RNA-encoding genes have been identified in diverse bacterial species, suggesting that 6S RNA plays a universal role. Perhaps the timing and magnitude of 6S RNA regulation of transcription and resulting growth phenotypes provide clues. During late-stationary phase (>24 h), nutrient pools are diminishing in quantity and quality. Long-term cell survival is dependent, at least in part, on appropriate use of the remaining resources. Long-term cell survival is enhanced by 6S RNA (29), suggesting that there may be a trade-off between survival of immediate stresses and long term survival. We propose that 6S RNA is one mechanism that places an upper limit on the extent of the psp response under suboptimal nutrient conditions. Such a constraint allows some level of stress protection while preventing overcommitment of resources. A similar trade-off between immediate needs during early-stationary phase and long-term survival may be exemplified by many GASP (growth advantage in stationary phase) alleles that result in attenuated σS activity (10). σS mediates a general stress response and is important in transition from exponential to early-stationary-phase growth (13). However, long-term survival and fitness for competitive survival are dramatically enhanced in strains with decreased σS activity (10).
Intriguingly, medium pH did not alter the competitive disadvantage of 6S RNA-deficient cells. This observation may illustrate the complexity of gene expression changes mediated by 6S RNA function and the complexity of interacting activities and pathways which contribute to cell survival. Understanding the details responsible for the lack of an additive effect between pH response and competitive fitness changes in 6S RNA-deficient cells will await elucidation of the molecular mechanisms responsible for the competitive growth advantage conferred by 6S RNA in wild-type cells.
6S RNA regulation of transcription and the psp response.
6S RNA-mediated changes in transcription are rather moderate (two- to fivefold) compared to changes regulated by most protein transcription factors. This type of regulation may represent a role for 6S RNA in modulating and balancing appropriate transcriptional levels independently of the specific response pathways mediated by traditional transcription factors. Consistent with a modulating role for 6S RNA, changes in growth associated with loss of 6S RNA are likewise rather modest: 3- to 50-fold, depending on the conditions of growth and time frames examined. However, even these moderate growth phenotypes are significant when their impact within a natural, competitive environment is considered.
Our data demonstrate that transcription of pspF is increased in the absence of 6S RNA. pspF contains three σ70-dependent promoters (14); the primary promoter (P1) contains an extended −10 element. One common property of identified σ70-dependent promoters inhibited by 6S RNA is the presence of an extended −10 element (29), suggesting that the pspFP1 promoter is inhibited by 6S RNA. The pspF-lacZ reporter examined here contained all three pspF promoters, and therefore, whether or not all three promoters are regulated by 6S RNA, it is clear that the overall expression of pspF is increased in the absence of 6S RNA.
The 6S RNA-dependent increase in PspF activates transcription of pspABCDE and pspG. PspA has been suggested to function in the maintenance of proton motive force (15). Current models focus on the complex regulation of PspA through interactions with PspF, PspB, and PspC in stress response (1, 9; reviewed in references 7 and 23). A more recent addition to the psp response, pspG, clearly plays a role as well, although details of its function await further investigation (12, 17). Under noninducing conditions, PspA activity may be regulated primarily through interaction with PspF. 6S RNA down-regulation of transcription of pspF results in decreased expression of both pspA and pspG, presumably altering the ground state level of PspA and PspG activities under noninducing conditions.
PspA activity is induced by a number of environmental conditions, including increased pH. It has been suggested that induction is mediated through changing interactions with PspF, PspB, and PspC (1, 7, 19, 34), in addition to increased expression of pspA upon release of PspF (5) (Fig. 3). Induction of PspA is not due to increased PspF levels, as pspF expression remains fairly constant throughout growth and under inducing conditions (14; A. E. Trotochaud and K. M. Wassarman, unpublished data) (Fig. 2A). Likewise, PspA induction is not dependent on 6S RNA; the relative ratio of induced to uninduced levels of pspA-lacZ remains ∼4- to 5-fold in the wild-type or ssrS1 strain background. Other mechanisms are presumed to limit the extent of PspA induction by regulating protein levels or protein activity, perhaps through feedback regulation by other Psp proteins. Most likely, such constraints account for the observations that overexpression of PspF from the plasmid (pKK-pspF*) in the ssrS1 background did not lead to additive phenotypic changes, even though there were additive effects on pspA-lacZ expression. In addition, the psp response is only one of many pathways that are induced by elevated pH environments (27, 28).
The psp response has been implicated in a number of stress responses, as well as in protein secretion, in motility, and in virulence (7, 23) suggesting that 6S RNA may modulate the psp response more broadly. 6S RNA-deficient cells exhibit decreased motility. However, this phenotype appears to result from 6S RNA regulation of other genes in addition to pspF, as changes in pspF expression alone were not able to fully recapitulate observed phenotypes (Trotochaud and Wassarman, unpublished).
Conclusion.
The identification of pspF as a direct target for 6S RNA regulation has made the first connection between 6S RNA-dependent changes in transcription and growth phenotypes of cells lacking 6S RNA. The proposed role for 6S RNA in controlling the extent of the psp response under nutrient-limiting conditions may represent a global role for 6S RNA in rationing available resources during stationary phase. In addition to altered growth at high pH, 6S RNA-deficient cells exhibit phenotypes under several other stress conditions in stationary phase (Trotochaud and Wassarman, unpublished), suggesting that this RNA may have a similar modulatory role in other environmental responses. Such a function for 6S RNA in conservation of resources during stationary phase may ultimately be the way in which 6S RNA contributes to long-term cell survival. In any event, understanding the connection between 6S RNA and cell responses to high-pH environments has highlighted the importance of 6S RNA-mediated coordination between different environmental responses.
Acknowledgments
We thank S. E. Finkel and R. L. Gourse for providing strains.
This work was supported by the National Institutes of Health (GM67955).
REFERENCES
- 1.Adams, H., W. Teertstra, J. Demmers, R. Boesten, and J. Tommassen. 2003. Interactions between phage-shock proteins in Escherichia coli. J. Bacteriol. 185:1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, M. M., and R. L. Gourse. 2001. Regulation of rRNA transcription correlates with nucleoside triphosphate sensing. J. Bacteriol. 183:6315-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barne, K. A., J. A. Bown, S. J. Busby, and S. D. Minchin. 1997. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the ′extended −10′ motif at promoters. EMBO J. 16:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrick, J. E., N. Sudarsan, Z. Weinberg, W. L. Ruzzo, and R. R. Breaker. 2005. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11:774-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brissette, J. L., M. Russel, L. Weiner, and P. Model. 1990. Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connell, N., Z. Han, F. Moreno, and R. Kolter. 1987. An E. coli promoter induced by the cessation of growth. Mol. Microbiol. 1:195-201. [DOI] [PubMed] [Google Scholar]
- 7.Darwin, A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621-628. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elderkin, S., S. Jones, J. Schumacher, D. Studholme, and M. Buck. 2002. Mechanism of action of the Escherichia coli phage shock protein PspA in repression of the AAA family transcription factor PspF. J. Mol. Biol. 320:23-37. [DOI] [PubMed] [Google Scholar]
- 10.Finkel, S. E., E. R. Zinser, and R. Kolter. 2000. Long-term survival and evolution in the stationary phase in Escherichia coli, p.231-238. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 11.Gaal, T., W. Ross, S. T. Estrem, L. H. Nguyen, R. R. Burgess, and R. L. Gourse. 2001. Promoter recognition and discrimination by EσS RNA polymerase. Mol. Microbiol. 42:939-954. [DOI] [PubMed] [Google Scholar]
- 12.Green, R. C., and A. J. Darwin. 2004. PspG, a new member of the Yersinia enterocolitica phage shock protein regulon. J. Bacteriol. 186:4910-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 14.Jovanovic, G., L. Weiner, and P. Model. 1996. Identification, nucleotide sequence, and characterization of PspF, the transcriptional activator of the Escherichia coli stress-induced psp operon. J. Bacteriol. 178:1936-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleerebezam, M., W. Crielaard, and J. Tommassen. 1996. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the proton motive force under stress conditions. EMBO J. 15:162-171. [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, C. A., M. J. Fournier, and J. Beckwith. 1985. Escherichia coli 6S RNA is not essential for growth or protein secretion. J. Bacteriol. 161:1156-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd, L. J., S. E. Jones, G. Jovanovic, P. Gyaneshwar, M. D. Rolfe, A. Thompson, J. C. Hinton, and M. Buck. 2004. Identification of a new member of the phage shock protein response in Escherichia coli, the phage shock protein G (PspG). J. Biol. Chem. 279:55707-55714. [DOI] [PubMed] [Google Scholar]
- 18.Luli, G. W., and W. R. Strohl. 1990. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 56:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxson, M. E., and A. J. Darwin. 2006. PspB and PspC of Yersinia enterocolitica are dual function proteins: regulators and effectors of the phage-shock-protein-response. Mol. Microbiol. 59:1610-1623. [DOI] [PubMed] [Google Scholar]
- 20.McFall, E., and E. B. Newman. 1996. Amino acids as carbon sources, p. 358-379. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Minakhin, L., and K. Severinov. 2003. On the role of the Escherichia coli RNA polymerase σ70 region 4.2 and the α-subunit C-terminal domains in promoter complex formation on the extended −10 galP1 promoter. J. Biol. Chem. 278:29710-29718. [DOI] [PubMed] [Google Scholar]
- 23.Model, P., G. Jovanovic, and J. Dworkin. 1997. The Escherichia coli phage-shock-protein (psp) operon. Mol. Microbiol. 24:255-261. [DOI] [PubMed] [Google Scholar]
- 24.Rao, L., W. Ross, J. A. Appleman, T. Gaal, S. Leirmo, P. J. Schlax, M. T. Record, Jr., and R. L. Gourse. 1994. Factor independent activation of rrnBP1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol. 235:1421-1435. [DOI] [PubMed] [Google Scholar]
- 25.Sanderson, A., J. E. Mitchell, S. D. Minchin, and S. J. Busby. 2003. Substitutions in the Escherichia coli RNA polymerase σ70 factor that affect recognition of extended −10 elements at promoters. FEBS Lett. 544:199-205. [DOI] [PubMed] [Google Scholar]
- 26.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Slonczewski, J. L., and J. W. Foster. 1996. pH-regulated genes and survival at extreme pH, p. 1539-1552. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 28.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trotochaud, A. E., and K. M. Wassarman. 2004. 6S RNA function enhances long-term cell survival. J. Bacteriol. 186:4978-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trotochaud, A. E., and K. M. Wassarman. 2005. A highly conserved 6S RNA structure is required for regulation of transcription. Nat. Struct. Mol. Biol. 12:313-319. [DOI] [PubMed] [Google Scholar]
- 31.Wassarman, K. M. 2002. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell 109:141-144. [DOI] [PubMed] [Google Scholar]
- 32.Wassarman, K. M., and G. Storz. 2000. 6S RNA regulates E. coli RNA polymerase activity. Cell 101:613-623. [DOI] [PubMed] [Google Scholar]
- 33.Weiner, L., and P. Model. 1994. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc. Natl. Acad. Sci. USA 91:2191-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiner, L., J. L. Brissette, and P. Model. 1991. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on σ54 and modulated by positive and negative feedback mechanisms. Genes Dev. 5:1912-1923. [DOI] [PubMed] [Google Scholar]
- 35.Willkomm, D. K., J. Minnerup, A. Huttenhofer, and R. K. Hartmann. 2005. Experimental RNomics in Aquifex aeolicus: identification of small non-coding RNAs and the putative 6S RNA homolog. Nucleic Acids Res. 33:1949-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]