Abstract
Ehrlichia canis, a small obligately intracellular, tick-transmitted, gram-negative, α-proteobacterium, is the primary etiologic agent of globally distributed canine monocytic ehrlichiosis. Complete genome sequencing revealed that the E. canis genome consists of a single circular chromosome of 1,315,030 bp predicted to encode 925 proteins, 40 stable RNA species, 17 putative pseudogenes, and a substantial proportion of noncoding sequence (27%). Interesting genome features include a large set of proteins with transmembrane helices and/or signal sequences and a unique serine-threonine bias associated with the potential for O glycosylation that was prominent in proteins associated with pathogen-host interactions. Furthermore, two paralogous protein families associated with immune evasion were identified, one of which contains poly(G-C) tracts, suggesting that they may play a role in phase variation and facilitation of persistent infections. Genes associated with pathogen-host interactions were identified, including a small group encoding proteins (n = 12) with tandem repeats and another group encoding proteins with eukaryote-like ankyrin domains (n = 7).
Ehrlichia spp., α-proteobacteria that belong to the order Rickettsiales, cause diseases of veterinary importance and are also responsible for emerging life-threatening anthropozoonoses. E. canis is a small obligately intracellular, gram-negative, dimorphic bacterium transmitted by the brown dog tick, Rhipicephalus sanguineus, that resides as a microcolony within a membrane-lined intracellular vacuole (morula), primarily within monocytes and macrophages of mammalian hosts (20, 26, 58). E. canis is the primary etiologic agent of canine monocytic ehrlichiosis, has a complex life cycle involving ticks, and is maintained in nature by persistent infection of wild and domestic canids (25). E. canis was first described in 1935 in Algeria (16) and is now recognized to have global distribution, including the United States, Europe, South America, and Asia. E. canis was reported in the United States in 1963 and received more recognition as a pathogen of veterinary importance after outbreaks in British military dogs in Singapore in 1963 and in United States military dogs in Vietnam that resulted in approximately 200 deaths over a 4-year period.
Clinically, E. canis infections progress in three phases: the acute, the subclinical, and the chronic (59, 62). With adequate treatment, dogs typically recover from the acute infections, but untreated or inappropriately treated dogs may develop subclinical persistent infections and thus can become asymptomatic carriers of the organism for years (20, 31). Dogs that do not eliminate the infection can develop a severe chronic form of the disease, where bone marrow failure with anemia leads to opportunistic infections, poor response to treatment, and death from massive hemorrhage.
Recent molecular characterization of E. canis has identified a limited set of major immunoreactive proteins that include glycoproteins and a major outer membrane protein family containing 25 paralogous genes that could be differentially expressed in the tick and mammalian hosts, contributing to persistent infections of the natural hosts (42, 50). Within the group of known major immunoreactive proteins, three glycoproteins have been identified in E. canis (gp36, gp140, and gp200) with corresponding orthologs in the human pathogen, E. chaffeensis (18, 43, 47). These glycoproteins are among the first such proteins described in pathogenic bacteria, and appear to be important targets of the host immune response, attachment to the host cell, and other potentially significant roles in ehrlichial pathobiology (52). Genes for a type IV secretion system have been identified in E. canis (VirB and VirD). Vir proteins play a key role in pathogen-host cell interactions and are associated with protein secretion and inhibition of bacterial inclusion trafficking to the lysosomes; among them VirB9 is a highly antigenic protein expressed in both mammalian and tick cells (21, 51).
The genus Ehrlichia is closely related to the genera Rickettsia, Anaplasma, and Wolbachia, whose members have a similar obligately intracellular existence (19). The genomes of three strains of E. ruminantium (13), and several related organisms, namely, Rickettsia marginale (6), R. conorii (49), R. typhi (44), R. prowazekii (3), R. akari, R. sibirica, Wolbachia sp. (from Drosophila melanogaster) (66), and Wolbachia sp. strain TRS (from Brugia malayi) (22), have been sequenced previously. They are all characterized by small gene numbers and a low coding density as a result of convergent reductive evolution in response to their intracellular existence.
The pathogenesis of canine and human ehrlichioses, as well as the host response to Ehrlichia spp., remains poorly understood. The E. canis genome was sequenced to advance understanding of the pathobiology of this obligately intracellular pathogen and its evolutionary relationship with related organisms and to identify potentially immunoprotective antigens that may be critical for development of effective vaccines.
MATERIALS AND METHODS
Genome sequencing and assembly.
The complete genome of E. canis was sequenced at the Joint Genome Institute (JGI) by using a combination of 3-kb, 8-kb, and fosmid (40-kb) libraries. Due to contamination complexities inherent in isolating genomic DNA from obligately intracellular microbes, four libraries had to be constructed, each with various amounts of host DNA contamination, and >40,000 reads were generated (roughly 20-fold coverage), though only 61% of the reads were incorporated into the final E. canis genome assembly. Library construction, sequencing, finishing, and automated annotation steps were used as described previously (9). Predicted coding sequences were subjected to manual analysis using JGI's gene models quality assessment pipeline (available at Genome Biology Program's Web site). The predicted functions were further analyzed by using the Integrated Microbial Genomes (IMG) annotation pipeline (http://img.jgi.doe.gov) (38).
Genome analysis.
Comparative analysis of E. canis and related organisms (Ehrlichia, Anaplasma, and Rickettsia spp.) was performed by using a set of tools available in IMG. Unique and orthologous E. canis genes were identified by using BLASTp (cutoff scores of E < 10−2 and 20% identity and reciprocal hits with cutoffs of E < 10−5 and 30%, respectively). Signal peptides and transmembrane helices were predicted by using SignalP 3.0 (4) and TMHMM 2.0 (32) set at default values. Nuclear localization signal (NLS) sequences were predicted by using PredictNLS server (12); glycosylation sites were predicted by using a eukaryotic prediction server, NetOGlyc 3.1 (30); and repeats were identified by using MUMmer 3.0 (33). Ser/Thr clusters were considered to be regions of 15 tandem amino acids containing at least five Ser/Thr residues. In order to identify proteins with an increased Ser/Thr or Cys content, we calculated the average content of all of the proteins of the genome in these residues. Proteins that had a percent content of more than the average plus two standard deviations were considered to be rich in these residues. Riboswitch sequences were checked by using the RibEx Web server (1).
GenBank accession numbers.
The sequence data described here have been deposited in GenBank (CP000107).
RESULTS AND DISCUSSION
General features of the genome.
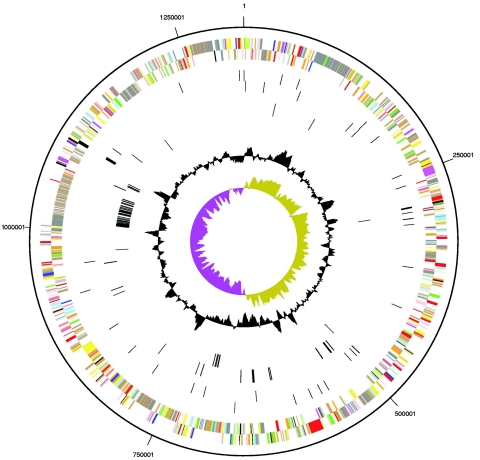
The genome of E. canis consists of a single circular chromosome spanning 1,315,030 nucleotides and has a G+C content of 28.96%. The origin of replication was mapped across a region between kb 630 and 670, which exhibits characteristics that are frequently associated with the bacterial termination of replication, such as GC-skew and a large number of duplications and rearrangements. dnaA is usually associated with origin of replication, but in E. canis, dnaA is located 250 kb downstream, a finding consistent with its location in other Ehrlichia spp. genomes (13). The E. canis genome is smaller than other ehrlichiae, which are approximately 1.5 Mb, but it has a similar number of predicted genes. Hence, E. canis has a higher density of predicted functional genes, but the ratio of coding to noncoding sequence still remains substantially lower than the average bacterium (Table 1). A total of 984 genes were identified in the E. canis genome, including one copy of each of the rRNA genes (5S, 16S, and 23S). The 5S and 23S rRNA genes form an operon, while the 16S rRNA gene is separated by ∼0.8 Mb. This characteristic is unusual for bacterial genomes, which typically have one to multiple copies of rRNAs contained in 16S-23S-5S operons, but is a common feature of rickettsial organisms of genera Ehrlichia, Rickettsia, Anaplasma, and Wolbachia. Thirty-six tRNA genes were identified, which include cognates for all amino acids. A probable function was assigned to 654 genes, and 290 were annotated as coding sequences without function assignment (Fig. 1).
TABLE 1.
Comparison of the genome properties of members of the Rickettsialesa
| Organism name | No of genes | %G+C | No. of bases | No. of coding bases | Coding bases/total bases (%) |
|---|---|---|---|---|---|
| Anaplasma marginale strain Maries | 986 | 49.76 | 1197,687 | 1024,175 | 85 |
| Ehrlichia canis strain Jake | 967 | 28.96 | 1315,030 | 955,386 | 73 |
| Ehrlichia chaffeensis sapulpa | 833 | 29.98 | 1,005,812 | 731,291 | 73 |
| Ehrlichia ruminantium strain Welgevonden (CIRAD) | 997 | 27.48 | 1,512,977 | 967,368 | 64 |
| Ehrlichia ruminantium strain Gardel | 989 | 27.51 | 1,499,920 | 968,505 | 64 |
| Ehrlichia ruminantium strain Welgevonden (ARC-OVI) | 959 | 27.48 | 1,516,355 | 960,154 | 63 |
| Rickettsia akari strain Hartford | 1,217 | 32.33 | 1,231,060 | 946,961 | 77 |
| Rickettsia conorii strain Malish 7 | 1,410 | 32.44 | 1268,755 | 1,025,607 | 81 |
| Rickettsia prowazekii Madrid E | 870 | 29.00 | 1,111,523 | 845,091 | 76 |
| Rickettsia sibirica 246 | 1,234 | 32.47 | 1,250,021 | 969,726 | 77 |
| Rickettsia typhi strain Wilmington | 874 | 28.92 | 1,111,496 | 843,854 | 76 |
| Wolbachia endosymbiont strain TRS of Brugia malayi | 842 | 34.18 | 1,080,084 | 730,178 | 68 |
| Wolbachia pipientis wMel | 1,232 | 35.23 | 1,267,782 | 1,021,568 | 81 |
| Bacteria in IMG (v1.1) | 1,180,331 | 1,333,547,111 | 1,121,629,579 | 84 |
Data obtained from IMG (v1.1; http://img.jgi.doe.gov).
FIG. 1.
Circular representation of the genome of E. canis. From outside to inside, the first two circles represent COG assignment for predicted coding sequences on the plus and minus strands, respectively. Colors indicate the following: dark gray, hypothetical proteins; light gray, conserved hypothetical and unknown function; brown, general function prediction; red, replication and repair; green, energy metabolism; blue, carbon and carbohydrate metabolism; cyan, lipid metabolism; magenta, transcription; yellow, translation; orange, amino acid metabolism; pink, metabolism of cofactors and vitamins; light red, purine and pyrimidine metabolism; lavender, signal transduction; and sky blue, cellular processes. The third and fourth circles represent RNA genes. The fifth and sixth circles represent unique coding sequences. The two innermost circles represent the %G+C content and G+C skew values, respectively.
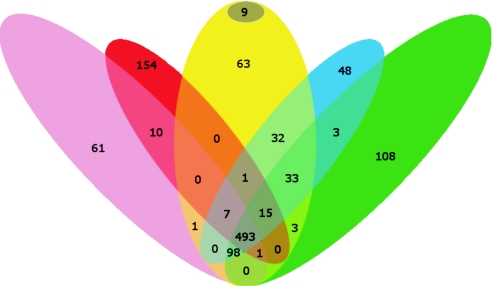
This genome has undergone a severe loss of metabolic pathway enzymes, as a result of reductive evolution (45). A basic core of biochemical functions shared by other members of the same genus is dictated by their intracellular lifestyle (see Fig. 3).
FIG. 3.
Venn diagram showing comparison of conserved and unique genes in Ehrlichia canis (yellow), Ehrlichia spp. (blue), Wolbachia spp. (pink), Rickettsia spp. (red), and Anaplasma spp. (green). The dark circle represents genes unique in E. canis.
Detailed information about the genome properties and genome annotation can be obtained at http://img.jgi.doe.gov/pub/main.cgi?page = taxonDetail&taxon_oid = 623500000.
Information transfer.
Genes encoding enzymes for DNA replication and repair, RNA synthesis and degradation, ribosomal proteins (except L30 and S22), and UV excision nucleases UvrA and D, as well as the RecA, F, O, R, G recombination synaptic, the RuvABC postsynaptic pathways, and a gene for tmRNA, responsible for tagging incomplete proteins for hydrolysis on stalled ribosomes, were identified. Several enzymes present in Rickettsia spp. were absent from E. canis and other Ehrlichia spp. (e.g., DNA polymerase III, chi subunit, recombination protein N [RecN], and RNase PH).
In addition, E. canis does not have a formamidopyrimidine DNA glycosidase, an enzyme that mediates recovery from mutagenesis or lethal cell injury caused by alkylating agents, and removes an oxidatively damaged form of guanine (36). This gene was identified close to the termination of the replication site in E. ruminantium strains, but the high degree of shuffling of this region may be responsible for the loss of this gene in E. canis. This gene is present in other rickettsiae, and the loss of this enzyme and potential for an increased A/T mutational pressure and sensitivity to mutagenic agents could be an area of further investigation.
A set of common chaperonins was also identified in the genome (groEL, groES, dnaK, dnaJ, hslU, hslV, and htpG), and secA protein-dependent and secA-independent (type I) protein secretion systems for protein translocation were also present in the organism. In addition, genes for twin arginine translocases (tatA and tatC) were also identified.
Transcriptional regulation.
Members of the order Rickettsiales have a small number of transcriptional regulators, a trait also observed in other intracellular pathogens. In E. canis, six transcriptional regulators were identified belonging to the COG groups COG1396, COG1475, COG0789, COG0745, COG5606, and COG2932. E. ruminantium has one additional transcriptional regulator (COG1959) that is not present in E. canis. Rickettsia, Anaplasma, and Wolbachia spp. also have a small number of transcriptional regulators, but these are not necessarily representatives of the same groups (Table S1 in the supplemental material).
It is known that posttranscriptional regulation based on antisense RNA-mRNA interactions also plays a significant role in gene regulation in bacteria and is mediated by the Sm -like protein Hfq (39) or the presence of metabolite-binding domains in certain mRNA molecules (riboswitches) (37, 65). However, an hfq ortholog was not identified in any of the rickettsial genomes, indicating that a similar mechanism does not exist or that posttranscriptional regulation occurs by a different mechanism in these organisms; furthermore, sequences similar to riboswitches were not identified.
The small number of transcription regulators and the absence of alternative regulation methods, common features among these organisms, appear to be a result of the reductive evolution coupled to the diminished demand for regulation due to the small genome size, as well as the relatively static conditions provided by the intracellular environment of the host cell (7).
Central metabolic pathways and transporters.
E. canis is an aerobic organism that is unable to use glucose or fructose as carbon or energy source, since transport systems and essential enzymes for the utilization of these substrates were not identified. However, amino acid transporters and enzymes for the utilization of aspartate, proline, glutamate, glutamine, and arginine were identified, indicating that amino acids constitute the main energy and carbon source for E. canis. All enzymes of the tricarboxylic acid pathway were present, as well as enzymes that allow the transfer of amino acids to the tricarboxylic acid cycle. A gluconeogenesis pathway exists, which terminates at 6P-fructose and a complete nonoxidative pentose-phosphate pathway. E. canis has biosynthetic pathways for proline, glutamate, glutamine, aspartate, lysine, and arginine. A complete set of enzymes involved in the biosynthesis of purine samples and pyrimidines, as well as sets of enzymes involved in the biosynthesis of lipids and phospholipids, enzymes for the metabolism of cofactors (i.e., riboflavin, nicotinate and nicotinamide, biotin, folate, and porphyrin) and a partial pathway for ubiquinone biosynthesis, and genes for enzyme complexes typical of aerobic respiration (i.e., ATP synthase complex, NADH dehydrogenase complex, the cytochrome oxidase complex, and a complete succinate dehydrogenase complex) were also present. Unlike Rickettsia spp., E. canis does not seem to have translocases for ATP or NADH, so it appears to rely on its own intracellular ATP production (3, 24). The genome contains several orthologs involved in membrane transport systems that can supply the necessary metabolites for the absent or incomplete pathways.
Cell wall components.
Enzymes relevant to the biosynthesis of lipid A and murein sacculus and to the metabolism of peptidoglycans and amino sugars are not present. This finding is in agreement with experimental data suggesting that peptidoglycan and lipopolysaccharide are absent from the outer membranes of Ehrlichia spp. E. chaffeensis and A. phagocytophilum acquire cholesterol from the host and incorporate it into the outer membrane possibly to compensate for the decreased membrane stability due to the lack of peptidoglycan (34). Enzymes involved in production or biosynthetic modification of cholesterol were not identified in E. canis, suggesting that cholesterol is acquired from the host.
A total of 310 proteins (33% of the proteome) were predicted to be secreted or contain transmembrane helices, indicating that these proteins could interact to form a complex, protein dominated, cell wall structure to compensate for the absence of the peptidoglycan component. A large number of these proteins (n = 121) had an unknown function, similar to what was observed in other Ehrlichia spp. A large subset (n = 179) were predicted to have a signal peptide at the N terminus; however, many of these proteins (n = 39) did not have predicted transmembrane helices, suggesting that they are secreted.
It is likely that ehrlichial outer membranes are primarily dependent on covalent and noncovalent association between outer membrane proteins for structural rigidity (41, 64). Disulfide bond formation linkages have been demonstrated between major surface proteins (Msp) in Anaplasma spp. (64). A group of 36 proteins appears to contain an increased number of cysteine residues, 15 of which are predicted to be either secreted or membrane associated. Their increased potential to create disulfide bonds suggests involvement in the formation of a protein envelope around the cell (Table S2 in the supplemental material) (41, 64).
Proteins with tandem repeats play important roles in pathogenicity and pathogen-host cell interactions (10). Twelve proteins containing tandem repeats were identified (Ecaj_0017, Ecaj_0060, Ecaj_0062, Ecaj_0109, Ecaj_0220, Ecaj_0221, Ecaj_0387, Ecaj_0472, Ecaj_0529, Ecaj_0530, Ecaj_07, and Ecaj_0716). Most of these proteins (except Ecaj_0062, Ecaj_0220, Ecaj_0221, and Ecaj_0715) have orthologs in other genomes from Rickettsiales that also contain tandem repeats.
Adaptation of the pathogen to the host, especially in persistent infections, dictates that the organism has mechanisms that allow it to evade host defenses. Most likely these are related to reduction in the host innate and adaptive immune responses, for example, by alteration of the surface architecture and/or the expression of different protein variants. Ehrichia spp. lack peptidoglycan and lipopolysaccharide, major pathogen-associated molecular patterns found in the cell walls of other gram-negative bacteria. This condition suggests that ehrlichial cell walls have fundamental structural and composition differences and thus may not be recognized by innate pattern recognition receptors such as Toll-like receptors 2 and 4 (35).
Paralogous protein families and repeats.
Compared to other Ehrlichia and Anaplasma spp., E. canis exhibits a smaller number of short sequence repeats, which are distributed evenly within the genome. Conversely, E. canis has a similar number of paralogous protein families (15) compared to E. ruminantium strains (strains Welgevonden [CIRAD] 21, Gardel 19, and Welgevonden [ARC-OVI] 13) and a similar number of paralogous genes.
The production of different allelic forms of proteins with tandem repeats could account for the surface diversity and avoidance of immune recognition. Similar proteins with tandem repeats exist in other rickettsial genomes, suggesting a similar role and possible function in pathogenicity. Two of these proteins, gp140 and gp36 (Ecaj_0017 and Ecaj_0109), are glycosylated and predicted to act as adhesins on the basis of the sequence similarity with proteins of E. chaffeensis and E. ruminantium, respectively (15, 43, 67), and a similar mode of action for the others cannot be excluded. Furthermore, Ecaj_0062 is similar to WASP family proteins, which interact with the cytoskeleton facilitating bacterial motility (23, 29). Although motility has not been demonstrated with Ehrlichia spp., this protein could mediate a different type of interaction between the pathogen and the host cell.
An operon of 22 paralogous membrane proteins was identified. These are major antigenic proteins that have been previously identified and molecularly characterized, and their role in antigenic diversity and immunoprotection has been investigated (42, 50, 54, 55). Notably, E. canis contains more members (25) in this family than other Ehrlichia spp. (50). This appears to be a result of a recent duplication that created a new locus containing three additional p28/p30 genes. The large number of major antigenic proteins certainly could facilitate the antigenic variation observed with these organisms; the differential expression of these proteins could allow the organism to avoid host immune surveillance and establish persistent infections (40, 50, 55, 67). Only a few of these proteins appear to be predominantly expressed in cultured tick and mammalian cells (54), and one p28/30 gene transcript was detected in E. canis-infected ticks; however, multiple transcripts were detected in infected dogs, suggesting that differential expression may not occur in the mammalian host (63), and thus the p28/p30 proteins may not be involved in immune evasion.
Ankyrin domain and actin polymerization proteins.
Ankyrin repeats have been found in numerous proteins mediating specific protein-protein interactions (46). Five proteins that contain ankyrin domains were identified in the genome (Ecaj_0052, Ecaj_0221, Ecaj_0365, Ecaj_0387, and Ecaj_0627), which have no assigned function. Three of these proteins (Ecaj_0221, Ecaj_0365, and Ecaj_0387) also exhibited a high Ser/Thr content.
One of these proteins, gp200 (Ecaj_0365), is translocated to the nucleus, where it interacts with the chromatin (47), potentially modulating host cell gene expression, an observation similarly reported with an ankyrin domain-containing protein (AnkA) of A. phagocytophilum (8). Interestingly, the Ehrlichia spp. proteins do not have classic NLS sequences. This absence supports a different mechanism for protein translocation to the host cell nucleus.
It has been reported that some members of the Rickettsiales utilize host cell actin polymerization to achieve bacterial motility (23, 29). The actin polymerization depends on activation of the host Arp2/3 complex by the WASP family protein RickA. Although motility has not been demonstrated with Ehrlichia spp., Ecaj_0062 exhibits similarity (E = 5 × 10−5) to RickA proteins, suggesting that a mechanism of interacting with the cytoskeleton exists in this pathogen.
Furthermore, it has been shown in Pseudomonas aeruginosa that AnkB, an ankyrin repeat-containing protein, is essential for optimal activity of periplasmic catalase (28). It is therefore reasonable to propose that these ankyrin-containing proteins may play a significant role in the adaptation of the pathogen to the host cell, mediating a series of interactions or reprogramming of the host cell functions.
Poly(G-C) tracts.
Poly(G-C) tracts which exhibit variation in their length between otherwise identical clones were identified in seven predicted E. canis protein-coding genes; conversely, E. ruminantium appears to have only one (13). Five of the poly(G-C) tract genes (Ecaj_0063, Ecaj_0065, Ecaj_0069, Ecaj_0070, and Ecaj_0072) appear to be positioned in a cluster of 12 paralogous proteins unique to Rickettsiales, whereas the other two (Ecaj_0423 and Ecaj_0496) are in isolated loci unique to E. canis. Among these proteins, Ecaj_0063, Ecaj_0069, and Ecaj_0070 are predicted to contain transmembrane helices (Table S2 in the supplemental material). Thus, they can act as membrane-bound antigens, and modulation of their expression or production of variants due to the homopolymeric tracts could increase the diversity of the exposed surface. However, to date, none of these proteins has been identified as such in E. canis, E. chaffeensis, or E. ruminantium, and their role in immune evasion remains to be elucidated.
Host interaction and pathogenicity-associated genes.
In the genome of E. canis we were able to identify two clusters of Vir homologous proteins. The first cluster contains virB8/virB9/virB10/virB11/virD4, while the second one contains virB3/virB4/virB6 and three large virB6-related genes. The organization of virB genes and the presence of the three large genes appears to be common among Ehrlichia and Rickettsia spp., and they have been associated with the secretion of toxins and intracellular existence (21, 51, 57).
Serine-threonine bias and ehrlichial glycoproteins.
Glycosylated proteins can play a role in the pathogen-host interaction, as well as in the assembly of the protein envelope, and could be candidates for a vaccine and therapeutic interventions (15, 61). In recent years, protein glycosylation pathways of bacteria have been increasingly studied (5, 53, 56, 60, 61).
Glycoproteins have been identified in E. canis, and these proteins exhibit high serine/threonine content, suggesting that glycans are attached to these residues by O-linkages. Our analysis revealed 180 proteins that exhibit either clustered Ser/Thr residues, or an increased Ser/Thr content. This group contains proteins that have already been identified as glycoproteins (gp36/Ecaj_0109, gp140/Ecaj_0017, gp200/Ecaj_0365, and p28/Ecaj_0917). Many (n = 69) of these are also predicted to be secreted or contain transmembrane helices. These proteins could appear on the surface of the organism, exhibiting different patterns of glycosylation, thus creating a large number of variants. Interestingly, almost all of the proteins with tandem repeats (except Ecaj_0062 and Ecaj_0472) appear also to be putative glycoproteins. Similarly, proteins with homopolymeric repeats (Ecaj_0063, Ecaj_0070, and Ecaj_0072), as well as VirB components, also exhibit Ser/Thr clusters. Compared to other genera in the Rickettsiales, Ehrlichia, and Anaplasma spp. possess a significantly higher proportion of Ser/Thr-rich proteins. This observation, combined with the absence of the peptidoglycan cell wall content, implies a significant role for glycosylated proteins as cell wall constituents and a potential for antigenic variability of the outer cell surface.
Some of the ehrlichial proteins are predicted to have glycosylation sites based on the eukaryotic glycoprotein database, suggesting that they can be glycosylated by the host machinery when secreted; however, glycosylation of recombinant forms of these proteins is also performed by Escherichia coli, demonstrating that protein glycosylation patterns are also recognized by the bacterial glycosylation machinery (17). Therefore, it is possible that Ehrlichia spp. have enzymes that mediate protein glycosylation.
Several proteins that participate in protein glycosylation have been identified in Campylobacter jejuni, Neisseria meningitidis, and other species, whereas a large number of enzymes are still unknown (61). In several cases, ehrlichial proteins that were annotated by using a general function prediction exhibit similarity to genes related to protein glycosylation (ABC transporter similar to WlaB and dehydrogenase similar to PtmA). Furthermore, in C. jejuni several proteins belonging to protein glycosylation pathways appear to be positionally clustered with their target genes (60). Although we were not able to conclusively identify proteins that participate in protein glycosylation pathways in the E. canis genome, there are three proteins without any assigned function (Ecaj_0897, Ecaj_0900, and Ecaj_0902) that exhibit low similarity to protein glycosylation-related enzymes (Table S3 in the supplemental material) and are present in a locus adjacent to Ecaj_0917, which has been shown to be glycosylated (55). These proteins are candidates for further research.
Comparison with other rickettsial genomes.
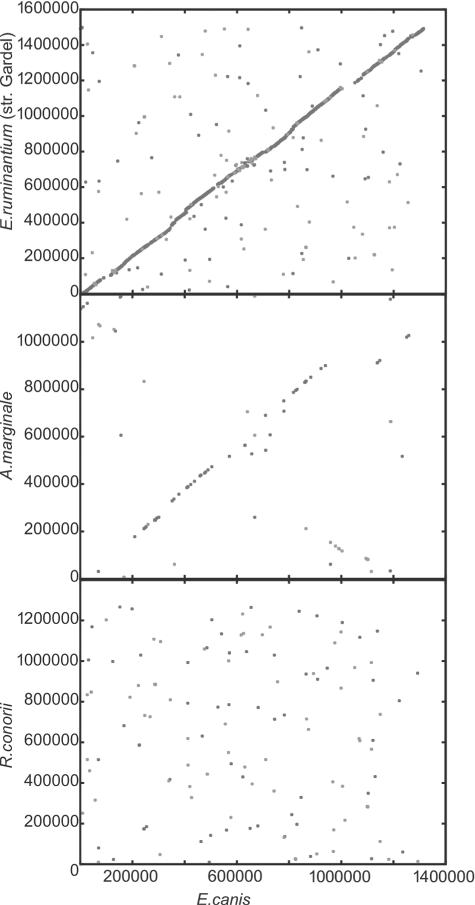
The E. canis genome exhibits almost complete synteny with Ehrlichia ruminantium (Welgevoden and Gardel strains) and, to a lower extent, with A. marginale (St. Maries strain), whereas no significant synteny is observed compared to other members of the Rickettsiales (Fig. 2). Comparison with the completely sequenced rickettsial genomes is illustrated in Fig. 3. E. canis shares a core of 834 proteins (88% of the proteome) with E. ruminantium strains and 493 proteins (52% of the proteome) with other complete genomes of the Rickettsiales. Thirty-two proteins corresponding to 3.1% of the proteome are common among Ehrlichia, but orthologs for these proteins are not present in the other members of Rickettsiales. Among them Ecaj_0367, Ecaj_368, Ecaj_0541, and Ecaj_0628 have assigned functions and point to acquired functions in this genus or functions lost from the Rickettsiales. E. canis contains 72 (7.6% of the proteome) genes that are unique to members of the Rickettsiales, 9 of which (0.9% of the proteome) do not exhibit any similarity to other proteins of bacterial origin. The function of all of these proteins is unknown.
FIG. 2.
Synteny plots between E. canis (horizontal axis) and E. ruminantium (strain Gardel), A. marginale, and R. conorii. The numbers represent base pairs.
A small number of insertions and deletions, as well as local inversions, were identified by comparing those genomes. The first insertion in E. canis includes genes Ecaj_0479 to Ecaj_0485, the second includes genes Ecaj_0716 to Ecaj_0754, which are predicted to be either secreted or membrane proteins. Finally, Ecaj_0831 to Ecaj_0833 represents a unique region in E. canis and includes three major antigenic proteins from the map locus that appear to be duplicated. This duplication is identical to a region in the primary p28/p30 locus, suggesting that this is a recent event. Comparison of E. canis with other rickettsial genomes revealed 53 proteins of unknown function unique among the Rickettsiales, and 2 proteins that were unique to E. canis (Fig. 1). Most of these proteins are members of two groups of hypothetical proteins (Eca_479 to Ecaj_0485 and Ecaj_0716 to Ecaj_0772).
Pseudogenes.
Inspection of the genome of E. canis revealed a relatively small number of pseudogenes. Seventeen fragments that correspond to 11 genes were identified. These pseudogenes correspond to full-length functional genes present in the E. canis genome. In the three strains of E. ruminantium, the pseudogenes appear to be consistent and homologous. In contrast, pseudogenes in the E. canis genome are found in different relative positions and correspond to fragments from different genes, an indication that this genome has reached a minimum number of genes and is not currently losing genes. It appears that duplication events have led to the expansion of the genome, competing with gene loss as a result of reductive evolution, creating paralogous genes and gene fragments identified as pseudogenes (13). The fact that these fragments display no homology between ehrlichial genomes suggests that duplication events occurred independently in Ehrlichia after the separation of the species.
The intracellular lifestyle, which restricts access of this bacterium to new genes, combined with the absence of any apparent transposable elements, results in low numbers of horizontally transferred genes (2). Although there are genes in E. canis without orthologs among other Rickettsiales members, but with homologs in other organisms, we were not able to identify any genes that showed higher similarities to counterparts in organisms from other genera as candidates for horizontal transfer events.
Evolutionary rates of substitution and duplication.
Considerations of the specific associations of Anaplasma and Ehrlichia spp. with their tick hosts allow us to use the tick fossil record to calibrate the frequency of appearance of duplicated genes and gene fragments within E. canis. Given that all species in the genera Anaplasma and Ehrlichia, as recently redefined (19) on the basis of 16S rRNA phylogeny, are associated with hard tick hosts, we assume that the phylogenetic clade containing these two genera is not older than the earliest appearance of hard ticks in the fossil record, which has been established to be 120 million years ago (MYA) (14). Moreover, Anaplasma spp. are found within tick hosts belonging to the two existing groups of hard ticks, whereas Ehrlichia spp. are restricted to Metastriata ticks (19), which appear in the fossil record no earlier than 40 MYA (14). Therefore, we assume that the Ehrlichia genus cannot be older than 40 MY and that this is the oldest possible divergence date for E. canis and E. ruminantium. This assumption can be corroborated by analysis of 16S rRNA divergence of A. marginale, E. ruminantium, and E. canis. The distance (uncorrected p or number of nucleotide differences per site) between E. canis and A. marginale 16S rRNA is 0.077, whereas the average distance between E. canis and the E. ruminantium strains is 0.026, as would be expected for a time of divergence three times shorter, assuming equal rates of 16S rRNA evolution.
In comparing the E. canis genome to those of the three sequenced strains of E. ruminantium and to A. marginale, we have detected 16 gene paralogs that have appeared by gene duplication within the lineage leading to E. canis after its divergence from E. ruminantium. Given a maximal divergence time of 40 MY for the two Ehrlichia species, this represents a minimal rate of generation of duplicate genes of 0.4 new paralogs per MY. This can be compared to the rate of fixation of gene duplicates in E. coli, which has been estimated to be around 2 per MY (27) or five times higher. Similarly, the rate of 16S rRNA evolution in Ehrlichia (0.48 changes per MY or 1% sequence divergence in 300 MY) is around six times slower than the estimated rate for E. coli (1% 16S rRNA divergence per 50 MY) (48). The similar decrease of 16S rRNA divergence and the rate of fixation of duplicates in Ehrlichia suggest that both of these rates are responding to common factors. An important factor that could affect both rates is generation time and, remarkably, the number of generations per year has been estimated to be six times less in obligately intracellular bacteria than in E. coli (11). In addition, we detected 17 pseudogenic fragments that have originated by gene duplication within the lineage leading to E. canis. These fragments total 3,459 bp and represent a minimal rate of generation of duplicated base pairs of 86.5 per MY. This rate can be interpreted as a lower boundary to the rate of neutral duplication for coding bases, since it only accounts for bases that have survived deletion pressure after duplication.
Conclusions.
The genomic sequence of E. canis provides the resources necessary for a detailed analysis of this pathogen and for thorough understanding of the molecular basis of host-pathogen interactions. Ehrlichia spp. are gram-negative bacteria but are deficient in structural components, including peptidoglycan and lipopolysaccharide. The lack of these constituents has resulted in the development of complex protein structures in the outer membrane. Proteins that have a potential role in the adaptive strategy of this organism by assisting immune evasion, as well as proteins that mediate host cell -pathogen interactions, were identified, providing subjects for further research for vaccine and drug development.
Supplementary Material
Acknowledgments
This study was performed under the auspices of the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Livermore National Laboratory, under contract W-7405-Eng-48, the Lawrence Berkeley National Laboratory under contract DE-AC02-05CH11231, and the Los Alamos National Laboratory under contract W-7405-ENG-36; the Clayton Foundation for Research; and the University of Texas Medical Branch Sealy Center for Vaccine Development.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abreu-Goodger, C., and E. Merino. 2005. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 33:W690-W692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, S. G., and C. G. Kurland. 1998. Reductive evolution of resident genomes. Trends Microbiol. 6:263-268. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Benz, I., and M. A. Schmidt. 2002. Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45:267-276. [DOI] [PubMed] [Google Scholar]
- 6.Brayton, K. A., L. S. Kappmeyer, D. R. Herndon, M. J. Dark, D. L. Tibbals, G. H. Palmer, T. C. McGuire, and D. P. Knowles, Jr. 2005. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc. Natl. Acad. Sci. USA 102:844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cases, I., V. de Lorenzo, and C. A. Ouzounis. 2003. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 11:248-253. [DOI] [PubMed] [Google Scholar]
- 8.Caturegli, P., K. M. Asanovich, J. J. Walls, J. S. Bakken, J. E. Madigan, V. L. Popov, and J. S. Dumler. 2000. ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect. Immun. 68:5277-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chain, P., J. Lamerdin, F. Larimer, W. Regala, V. Lao, M. Land, L. Hauser, A. Hooper, M. Klotz, J. Norton, L. Sayavedra-Soto, D. Arciero, N. Hommes, M. Whittaker, and D. Arp. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Citti, C., M. F. Kim, and K. S. Wise. 1997. Elongated versions of Vlp surface lipoproteins protect Mycoplasma hyorhinis escape variants from growth-inhibiting host antibodies. Infect. Immun. 65:1773-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, M. A., N. A. Moran, and P. Baumann. 1999. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol. Biol. Evol. 16:1586-1598. [DOI] [PubMed] [Google Scholar]
- 12.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins, N. E., J. Liebenberg, E. P. de Villiers, K. A. Brayton, E. Louw, A. Pretorius, F. E. Faber, H. van Heerden, A. Josemans, M. van Kleef, H. C. Steyn, M. F. van Strijp, E. Zweygarth, F. Jongejan, J. C. Maillard, D. Berthier, M. Botha, F. Joubert, C. H. Corton, N. R. Thomson, M. T. Allsopp, and B. A. Allsopp. 2005. The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proc. Natl. Acad. Sci. USA 102:838-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Fuente, J. 2003. The fossil record and the origin of ticks (Acari: Parasitiformes: Ixodida). Exp. Appl. Acarol. 29:331-344. [DOI] [PubMed] [Google Scholar]
- 15.de la Fuente, J., J. C. Garcia-Garcia, A. F. Barbet, E. F. Blouin, and K. M. Kocan. 2004. Adhesion of outer membrane proteins containing tandem repeats of Anaplasma and Ehrlichia species (Rickettsiales: Anaplasmataceae) to tick cells. Vet. Microbiol. 98:313-322. [DOI] [PubMed] [Google Scholar]
- 16.Donatein, A., and F. Lestoquard. 1935. Existence and Algerie d'une rickettsia du chien. Bull. Soc. Pathol. Exot. 28:418-419. [Google Scholar]
- 17.Doyle, C. K., K. A. Nethery, V. L. Popov, and J. W. McBride. 2006. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 74:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle, C. K., X. Zhang, V. L. Popov, and J. W. McBride. 2005. An immunoreactive 38-kilodalton protein of Ehrlichia canis shares structural homology and iron-binding capacity with the ferric ion-binding protein family. Infect. Immun. 73:62-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dumler, J. S., A. F. Barbet, C. P. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 20.Ewing, S. A. 1963. Observations on leukocytic inclusion bodies from dogs infected with Babesia canis. J. Am. Vet. Med. Assoc. 143:503-506. [PubMed] [Google Scholar]
- 21.Felek, S., H. Huang, and Y. Rikihisa. 2003. Sequence and expression analysis of virB9 of the type IV secretion system of Ehrlichia canis strains in ticks, dogs, and cultured cells. Infect. Immun. 71:6063-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster, J., M. Ganatra, I. Kamal, J. Ware, K. Makarova, N. Ivanova, A. Bhattacharyya, V. Kapatral, S. Kumar, J. Posfai, T. Vincze, J. Ingram, L. Moran, A. Lapidus, M. Omelchenko, N. Kyrpides, E. Ghedin, S. Wang, E. Goltsman, V. Joukov, O. Ostrovskaya, K. Tsukerman, M. Mazur, D. Comb, E. Koonin, and B. Slatko. 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouin, E., C. Egile, P. Dehoux, V. Villiers, J. Adams, F. Gertler, R. Li, and P. Cossart. 2004. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature 427:457-461. [DOI] [PubMed] [Google Scholar]
- 24.Greub, G., and D. Raoult. 2003. History of the ADP/ATP-translocase-encoding gene, a parasitism gene transferred from a Chlamydiales ancestor to plants 1 billion years ago. Appl. Environ. Microbiol. 69:5530-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groves, M. G., G. L. Dennis, H. L. Amyx, and D. L. Huxsoll. 1975. Transmission of Ehrlichia canis to dogs by ticks (Rhipicephalus sanguineus). Am. J. Vet. Res. 36:937-940. [PubMed] [Google Scholar]
- 26.Hildebrandt, P. K., J. D. Conroy, A. E. McKee, M. B. Nyindo, and D. L. Huxsoll. 1973. Ultrastructure of Ehrlichia canis. Infect. Immun. 7:265-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hooper, S. D., and O. G. Berg. 2003. On the nature of gene innovation: duplication patterns in microbial genomes. Mol. Biol. Evol. 20:945-954. [DOI] [PubMed] [Google Scholar]
- 28.Howell, M. L., E. Alsabbagh, J. F. Ma, U. A. Ochsner, M. G. Klotz, T. J. Beveridge, K. M. Blumenthal, E. C. Niederhoffer, R. E. Morris, D. Needham, G. E. Dean, M. A. Wani, and D. J. Hassett. 2000. AnkB, a periplasmic ankyrin-like protein in Pseudomonas aeruginosa, is required for optimal catalase B (KatB) activity and resistance to hydrogen peroxide. J. Bacteriol. 182:4545-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeng, R. L., E. D. Goley, J. A. D'Alessio, O. Y. Chaga, T. M. Svitkina, G. G. Borisy, R. A. Heinzen, and M. D. Welch. 2004. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 6:761-769. [DOI] [PubMed] [Google Scholar]
- 30.Julenius, K., A. Molgaard, R. Gupta, and S. Brunak. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O glycosylation sites. Glycobiology 15:153-164. [DOI] [PubMed] [Google Scholar]
- 31.Kelch, W. J. 1984. The canine ehrlichiosis (tropical canine pancytopenia) epizootic in Vietnam and its implications for the veterinary care of military working dogs. Mil. Med. 149:327-331. [PubMed] [Google Scholar]
- 32.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 33.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, M., and Y. Rikihisa. 2003. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect. Immun. 71:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, M., and Y. Rikihisa. 2004. Ehrlichia chaffeensis downregulates surface Toll-like receptors 2/4, CD14 and transcription factors PU.1 and inhibits lipopolysaccharide activation of NF-κB, ERK 1/2 and p38 MAPK in host monocytes. Cell Microbiol. 6:175-186. [DOI] [PubMed] [Google Scholar]
- 36.Lu, A. L., X. Li, Y. Gu, P. M. Wright, and D. Y. Chang. 2001. Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem. Biophys. 35:141-170. [DOI] [PubMed] [Google Scholar]
- 37.Mandal, M., B. Boese, J. E. Barrick, W. C. Winkler, and R. R. Breaker. 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113:577-586. [DOI] [PubMed] [Google Scholar]
- 38.Markowitz, V. M., F. Korzeniewski, K. Palaniappan, E. Szeto, G. Werner, A. Padki, X. Zhao, I. Dubchak, P. Hugenholtz, I. Anderson, A. Lykidis, K. Mavromatis, N. Ivanova, and N. C. Kyrpides. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34:D344-D348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masse, E., N. Majdalani, and S. Gottesman. 2003. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6:120-124. [DOI] [PubMed] [Google Scholar]
- 40.McBride, J. W., R. E. Corstvet, S. D. Gaunt, C. Boudreaux, T. Guedry, and D. H. Walker. 2003. Kinetics of antibody response to Ehrlichia canis immunoreactive proteins. Infect. Immun. 71:2516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McBride, J. W., L. M. Ndip, V. L. Popov, and D. H. Walker. 2002. Identification and functional analysis of an immunoreactive DsbA-like thio-disulfide oxidoreductase of Ehrlichia spp. Infect. Immun. 70:2700-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride, J. W., X. J. Yu, and D. H. Walker. 2000. A conserved, transcriptionally active p28 multigene locus of Ehrlichia canis. Gene 254:245-252. [DOI] [PubMed] [Google Scholar]
- 43.McBride, J. W., X. J. Yu, and D. H. Walker. 2000. Glycosylation of homologous immunodominant proteins of Ehrlichia chaffeensis and Ehrlichia canis. Infect. Immun. 68:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLeod, M. P., X. Qin, S. E. Karpathy, J. Gioia, S. K. Highlander, G. E. Fox, T. Z. McNeill, H. Jiang, D. Muzny, L. S. Jacob, A. C. Hawes, E. Sodergren, R. Gill, J. Hume, M. Morgan, G. Fan, A. G. Amin, R. A. Gibbs, C. Hong, X. J. Yu, D. H. Walker, and G. M. Weinstock. 2004. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol. 186:5842-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moran, N. A. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosavi, L. K., T. J. Cammett, D. C. Desrosiers, and Z. Y. Peng. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13:1435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nethery, K., C. Doyle, B. Elsom, N. Herzog, V. Popov, and J. McBride. 2005. Ankyrin repeat containing immunoreactive 200 kDa glycoprotein (gp200) orthologs of Ehrlichia chaffeensis and E. canis are translocated to the nuclei of infected monocytes, abstr. O-60. Fourth International Conference on Rickettsiae and Rickettsial Diseases, Logrono, Spain, 18 to 21 June.
- 48.Ochman, H., and A. C. Wilson. 1987. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 26:74-86. [DOI] [PubMed] [Google Scholar]
- 49.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournier, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 50.Ohashi, N., Y. Rikihisa, and A. Unver. 2001. Analysis of transcriptionally active gene clusters of major outer membrane protein multigene family in Ehrlichia canis and E. chaffeensis. Infect. Immun. 69:2083-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohashi, N., N. Zhi, Q. Lin, and Y. Rikihisa. 2002. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect. Immun. 70:2128-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Popov, V. L., V. C. Han, S. M. Chen, J. S. Dumler, H. M. Feng, T. G. Andreadis, R. B. Tesh, and D. H. Walker. 1998. Ultrastructural differentiation of the genogroups in the genus Ehrlichia. J. Med. Microbiol. 47:235-251. [DOI] [PubMed] [Google Scholar]
- 53.Power, P. M., and M. P. Jennings. 2003. The genetics of glycosylation in gram-negative bacteria. FEMS Microbiol. Lett. 218:211-222. [DOI] [PubMed] [Google Scholar]
- 54.Reddy, G. R., and C. P. Streck. 1999. Variability in the 28-kDa surface antigen protein multigene locus of isolates of the emerging disease agent Ehrlichia chaffeensis suggests that it plays a role in immune evasion. Mol. Cell. Biol. Res. Commun. 1:167-175. [DOI] [PubMed] [Google Scholar]
- 55.Reddy, G. R., C. R. Sulsona, A. F. Barbet, S. M. Mahan, M. J. Burridge, and A. R. Alleman. 1998. Molecular characterization of a 28 kDa surface antigen gene family of the tribe Ehrlichiae. Biochem. Biophys. Res. Commun. 247:636-643. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt, M. A., L. W. Riley, and I. Benz. 2003. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 11:554-561. [DOI] [PubMed] [Google Scholar]
- 57.Sexton, J. A., and J. P. Vogel. 2002. Type IVB secretion by intracellular pathogens. Traffic 3:178-185. [DOI] [PubMed] [Google Scholar]
- 58.Simpson, C. F. 1972. Structure of Ehrlichia canis in blood monocytes of a dog. Am. J. Vet. Res. 33:2451-2454. [PubMed] [Google Scholar]
- 59.Skotarczak, B. 2003. Canine ehrlichiosis. Ann. Agric. Environ. Med. 10:137-141. [PubMed] [Google Scholar]
- 60.Szymanski, C. M., S. M. Logan, D. Linton, and B. W. Wren. 2003. Campylobacter: a tale of two protein glycosylation systems. Trends Microbiol. 11:233-238. [DOI] [PubMed] [Google Scholar]
- 61.Szymanski, C. M., and B. W. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3:225-237. [DOI] [PubMed] [Google Scholar]
- 62.Troy, G. C., and S. D. Forrester. 1990. Canine ehrlichiosis, p. 404-415. In C. E. Greene (ed.), Viral, rickettsial, and mycoplasmal infections. W. B. Saunders Co., Philadelphia, Pa.
- 63.Unver, A., N. Ohashi, T. Tajima, R. W. Stich, D. Grover, and Y. Rikihisa. 2001. Transcriptional analysis of p30 major outer membrane multigene family of Ehrlichia canis in dogs, ticks, and cell culture at different temperatures. Infect. Immun. 69:6172-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vidotto, M. C., T. C. McGuire, T. F. McElwain, G. H. Palmer, and D. P. Knowles, Jr. 1994. Intermolecular relationships of major surface proteins of Anaplasma marginale. Infect. Immun. 62:2940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vitreschak, A. G., D. A. Rodionov, A. A. Mironov, and M. S. Gelfand. 2004. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet. 20:44-50. [DOI] [PubMed] [Google Scholar]
- 66.Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy, J. C. Brownlie, E. A. McGraw, W. Martin, C. Esser, N. Ahmadinejad, C. Wiegand, R. Madupu, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, J. F. Kolonay, W. C. Nelson, Y. Mohamoud, P. Lee, K. Berry, M. B. Young, T. Utterback, J. Weidman, W. C. Nierman, I. T. Paulsen, K. E. Nelson, H. Tettelin, S. L. O'Neill, and J. A. Eisen. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu, X. J., J. W. McBride, C. M. Diaz, and D. H. Walker. 2000. Molecular cloning and characterization of the 120-kilodalton protein gene of Ehrlichia canis and application of the recombinant 120-kilodalton protein for serodiagnosis of canine ehrlichiosis. J. Clin. Microbiol. 38:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.