Abstract
The presence of l-rhamnose (Rha) branches in the coaggregation receptor polysaccharides (RPS) of Streptococcus gordonii 38 and Streptococcus oralis J22 was eliminated by replacement of wefB with ermAM in these strains. The expression of this gene in S. oralis 34 did not, however, result in the addition of Rha branches to the linear RPS of this strain, which is identical to that produced by the wefB-deficient mutant of S. gordonii 38. This paradoxical finding was explained by a subtle difference in acceptor specificity of the galactose-1-phosphotransferases encoded by downstream wefC in S. gordonii 38 and wefH in S. oralis 34. These genes were distinguished by the unique ability of WefC to act on the branched acceptor formed by the action of WefB.
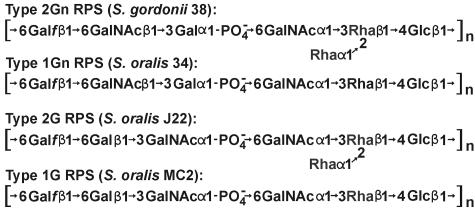
Interactions of viridans group streptococci with other oral species contribute to early biofilm formation during microbial colonization of the human tooth surface (10, 15, 16). These interactions generally depend on the binding of lectin-like adhesins, such as those associated with the type 2 fimbriae of Actinomyces naeslundii (7), to different streptococcal coaggregation receptor polysaccharides (RPS), including type 1Gn of Streptococcus oralis 34 (3, 13), type 2Gn of Streptococcus gordonii 38 (17), and type 2G of S. oralis J22 (2) (Fig. 1). Adhesin-mediated recognition of these polysaccharides depends on the presence of host-like features, either GalNAcβ1-3Gal (Gn) or Galβ1-3GalNAc (G), in the repeating units of these molecules (5). In contrast, the reactions of these polysaccharides as antigens reflect other features such as the l-rhamnose (Rha) branch of type 2Gn RPS, which forms the major epitope of this polysaccharide (17) at the expense of immunodominant α-GalNAc in type 1Gn RPS (14).
FIG. 1.
Structural types of RPS produced by different streptococci in the present study.
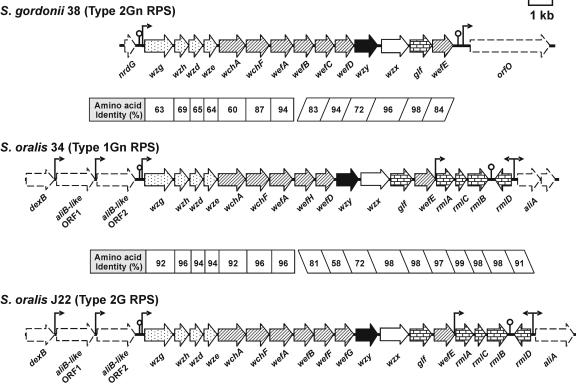
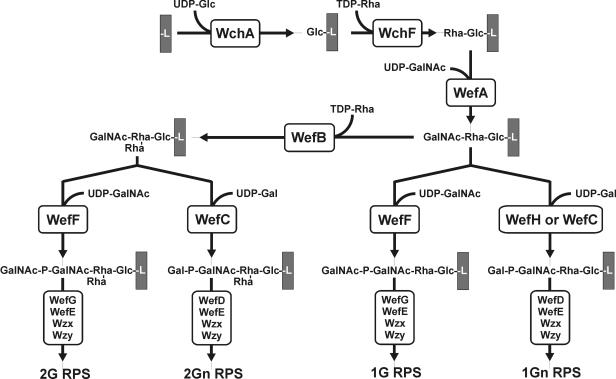
The structural difference between type 2Gn RPS of S. gordonii 38 and type 2G RPS of S. oralis J22 (Fig. 1) depends on two of the seven genes for glycosyltransferases in the RPS gene cluster of each strain (Fig. 2), wefC and wefD in strain 38 versus wefF and wefG in strain J22 (21). Three of the remaining genes (i.e., wchA, wchF, and wefE) for glycosyltransferases in each RPS gene cluster have also been associated with individual biosynthetic steps (Fig. 3), leaving wefA and wefB as likely candidates for the transfer of α-GalNAc and α-Rha to Rhaβ1-4Glc (20). The specific roles of wefA and wefB in synthesis of the resulting branched structure is, however, unclear, as neither gene has a well-studied homologue in the database.
FIG. 2.
ORF diagrams of RPS gene clusters indicating the homology between strains and the predicted roles of different genes in RPS biosynthesis. Each cluster contains four common regulatory genes (stippled arrows), six or seven genes for glycosyl or glycosyl-1-phosphotransferases (hatched arrows), and additional genes for a polysaccharide polymerase (solid arrows), a repeat unit transporter (open arrows), and enzymes for nucleotide sugar biosynthesis (brick-pattern arrows). Flanking genes (arrows with dashed outlines) are also identified, as are the positions of putative promoters (bent arrows) and rho-independent terminators (lollipops).
FIG. 3.
Proposed pathway for biosynthesis of types 2G, 2Gn, 1G, and 1Gn RPS beginning with the WchA-catalyzed transfer of Glc-1-PO4− to carrier lipid (L). The present findings define the roles of WefA and WefB in RPS biosynthesis and reveal a difference in the specificities of WefC and WefH for branched versus linear acceptors.
Type 2Gn RPS of S. gordonii 38 is identical to type 1Gn RPS of S. oralis 34 except for the presence of Rha branches in the former polysaccharide (Fig. 1). Therefore, to gain insight into the role of wefA or wefB in branch formation, we identified and sequenced the S. oralis 34 RPS gene cluster and flanking regions (GenBank accession no. AB181234), following methods essentially identical to those previously described in studies of S. oralis J22 (21). The RPS gene cluster of S. oralis 34, which was PCR amplified from genomic DNA of this strain, more closely resembled the RPS gene cluster of S. oralis J22 than that of S. gordonii 38 (Fig. 2), both in terms of its overall location between the genes for dextranase (dexB) and an oligopeptide-binding protein (aliA) and in terms of its association with the four rml genes for dTDP-l-Rha biosynthesis. Of greater interest was the presence of six rather than seven genes for putative glycosyltransferases in the RPS gene cluster of S. oralis 34, including wefA but not wefB (Fig. 2). Thus, wefA may be associated with the transfer of α-GalNAc in types 1Gn, 2Gn, and 2G RPS (Fig. 1) and wefB with synthesis of the Rha branches in type 2Gn and 2G RPS.
To test these hypotheses, we replaced various specific genes in S. gordonii 38, S. oralis 34, or S. mitis J22 as previously described (21) by transforming these bacteria with DNA constructs containing the ermAM cassette (12) flanked by appropriate 0.5- to 1-kb gene-targeting sequences. The constructs used as transforming DNA were prepared by overlap extension PCR (9, 11). The location of the cassette in the resulting erythromycin-resistant transformants (Table 1) was verified by amplification of specific PCR products across the upstream and downstream boundaries of the ermAM insertion (results not shown). Cell surface RPS production by wild-type and mutant strains was characterized and compared by the binding of different RPS-specific probes to decreasing numbers of bacteria immobilized on nitrocellulose membranes (21). The probes utilized included anti-type 1 RPS specific immunoglobulin G (IgG) from antiserum against S. oralis 34, which was purified by elution from coupled type 1Gn RPS (16), anti-type 2 RPS specific IgG from antiserum R103 against S. gordonii 38 (6), which was purified by elution (20) from coupled type 2G RPS of S. oralis J22, and biotin-labeled A. naeslundii 12104 (4, 18), which binds both Gn and G types of streptococcal cell surface RPS (5, 21).
TABLE 1.
Streptococci and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strainsa | ||
| S. oralis 34 | Wild-type strain (type 1Gn RPS) | 3 |
| S. oralis OC1 | S. oralis 34 containing ermAM in place of wchA | This study |
| S. oralis OC5 | S. oralis 34 containing wefB of S. gordonii 38 and ermAM between wefA and wefH | This study |
| S. gordonii 38 | Wild-type strain (type 2Gn RPS) | 17 |
| S. gordonii GC10 | S. gordonii 38 containing ermAM in place of wchF | This study |
| S. gordonii GC12 | S. gordonii 38 containing ermAM in place of wefB | This study |
| S. gordonii GC13 | S. gordonii 38 containing ermAM in place of wefC | 21 |
| S. gordonii GC20 | S. gordonii 38 containing ermAM in place of wzy | This study |
| S. gordonii GC21 | S. gordonii 38 containing ermAM in place of wefA | This study |
| S. gordonii GC22 | S. gordonii 38 containing ermAM in place of wzx | This study |
| S. gordonii GC45 | S. gordonii GC20 containing wzy of S. oralis 34 in place of ermAM | This study |
| S. gordonii GC48 | S. gordonii GC13 containing wefH of S. oralis 34 in place of ermAM | This study |
| S. gordonii GC49 | S. gordonii GC22 containing wzx of S. oralis 34 in place of ermAM | This study |
| S. gordonii GC50 | S. gordonii GC21 containing wefA of S. oralis 34 in place of ermAM | This study |
| S. oralis J22 | Wild-type strain (type 2G RPS) | 2 |
| S. oralis MC2 | S. oralis J22 containing ermAM in place of wefB | This study |
| Plasmids | ||
| pYY101 | pCM18 lacking gfp; confers Emr | 8; this study |
| pYY110 | pYY101 containing wefH from S. oralis 34 | This study |
Streptococci were grown in Todd-Hewitt broth (Difco) containing erythromycin (10 μg/ml) as needed for the maintenance of ermAM.
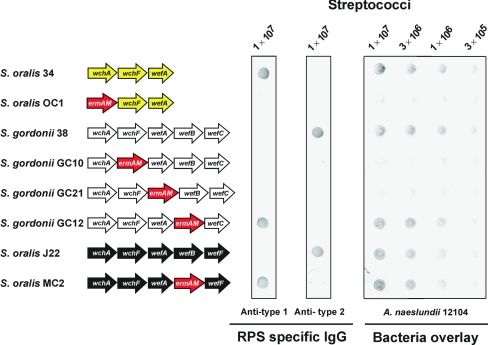
Cell surface RPS production was abolished by ermAM replacement of either wchA in S. oralis 34 or wchF or wefA in S. gordonii 38, the genes for the first three predicted steps in RPS biosynthesis (Fig. 3). Thus, the resulting mutants, S. oralis OC1, S. gordonii GC10, or S. gordonii GC21, respectively, failed to bind either RPS-specific antibody or A. naeslundii (Fig. 4). In contrast, ermAM replacement of wefB in S. gordonii 38 or S. oralis J22 altered the immunoreactivity of these bacteria without affecting the binding of A. naeslundii (Fig. 4). The parental strains 38 and J22 bound anti-type 2 but not anti-type 1 RPS specific IgG, whereas the wefB mutants (i.e., S. gordonii GC12 and S. oralis MC2, respectively) exhibited the opposite pattern of reactivity. In further studies, the ability of wefB to complement type 2Gn RPS production in trans was demonstrated following the cloning of this gene in pYY101 (Table 1) and transformation of an erythromycin-sensitive wefB deletion mutant of S. gordonii 38 with the resulting plasmid (results not shown).
FIG. 4.
RPS production by wild-type and mutant streptococci detected by dot immunoblotting with RPS-specific rabbit IgG or binding of biotin-labeled A. naeslundii 12104. Nitrocellulose membranes were spotted with streptococci, incubated with RPS-specific rabbit IgG or biotin-labeled A. naeslundii, washed, and developed with goat anti-rabbit IgG or horseradish peroxidase-conjugated avidin, followed by a substrate to detect bound IgG or A. naeslundii, respectively. Partial ORF diagrams of wild-type and mutant streptococci indicate the presence of genes from S. oralis 34 (yellow arrows), S. gordonii 38 (white arrows), S. oralis J22 (black arrows), or ermAM (red arrows).
To establish the structural basis for the switch in antigenicity associated with the deletion of wefB, we solubilized and purified the RPS of S. gordonii GC12 and S. oralis MC2 and recorded complete sets of homonuclear and heteronuclear (1H and 13C) NMR spectra at 500 MHz 1H frequency following previously described methods (1-3, 6, 21). The peaks in a heteronuclear single-quantum coherence (HSQC) spectrum of S. gordonii GC12 RPS (data not shown) corresponded exactly with the 1H and 13C resonances previously assigned for type 1Gn RPS of S. oralis 34 (3). Since the structure of S. oralis MC2 RPS was expected to be novel, a complete assignment of the 1H and 13C resonances was undertaken using standard homonuclear methods (correlation spectroscopy [COSY], total correlation spectroscopy [TOCSY], and nuclear Overhauser spectroscopy [NOESY]) and heteronuclear methods (HSQC and heteronuclear multiple bond correlation [HMBC]). The resulting assignments are reported in Fig. S1 and Table S1 in the supplemental material. Linkages between residues were determined using HMBC and NOESY as indicated in Table S2 in the supplemental material. These data identify the S. oralis MC2 polysaccharide as type 1G RPS (Fig. 1), a new structural type. In immunodiffusion experiments, this polysaccharide and type 1Gn RPS of S. oralis 34 reacted identically with different rabbit antisera against either S. oralis 34 or S. oralis MC2 (results not shown). Thus, the structural and corresponding antigenic difference between type 2Gn or 2G RPS and type 1Gn or 1G RPS clearly depends on the glycosyltransferase encoded by wefB.
The finding that wefB directs the synthesis of Rha branches in type 2Gn and 2G RPS leaves wefA for the transfer of α-GalNAc to Rhaβ1-4Glc in these polysaccharides. Experimental evidence supporting this role has been obtained in recent molecular studies of S. oralis 10557 type 3G RPS, which contains α-Gal rather than α-GalNAc as in type 2Gn RPS (Y. Yoshida et al., manuscript in preparation). While further studies are needed to verify the order of addition, we suspect that the WefA-catalyzed formation of GalNAcα1-3Rhaβ in linear type 1Gn RPS precedes the WefB-catalyzed addition of Rhaα to Rhaβ (Fig. 3), as the alternative would require that WefA utilize subterminal Rha as an acceptor in biosynthesis of type 2Gn and 2G RPS and terminal Rha as an acceptor in biosynthesis of type 1Gn RPS.
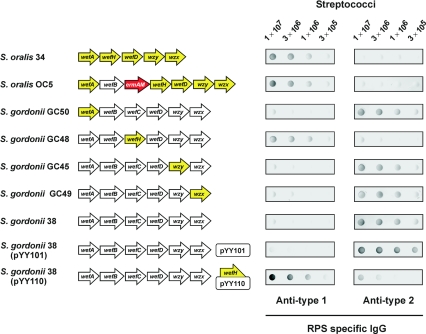
Based on the above findings, we expected that the expression of wefB in S. oralis 34 would switch RPS production from type 1Gn to type 2Gn. To examine this possibility, we isolated a wefB-inserted transformant of S. oralis 34 (i.e., S. oralis OC5) following transformation of this wild-type strain with wefB linked to the selectable ermAM cassette. Transforming DNA was prepared by two rounds of overlap extension PCR, the first to link wefB to ermAM and the second to link the resulting wefB-ermAM fragment to targeting sequences for insertion between wefA of S. oralis 34 and the downstream gene, which is identified below as wefH. The precise insertion of the wefB-ermAM fragment between wefA and wefH in S. oralis 34 was confirmed by DNA sequencing of this region in S. oralis OC5. Surprisingly, this mutant, which expresses wefB from the promoter at the 5′ end of the RPS gene cluster and downstream genes for RPS biosynthesis from the promoter in the ermAM cassette, as well a similar mutant in which the order of wefB and ermAM was reversed (results not shown), reacted with anti-type 1 but not anti-type 2 RPS specific IgG in dot immunoblotting (Fig. 5). Consistent with these reactions, the resonances in the HSQC NMR spectrum of the RPS purified from S. oralis OC5 corresponded exactly to the 1H and 13C assignment for S. oralis 34 type 1Gn RPS (3), with no indication (i.e., less than 5% [data not shown]) of Rha branches.
FIG. 5.
RPS production by wild-type and mutant streptococci detected by dot immunoblotting with anti-type 1 or anti-type 2 RPS specific rabbit IgG. Nitrocellulose membranes were spotted with decreasing numbers of streptococci, incubated with RPS-specific rabbit IgG, washed, and developed with goat anti-rabbit IgG followed by a substrate to detect bound IgG. Partial ORF diagrams of wild type and mutant streptococci strains indicate the presence of genes from S. oralis 34 (yellow arrows), S. gordonii 38 (white arrows), or ermAM (red arrows).
The above findings raise the possibility that the synthesis of Rha branches in S. gordonii 38 RPS depends on more than simply the expression of wefB in this strain. To explore this possibility, we replaced wefA, wefC, wzy, or wzx in S. gordonii 38 with the corresponding gene of S. oralis 34 by following a previously described strategy (21). The gene of interest in S. gordonii 38 was initially replaced by the ermAM cassette as described above to obtain S. gordonii GC21, GC13, GC20, and GC22, respectively (Table 1), which grew on erythromycin and were nonreactive with RPS-specific IgG. These strains were then transformed with overlap extension PCR products that contained individual genes of S. oralis 34 (i.e., wefA, wefH, wzy, or wzx, respectively) flanked by targeting sequences for the S. gordonii 38 genes present on either side of ermAM cassette. Replacement of the ermAM cassette with a complementary gene from S. oralis 34 restored cell surface RPS production, which was detected by colony immunoblotting (21), resulting in the isolation of S. gordonii GC50, GC48, GC45, and GC49, respectively (Table 1). The reactions of mutant S. gordonii GC50, GC45, and GC49 with anti-type 2 but not anti-type 1 RPS specific IgG were indistinguishable from those of wild-type S. gordonii 38 (Fig. 5), thereby indicating that wefA, wzy, and wzx of S. oralis 34 all support type 2Gn RPS production.
In contrast, S. gordonii GC48, obtained by replacement of wefC in S. gordonii 38 with the corresponding gene from S. oralis 34, reacted with anti-type 1 but not anti-type 2 RPS specific IgG (Fig. 5), thereby associating the production of type 1Gn RPS with the presence of a unique gene (i.e., wefH) in the latter strain. The antigenic identification of type 1Gn RPS on S. gordonii GC48 was confirmed by the HSQC NMR spectrum of the RPS purified from this strain (results not shown). In support of these findings, the plasmid-based expression of wefH in S. gordonii 38 altered the RPS-specific immunoreactivity of this strain. Whereas S. gordonii 38 harboring control plasmid pYY101 reacted with anti-type 2 but not anti-type 1 RPS specific IgG, strain 38 harboring pYY110, which expresses wefH, reacted strongly with anti-type 1 and weakly with anti-type 2 RPS specific IgG (Fig. 5). Thus, the WefH-mediated transfer of Galαl-PO4− to GalNAcα (Fig. 3) prevents WefB from acting, presumably by eliminating the acceptor of this enzyme. This effect may, in turn, limit the accumulation of lipid-linked branched tetrasaccharide in S. gordonii GC48, S. gordonii 38 (pYY110), and S. oralis OC5 by the action of WefB in these strains. Conversely, the branched acceptor formed by the WefB-catalyzed transfer of Rhaα to subterminal Rhaβ may not be recognized by WefH, a possibility consistent with the known ability of the Rha branch to block the binding of anti-type 1 antibody (14, 17) or the Codium fragile lectin (5) to adjacent GalNAcα of type 2Gn RPS.
WefC contributes to the synthesis of linear type 1Gn RPS in wefB-deficient S. gordonii GC12 (Fig. 4) and of branched-type 2Gn RPS in wefB-containing S. gordonii 38 (Fig. 3). The preference of WefC for the branched acceptor structure synthesized by strain 38 can, however, be inferred from the NMR spectra of type 2Gn RPS (17), which indicate the absence of detectable (<5%) linear hexasaccharide repeats in this polysaccharide. Likewise, WefF of S. oralis J22 utilizes a branched acceptor in synthesis of type 2G RPS and a linear acceptor in synthesis of type 1G RPS by wefB-deficient S. oralis MC2 (Fig. 3 and 4). The identification and structural characterization of type 1G RPS in the present investigation should facilitate the identification of wild-type streptococci that produce this polysaccharide. Once they are identified, it will be of interest to determine whether these bacteria utilize WefF to transfer GalNAcαl-PO4− to GalNAcα or a closely related enzyme that only acts on the linear acceptor formed in the absence of WefB (Fig. 3).
The proteins encoded by wefH of S. oralis 34, wefC of S. gordonii 38, and wefF of S. oralis J22 have pivotal roles in RPS biosynthesis, linking the recognition and antigenic regions of these polysaccharides. This biological role is a direct reflection of the donor specificities of these enzymes for Galαl-PO4− or GalNAcαl-PO4− and their acceptor specificities for linear or branched structures. In view of the relatively high homology that exists between these proteins (Fig. 2), their ability to discriminate between different substrates is likely to depend on minor differences in amino acid sequence. The identification of such sequences and their association with the synthesis of different types of RPS would contribute to the further characterization of these proteins as members of a recently recognized group of glycosyl-1-phosphotransferases (19) and of the corresponding genes as genetic markers of oral biofilm development.
Nucleotide sequence accession number.
The S. oralis 34 RPS gene cluster and flanking regions have been deposited in GenBank under accession no. AB181234.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NIDCR, by a fellowship from the Japanese Society for the Promotion of Science to Y.Y. and by grant 02-12702 from the National Science Foundation to C.A.B.
We thank Kelly Ten Hagen and John Thompson for helpful comments during preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abeygunawardana, C., and C. A. Bush. 1993. Determination of the chemical structure of complex polysaccharides by heteronuclear NMR spectroscopy. Adv. Biophys. Chem. 3:199-249. [Google Scholar]
- 2.Abeygunawardana, C., C. A. Bush, and J. O. Cisar. 1990. Complete structure of the polysaccharide from Streptococcus sanguis J22. Biochemistry 29:234-248. [DOI] [PubMed] [Google Scholar]
- 3.Abeygunawardana, C., C. A. Bush, S. S. Tjoa, P. V. Fennessey, and M. R. McNeil. 1989. The complete structure of the capsular polysaccharide from Streptococcus sanguis 34. Carbohydr. Res. 191:279-293. [DOI] [PubMed] [Google Scholar]
- 4.Cisar, J. O., V. A. David, S. H. Curl, and A. E. Vatter. 1984. Exclusive presence of lactose-sensitive fimbriae on a typical strain (WVU45) of Actinomyces naeslundii. Infect. Immun. 46:453-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cisar, J. O., A. L. Sandberg, C. Abeygunawardana, G. P. Reddy, and C. A. Bush. 1995. Lectin recognition of host-like saccharide motifs in streptococcal cell wall polysaccharides. Glycobiology 5:655-662. [DOI] [PubMed] [Google Scholar]
- 6.Cisar, J. O., A. L. Sandberg, G. P. Reddy, C. Abeygunawardana, and C. A. Bush. 1997. Structural and antigenic types of cell wall polysaccharides from viridans group streptococci with receptors for oral actinomyces and streptococcal lectins. Infect. Immun. 65:5035-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cisar, J. O., A. E. Vatter, W. B. Clark, S. H. Curl, S. Hurst-Calderone, and A. L. Sandberg. 1988. Mutants of Actinomyces viscosus T14V lacking type 1, type 2, or both types of fimbriae. Infect. Immun. 56:2984-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen, M. C., R. J. Palmer, Jr., C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 9.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 10.Hsu, S. D., J. O. Cisar, A. L. Sandberg, and M. Kilian. 1994. Adhesive properties of viridans group streptococcal species. Microb. Ecol. Health Dis. 7:125-137. [Google Scholar]
- 11.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lunsford, R. D., and J. London. 1996. Natural genetic transformation in Streptococcus gordonii: comX imparts spontaneous competence on strain wicky. J. Bacteriol. 178:5831-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIntire, F. C., C. A. Bush, S. S. Wu, S. C. Li, Y. T. Li, M. McNeil, S. S. Tjoa, and P. V. Fennessey. 1987. Structure of a new hexasaccharide from the coaggregation polysaccharide of Streptococcus sanguis 34. Carbohydr. Res. 166:133-143. [DOI] [PubMed] [Google Scholar]
- 14.McIntire, F. C., L. K. Crosby, A. E. Vatter, J. O. Cisar, M. R. McNeil, C. A. Bush, S. S. Tjoa, and P. V. Fennessey. 1988. A polysaccharide from Streptococcus sanguis 34 that inhibits coaggregation of S. sanguis 34 with Actinomyces viscosus T14V. J. Bacteriol. 170:2229-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 16.Palmer, R. J., Jr., S. M. Gordon, J. O. Cisar, and P. E. Kolenbrander. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 185:3400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy, G. P., C. Abeygunawardana, C. A. Bush, and J. O. Cisar. 1994. The cell wall polysaccharide of Streptococcus gordonii 38: structure and immunochemical comparison with the receptor polysaccharides of Streptococcus oralis 34 and Streptococcus mitis J22. Glycobiology 4:183-192. [DOI] [PubMed] [Google Scholar]
- 18.Ruhl, S., A. L. Sandberg, and J. O. Cisar. 2004. Salivary receptors for the proline-rich protein-binding and lectin-like adhesins of oral actinomyces and streptococci. J. Dent. Res. 83:505-510. [DOI] [PubMed] [Google Scholar]
- 19.Tiede, S., S. Storch, T. Lubke, B. Henrissat, R. Bargal, A. Raas-Rothschild, and T. Braulke. 2005. Mucolipidosis II is caused by mutations in GNPTA encoding the α/β GlcNAc-1-phosphotransferase. Nat. Med. 10:1109-1112. [DOI] [PubMed] [Google Scholar]
- 20.Xu, D. Q., J. Thompson, and J. O. Cisar. 2003. Genetic loci for coaggregation receptor polysaccharide biosynthesis in Streptococcus gordonii 38. J. Bacteriol. 185:5419-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida, Y., S. Ganguly, C. A. Bush, and J. O. Cisar. 2005. Carbohydrate engineering of the recognition motifs in streptococcal coaggregation receptor polysaccharides. Mol. Microbiol. 58:244-256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.