Abstract
In contrast to eukaryotes, bacteria such as Escherichia coli contain only one form of RNA polymerase (RNAP), which is responsible for all cellular transcription. Using an RNAP-green fluorescent protein fusion protein, we showed previously that E. coli RNAP is partitioned exclusively in the nucleoid and that stable RNA synthesis, particularly rRNA transcription, is critical for concentrating a significant fraction of RNAP in transcription foci during exponential growth. The extent of focus formation varies under different physiological conditions, supporting the proposition that RNAP redistribution is an important element for global gene regulation. Here we show that extra, plasmid-borne copies of an rRNA operon recruit RNAP from the nucleoid into the cytoplasmic space and that this is accompanied by a reduction in the growth rate. Transcription of an intact rRNA operon is not necessary, although a minimal transcript length is required for this phenotype. Replacement of the ribosomal promoters with another strong promoter, Ptac, abolished the effect. These results demonstrate that active synthesis from rRNA promoters is a major driving force for the distribution of RNAP in bacteria. The implications of our results for the regulation of rRNA synthesis and cell growth are discussed.
Escherichia coli RNA polymerase (RNAP) is a multisubunit enzyme consisting of α2ββ′σ. Although there are seven σ factors in E. coli, σ70 predominates and is responsible for transcription of most genes (5). Unlike eukaryotes, in which three different RNAPs (PolI, PolII, and PolIII) (23) synthesize three different RNA species (rRNA, mRNA, and tRNA/5S rRNA, respectively), the single E. coli RNAP synthesizes all RNA and does so differentially in response to growth conditions (4, 21). For example, under optimal growth conditions, the vast majority of RNAP molecules synthesize rRNA and tRNA (termed stable RNAs), although the corresponding genes represent less than 1% of the genome. The remaining RNAP molecules are responsible for synthesizing the appropriate mRNAs from the other ∼4,300 genes. Under suboptimal conditions, such as growth in nutrient-poor media, a far smaller fraction of the RNAP molecules is needed to synthesize sufficient amounts of stable RNAs, which allows expansion of new gene transcription. When cells are shifted from nutrient-rich conditions to starvation conditions, such as amino acid starvation, there is a stringent response, which results in dramatic reprogramming of transcription such that the expression of stable RNAs is inhibited while amino acid biosynthetic operons are activated (7). The redistribution of RNAP according to growth conditions has been proposed to be a key feature of global gene regulation during processes such as the stringent (nutrient starvation) response and carbon source limitation responses (1, 16, 29).
Recently, we studied the location and distribution of RNAP under different physiologic conditions by using a functional rpoC-gfp gene fusion on the E. coli chromosome and imaging RNAP-green fluorescent protein (GFP) in vivo by fluorescence microscopy (6). Indeed, while RNAP is located exclusively either within and/or surrounding the nucleoid (i.e., there is no RNAP-GFP signal in the cytoplasmic space), RNAP distribution is dynamic and sensitive to growth conditions. In particular, RNAP in fast-growing cells forms several transcription foci within the nucleoid. We proposed that these foci are transcription “factories” that actively synthesize stable RNAs, forming a structure(s) analogous to the eukaryotic nucleolus (8).
The study mentioned above indicated that synthesis of stable RNAs, particularly rRNA synthesis, is a driving force for RNAP distribution inside the cell. In this study, we determined the effect of extrachromosomal copies of an rRNA operon on the location and distribution of RNAP. We chose two plasmids for our initial study: pNO1301, which contains an intact rrnB operon, and pNO1302, which harbors an rrnB operon with a partial deletion. The pNO1301 and pNO1302 plasmids were used previously to study the effects of rRNA operon copy number on rRNA synthesis (14, 27). These plasmids are derivatives of pBR322, which has been reported to localize in the cytoplasm of E. coli cells (9, 20, 22). Our results demonstrate that active rRNA synthesis plays a pivotal role in the in vivo distribution of RNAP. In addition, our results suggest an additional cause for the inhibitory effect of extra rRNA gene copies in trans on chromosomal rRNA synthesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strain DJ2735 is a DJ2599 (MG1655 rpoC-gfp Ampr) derivative in which the Ampr allele was replaced by the cat (Cmr) gene using a high-efficiency λ Red recombination system (28). Briefly, a PCR fragment containing the Cmr gene was synthesized from plasmid pACYC184 with oligonucleotides JC233A (5′ ATG AGT ATT CAA CAT TTC CGT GTC GCC CTT ATT CCC TTT TTT GCG GCA TTT ACC TGT GAC GGA AGA TCA CTT CGC 3′) and JC233B (5′ TTA CCA ATG CTT AAT CAG TGA GGC ACC TAT CTC AGC GAT CTG TCT ATT TCT TAA GGG CAC CAA TAA CTG CC 3′) (nucleotides identical to nucleotides in the Ampr gene are underlined). This PCR fragment was recombined into the chromosome of cells carrying the rpoC-gfp Ampr allele and a copy of the λ Red recombination system. Like DJ2599, strain DJ2735 is temperature sensitive for growth at temperatures above 37°C, likely due to a problem with GFP folding at high temperatures (11, 15, 24). The doubling time of DJ2735 in Luria-Bertani (LB) medium at 30°C is approximately 45 min.
The basic bacterial techniques used have been described elsewhere (18). All cultures were grown with vigorous agitation in a water bath at 30°C. Fresh overnight cultures were diluted 1/500 into fresh media. Cells were grown in M63 medium supplemented with glucose (final concentration, 0.2%) or in LB medium. Samples used for microscope observation were removed at an optical density at 600 nm of approximately 0.4.
Plasmid construction.
The plasmids used in this work are listed in Table 1. All recombinant DNA techniques used were based on protocols described by Maniatis et al. (17). All PCRs were carried out using plasmid pNO1301 as a template.
TABLE 1.
Plasmids used
| Plasmid | Description | Reference or source |
|---|---|---|
| pBR322 | Cloning vector | 3 |
| pKK223-3 | Expression vector | Pharmacia |
| pNO1301 | Includes rrnB whole operon | 14 |
| pNO1302 | pNO1301 derivative with deletion of a SalI-SalI fragment in the rrn operon | 14 |
| pDJ2754-11 | pNO1301 derivative lacking rrnB P1 and rrnB P2 | This study |
| pJ2754-17 | pNO1301 derivative lacking rrnB P2 | This study |
| pDJ2754-15 | pNO1301 derivative lacking rrnB P1 | This study |
| pDJ2790 | pKK223-1 derivative lacking the tac promoter | This study |
| pDJ2791-A | Fragment of the rrnB operon including rrnB P1 and rrnB P2 plus a 1,800-nucleotide transcript cloned into pDJ2790 | This study |
| pDJ2791-B | Fragment of the rrnB operon including rrnB P1 and rrnB P2 plus a 1,000-nucleotide transcript cloned into pDJ2790 | This study |
| pDJ2845 | Promoterless fragment of the rrnB operon (encoding the whole 16S rRNA) cloned into the pKK223-1 vector | This study |
To construct plasmids carrying promoterless or one-promoter copies of the rrnB gene, a 1.6-kbp NaeI-BglII fragment that carried both the rrnB P1 and P2 promoters in pNO1301 was replaced with DNA fragments synthesized by PCR and digested with NgoMIV and BglII as described below. Plasmid pDJ2754-17 containing a PCR fragment with the rrnB P1 promoter was obtained in two steps. First, two independent DNA fragments (fragments A and B) which share a complementary sequence (indicated by boldface type) at their 3′ and 5′ ends were each amplified by PCR. The primers used to amplify fragment A were JC254A (5′ ATT CTG ACT GTA GCC GGC GAT TAA ACC TGC TGC ACG 3′) for the upper strand and JC254E (5′ TGC CGT TGT TCC GTG TCA GTG GTG GCG CAT TAT AGG GAG 3′) for the lower strand. The primers used to amplify fragment B were JC254F (5′ ACT GAC ACG GAA CAA CGG CAA AAT TGA AGA GTT TGA TCA TGG C 3′) for the upper strand and JC254B (5′ GGT ATT CCT CCA GAT CTC TAC GCA TTT CAC 3′) for the lower strand. Primers JC254E and JC254F are partially complementary (indicated by boldface type). The second step was PCR amplification with primers JC254A and JC254B and a mixture of fragments A and B as the DNA template. The resulting PCR fragment containing rrnB P1 was purified, cut with the NgoMIV and BglII restriction enzymes (restriction sites for these enzymes are underlined in the sequences above), and ligated with previously digested plasmid pNO1301. Similarly, plasmid pDJ2754-15 containing a PCR fragment with the rrnB P2 promoter was obtained by a single PCR using oligonucleotides JC254C (5′ ATT CTG ACT GTA GCC GGC ACT GAC ACG GAA CAA CGG C 3′) and JC254B, and plasmid pDJ2754-11 containing a promoterless PCR fragment was obtained by PCR using oligonucleotides JC254D (5′ ATT CTG ACT GTA GCC GGC AAA TTG AAG AGT TTG ATC ATG GC 3′) and JC254B.
To clone different lengths of the rrnB operon, we first generated a pKK223-3 (Pharmacia) derivative designated pDJ2790 that lacked the tac promoter by deleting a BamHI fragment containing the promoter. Vector pDJ2790 was then digested with the BamHI and PstI restriction enzymes and ligated with BamHI-PstI-digested PCR fragments containing different lengths of the rrnB operon. Plasmid pDJ2791A contained a PCR fragment which was obtained in a reaction using oligonucleotides JC291A (5′ GCC GGC GAT TAA GGA TTC TGC ACG TCT GAC GGC AAA TGG 3′) and JC291B (5′ CTT TCT ATC AGA CTG CAG GTG TGA GCA CTA CAA AGT ACG C 3′) (the recognition sites of the BamHI and PstI restriction enzymes are underlined). The pDJ2791B plasmid contained a PCR fragment that was obtained in a reaction using oligonucleotides JC291A and JC291C (5′ GAG CGT CAG TCT CTG CAG AGG GGG CCG CCT TCG CCA CCG G 3′).
Plasmid pDJ2845 was obtained by cloning a PCR product that encompassed the sequences immediately downstream of rrnB P2 to the end of the gene encoding the 16S rRNA into the PstI sites of pKK223-1. The oligonucleotides used to synthesize the DNA fragment were JC345A (5′ ATA AAT TTA ATA CTG CAG CCG CGC CGC TGA GAA AAA GCG 3′) and JC291A. The proper orientation of the cloned fragment was confirmed by restriction enzyme mapping.
Microscopy and image analysis.
All samples used for microscopy were fixed with formaldehyde. To fix the cells, 5-ml aliquots of cultures were removed from the culture flasks, and formaldehyde was added to a final concentration of 3.7%. Cells were fixed for 1 h at room temperature, centrifuged, and resuspended in 1 ml of 1× phosphate-buffered saline. Before the cell mixtures were mounted on slides, 15 μl of a 10-μg/ml solution of 4′,6′-diamidino-2-phenylindole (DAPI) was added to each mixture. The final mixtures of fixed cells were mounted on slides using 1% low-melting-point agarose. Microscopy was performed with a Zeiss Axiophot II microscope equipped with a Plan-Apo ×100 objective, epifluorescence filters, and a 2.5 optovar. Images were captured with a charge-coupled device camera (Micromax) working at 2X2 binning. The images were processed with Adobe Photoshop.
The percentage of RNAP-GFP fluorescence emitted from the cytoplasm in individual cells was calculated by adding the intensities of all the pixels corresponding to the nucleoid-free area, dividing the number obtained by the sum of the intensities of all the pixels of the cell, and then multiplying the result by 100. The background intensity, measured in cell-free areas of the micrograph, was subtracted during the calculations. The measurements were obtained using digital pictures of 100 different cells for each plasmid-carrying strain, and the values shown in Table 2 are averages and standard deviations. Pixel intensities were measured using IPLab 3.6 (Innovision). The percentage for the RNAP-GFP signal emitted from the cytoplasm of cells without a plasmid was about 15%, which represented either emission from β′-GFP molecules not associated with the nucleoid or just scattering of light from the nucleoid due to the small size of the cells and the limited resolution of the optical microscope.
TABLE 2.
Plasmid construct with rrn, growth rate reduction, RNAP distribution, and cytoplasmic RNAP-GFP signal
With the exception of pBR322, all constructs have the antitermination sequence boxA (2, 12) and two rrn terminators.
The cell wall is represented by a rounded rectangle, and the nucleoid is represented by an oval. The cytoplasmic space is between the cell wall and the nucleoid of the cell. Gray shading indicates the presence of RNAP-GFP fluorescence signal. When RNAP was detected in the ctyplasmic space, the transcription foci in the nucleoid were diminished.
The cytoplasmic RNAP-GFP signal is expressed as a percentage, which was determined by dividing the RNAP-GFP fluorescence in the cytoplasmic space by the fluorescence in the whole cell, as described in Materials and Methods. The values are averages ± standard deviations for analyses of 100 cells.
The transcription foci in the nucleoid were operationally defined as regions of the cell or nucleoid that contained RNAP-GFP signals whose intensity was at least 80% the intensity of the brightest pixel with areas covering less than 10% of the total area (measured in pixels). Because of the small size of the cytoplasmic space, it was very difficult technically to perform the previously described contrast analysis (6) for the RNAP-GFP concentration outside the nucleoid in images of cells carrying plasmids, and the results were not reliable.
RESULTS
Expression of extrachromosomal copies of an rrn operon is detrimental to cell growth.
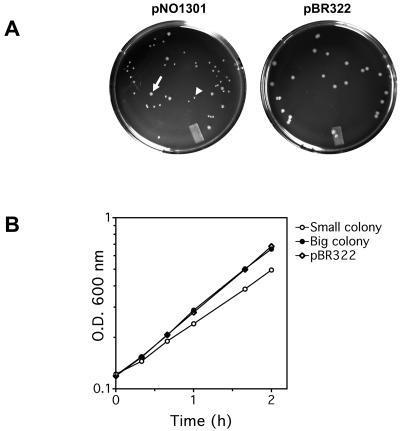
To study whether transcription of an rrn operon in trans affects the location and/or distribution of RNAP, the pBR322-based plasmid pNO1301, which carried an intact copy of the rrnB operon, including its two promoters, P1 and P2, and terminators, was introduced into DJ2735 cells by transformation. DJ2735 contains a functional rpoC-gfp allele as the sole source of β′ subunits, which allows tracking of RNAP by fluorescence microscopy (6). Intriguingly, two distinct sizes of colonies of Ampr transformants were observed after overnight growth (about 14 h); approximately 70% of the colonies were small, while the remaining 30% were large (Fig. 1A), suggesting that the population was heterogeneous. Indeed, the doubling times for cells from the large and small colonies when they were grown in LB medium containing ampicillin were 47 and 58 min, respectively (Fig. 1B). Note that the doubling time for cells from large colonies was the same as the doubling time for cells harboring the empty pBR322 vector. Restriction mapping of plasmid DNA isolated from the colonies that were two different sizes revealed that while the plasmids from the small colonies had an intact rrnB operon, all of the plasmids from the large colonies had either deletions or rearrangements of the operon (data not shown). As this phenotype has not been described previously, we performed the same experiments with MG1655 cells, and the same results were obtained. Thus, these results demonstrate that expression of extra plasmid-borne copies of the rrnB operon is harmful to a cell.
FIG. 1.
Presence of plasmid-borne rrnB (pNO1301) is detrimental to cell growth in rich media. (A) Heterogeneous colony sizes for cells transformed with plasmid pNO1301 compared to cells transformed with plasmid pBR322 on LB medium plates containing ampicillin. The arrow indicates a representative big colony that emerged in the otherwise small-colony background. A representative small colony is indicated by an arrowhead. (B) Growth curves for big and small transformants with plasmid pNO1301. For comparison, the growth curve for a transformant with pBR322 is included. Growth was monitored in cultures grown in LB medium containing ampicillin at 37°C. O.D. 600 nm, optical density at 600 nm.
Reasoning that reduced rRNA transcription from both chromosomal and plasmid-borne operons might eliminate the detrimental effect as rrn promoter activity is sensitive to nutrient richness (21), we selected for transformants on minimal glucose-ampicillin plates. Indeed, the sizes of the colonies on minimal glucose-ampicillin plates were similar, and the organisms had intact pNO1301 plasmids (data not shown). This indicates that the regulation of the plasmid-borne rrnB operon is the same as the regulation in the chromosome. Thus, in all experiments described below, starting cultures of DJ2735 containing pNO1301 or its derivatives were grown initially in minimal glucose medium in the presence of ampicillin to ensure the integrity of the rrnB construct.
RNAP localizes in the cytoplasm of cells harboring a plasmid with the rrnB operon.
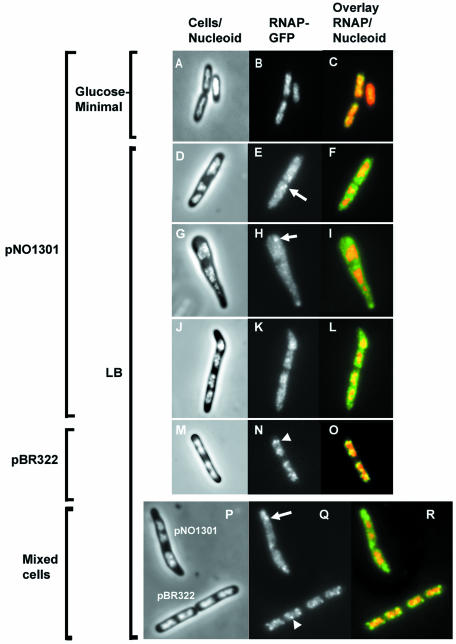
We examined the effect of pNO1301 on the location and distribution of RNAP under two different growth conditions (Fig. 2). When cells containing pNO1301 were grown in a nutrient-poor minimal glucose medium, the vast majority of RNAP was located in the nucleoid, and very little of the RNAP-GFP signal was detected in the cytoplasm (Fig. 2A, B, and C). In addition, the transcription foci under these conditions were less evident, as expected (6). Intriguingly, when the cells were grown in nutrient-rich LB medium, a significant amount of RNAP was observed in the cytoplasm in addition to the nucleoid (Fig. 2D to L). In contrast, RNAP was located exclusively in the nucleoids of cells transformed with pBR322 (Fig. 2M, N and O), just like RNAP in cells in the absence of a plasmid, as shown previously (6). Furthermore, while in rich media transcription foci were obvious in the nucleoids of cells harboring pBR322, there were fewer transcription foci in cells containing pNO1301 in these media. The difference in the intensities of the transcription foci in the nucleoids was evident when the two kinds of cells (cells containing pNO1301 and cells containing pBR322) were mixed prior to examination in the same microscope field (Fig. 2P, Q, and R). For the majority of cells, the RNAP-GFP signals recruited into the cytoplasmic space by plasmid-borne rrn operons were diffuse; however, about 8% of the cells containing pNO1301 showed aggregated RNAP-GFP signals in the cytoplasm (Fig. 2E, H, and Q), probably because of “clustering” of plasmids in the cell (9, 22). These results suggest that active transcription of a plasmid-borne rrn operon is able to recruit a significant amount of RNAP molecules.
FIG. 2.
Cell morphology and location and distribution of RNA polymerase in cells carrying the plasmid-borne rrnB operon in pNO1301 grown in rich and minimal media. (A, D, G, J, M, and P) Merged phase-contrast and DAPI fluorescence images. (B, E, H, K, N, and Q) Fluorescence from the RNAP-GFP fusion protein. (C, F, I, L, O, and R) Overlays of the fluorescence from the RNAP-GFP fusion protein (green) and the fluorescence of the DAPI-stained nucleoid (red). (P, Q, and R) Comparison of cell morphology and location and distribution of RNA polymerase in cells carrying either pNO1301 or pBR322 in the same microscope field. Cells carrying pNO1301 and pBR322 were grown in LB medium and mixed at a ratio of 1:1 before fixation, and this was followed by fluorescence microscopy. Representative images are shown. Note that while transcription foci are evident in the nucleoids of cells harboring pBR322 (arrowhead), they are diminished in the nucleoids of cells harboring pNO1301. Although most of the RNAP-GFP fluorescence signals in the cytoplasm are diffuse, sometimes concentrated signals are apparent in the cytoplasm of cells harboring pNO1301 (arrows). For simplicity, only one representative focus is indicated.
Additionally, a significant percentage (about 13%) of cells harboring pNO1301 were morphologically different than the normal rod-like cells, as shown in Fig. 2G and J. Along an imaginary axis connecting the cell poles, it was apparent that some pNO1301-containing cells were wider than wild-type cells. At present, we do not know the reasons for this phenotype; however, one possibility is that it was caused by extra rRNA made from additional plasmid-borne rRNA operons (25).
Localization of RNAP in the cytoplasm depends on the length of the plasmid-borne rrnB transcript but not on its integrity.
The complete rrnB operon in pNO1301 encodes a 5.4-kb transcript consisting of 16S, 5S, and 23S rRNAs plus a spacer tRNA (Table 2). To determine if transcription of the entire ribosomal operon is required to recruit RNAP molecules outside the nucleoid, a series of pNO1301 derivatives in which different portions of the rrnB operon were deleted were examined (Fig. 3 and Table 2).
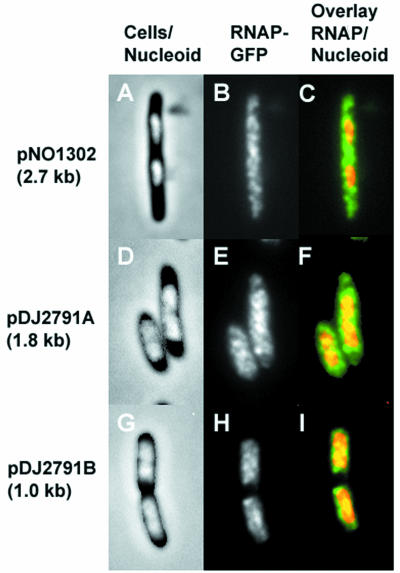
FIG. 3.
Cell morphology and location and distribution of RNA polymerase in cells carrying plasmid-borne rrnB operons that synthesize transcripts that are different lengths. (A, B, and C) Plasmid pNO1302 (2.67 kb). (D, E, and F) Plasmid pDJ2791A (1.8 kb). (G, H, and I) Plasmid pDJ2791B (1.0 kb). (A, D, and G) Merged phase-contrast and DAPI fluorescence images. (B, E, and F) Fluorescence from the RNAP-GFP fusion protein. (C, F, and I) Overlays of the fluorescence from the RNAP-GFP fusion protein (green) and the fluorescence of the DAPI-stained nucleoids (red).
pNO1302 was a derivative in which a 2.5-kbp SalI fragment encompassing approximately one-half of the 16S and 23S RNA-encoding genes, as well as the whole gene for the spacer tRNA, was removed. This resulted in a 2.7-kb rrnB transcript. Similar to the presence of pNO1301, the presence of pNO1302 was detrimental to cells grown in rich LB medium (data not shown). Intriguingly, cells harboring pNO1302 had a much lower growth rate (doubling time, 108 min) than cells containing either the pBR322 control (doubling time, 47 min) or pNO1301 (doubling time, 58 min) in LB broth, suggesting that the synthesis of a partially deleted rrnB transcript causes extra stress to the cells. Furthermore, a significant amount of RNAP was also localized in the cytoplasm of pNO1302-containing cells, similar to the effect of pNO1301 (Fig. 3A, B, and C).
Plasmid pDJ2791A, in which only the 16S RNA transcript (1.8 kb) (Table 2) was synthesized, yielded results similar to those obtained with pNO1301 and pNO1302 (Fig. 3D, E, and F) and caused a slow-growth phenotype. In contrast, cells harboring pDJ2791B, which expressed only approximately 1.0 kb of the 16S RNA gene (Table 2), were found to contain RNAP exclusively in the nucleoid (Fig. 3G, H, and I). Furthermore, pDJ2791B was stably maintained in the cells and was not detrimental to cell growth.
We observed a correlation between the length of the plasmid-borne synthesized transcript and the RNAP-GFP signal measured in the cytoplasm of cells carrying the plasmids (Table 2). In cells carrying plasmids that synthesized 5.4- and 2.7-kb transcripts more than 50% of the total cellular RNAP-GFP fluorescence signal was localized in the cytoplasm (plasmids pNO1301 and pNO1302) (Table 2). Cells carrying plasmids that synthesized shorter transcripts had weaker RNAP-GFP fluorescence signals in their cytoplasm, >30% for plasmid pDJ2791A (1.8-kb transcript) and <20% for plasmid pDJ2791B (1.0-kb transcript). In control cells carrying pBR322 or no plasmid at all less than 20% of the total cellular RNAP-GFP fluorescence signal was localized in the cytoplasm. This background signal likely resulted from scattering of light emitted from the nucleoid-bound RNAP-GFP, due to both the small size of the cell and the limited resolution of the optical microscope. We concluded from these experiments that synthesis of an rrnB transcript that is a certain length in trans is required to recruit RNAP into the cytoplasm.
Localization of RNAP in the cytoplasm depends on active transcription of a plasmid-borne rrnB operon.
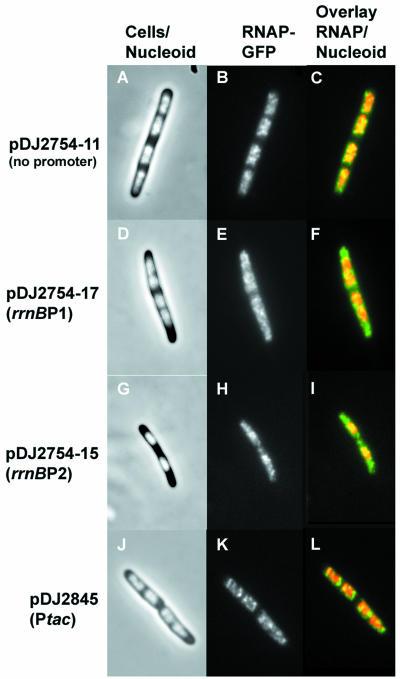
There are two promoters, rrnB P1 and rrnB P2, for the rrnB operon in pNO1301. To determine if either rrnB promoter alone is sufficient for cytoplasmic localization of RNAP, a series of pNO1301 derivatives were constructed in which one or both of the promoters were removed (Table 2). In cells transformed with a derivative lacking both rrnB P1 and rrnB P2 (plasmid pDJ2754-11), no RNAP-GFP signal above the background level was detected in the cytoplasm (Table 2), and all RNAP was localized exclusively in the nucleoid (Fig. 4A, B, and C), similar to the results for pBR322-containing cells. However, in cells containing derivatives that had either only rrnB P1 (plasmid pDJ2754-17) or only rrnB P2 (plasmid pDJ2754-15), the RNAP-GFP signals were detected in both the cytoplasm and the nucleoid (Fig. 4D to I). Image analysis of the micrographs showed that either rrnB P1or rrnB P2 was able to recruit about one-half of the total RNAP-GFP into the cytoplasm (Table 2). We concluded from these experiments that active transcription of a plasmid-borne rrnB operon from either rrnB P1 or rrnB P2 is sufficient to recruit RNAP molecules from the nucleoid into the cytoplasm.
FIG. 4.
Cell morphology and location and distribution of RNA polymerase in cells carrying a plasmid-borne fragment of the rrnB operon transcribed from different promoters. (A, B, and C) Plasmid pDJ2754-11 (no promoter). (D, E, and F) Plasmid pDJ2754-17 (rrnB P1 promoter). (G, H, and I) Plasmid pDJ2754-15 (rrnB P2 promoter). (J, K, and L) Plasmid pDJ2845 (tac promoter). (A, D, G, and J) Merged phase-contrast and DAPI fluorescence images. (B, E, H, and K) Fluorescence from the RNAP-GFP fusion protein. (C, F, I, and L) Overlays of the fluorescence from the RNAP-GFP fusion protein (green) and the fluorescence of the DAPI-stained nucleoids (red).
To determine whether any strong promoter had the same effect as the ribosomal promoters, we replaced both rrnB promoters in pDJ2791A with the Ptac promoter, resulting in plasmid pDJ2845. In contrast to the results obtained with pDJ2791A, RNAP-GFP was located exclusively in the nucleoids of cells containing pDJ2845 (Fig. 4J, K and L), and there were no cytoplasmic RNAP-GFP signals above the background level (Table 2). This result supports the hypotheis that rRNA transcription from ribosomal promoters in trans is uniquely able to recruit RNAP from the nucleoid into the cytoplasm.
DISCUSSION
In this study, we determined the effect of plasmid-borne rrnB on the location and distribution of RNAP in the cell. Our results provide direct visual evidence that active transcription of rrnB recruits a large amount of RNAP molecules. In cells without extrachromosomal copies of rrnB, RNAP is located exclusively in the nucleoid. However, in the presence of a plasmid-borne rrnB, RNAP is found both in the nucleoid and in the cytoplasm under some conditions. Also, we demonstrated that caution should be used to ensure the integrity of a plasmid-borne rRNA operon during experiments as active synthesis of extrachromosomal rRNA is detrimental to the cell. Our results are summarized in Table 2.
For RNAP to be recruited into the cytoplasm, two factors are required, as determined in this study. First, rrnB transcription must result from the rrnB P1 and/or rrnB P2 promoter. Furthermore, the effect is evident only when the rrnB promoters are fully active, such as during growth in rich medium, and is absent when cells are grown in a minimal medium in which rRNA expression is dramatically reduced. Also, another strong promoter, Ptac, was not able to replace the rrnB promoters, which is consistent with the extraordinary strength of the ribosomal promoters during rapid growth (12). Second, some minimal length of the rrnB transcript, but not necessarily an intact rrnB gene, must be present in the plasmid. One possibility is that the DNA sequence encoding the minimal length of the rrnB transcript contains necessary regulatory sites for the synthesis of rRNA. Alternatively, this requirement likely reflects the limitation of detection of the RNAP-GFP signals in our system. Assuming that an elongating RNAP molecule covers approximately 85 bp of DNA, as reported by French and Miller (10), at most about 21 RNAP-GFP molecules transcribe the 1.8-kb rrnB fragment in pDJ2791A, which is the minimal transcript size with which the effect was observed. However, the RNAP-GFP molecules transcribing a plasmid-borne rrnB are unlikely to be saturating (or maximal) as RNAP is probably limiting in the cell, as discussed below. Together, the results of this study demonstrate that active synthesis of ribosomal operons from rRNA promoters is a major driving force for the distribution of RNAP in E. coli. Our results are consistent with the results of numerous previous studies which showed that most RNAP molecules engage in rRNA synthesis in optimal growth conditions (4, 21).
Plasmids pNO1301 and pNO1302 were used previously to study rRNA regulation (14), and it was found that synthesis of chromosomal rRNA was reduced by pNO1301 (which contains an intact rrnB gene) but not by pNO1302 (which contains an rrnB gene with a partial deletion). Based on these results, Jinks-Robertson et al. proposed a “feedback” regulatory mechanism for rRNA expression by free rRNA. Also, it has been reported that fewer RNAP molecules transcribe the chromosomal rrn operons in cells harboring pNO1301 than in cells containing pBR322 or pNO1302 (27). While plasmids pNO1301 and pNO1302 had different effects in the previous studies, in our studies these plasmids behaved similarly in two respects, (i) recruiting RNAP into the cytoplasm and (ii) concomitantly decreasing the number of transcription foci in the nucleoid (Fig. 2Q and 3B). Thus, our results suggest that similar to chromosomal rRNA synthesis in cells harboring pNO1301, chromosomal rRNA synthesis in cells harboring pNO1302 is reduced also. The reasons for the apparent difference between the behavior of the two plasmids in our study and the behavior of the plasmids in the other studies mentioned above are not clear at present. One possibility is that the assays and conditions were different in the different studies.
Our study also has implications for cell growth and global gene regulation. Evidently, synthesis of rRNA from extrachromosomal copies of rrnB is a burden for the cell, and it could account for about 75% of the total rRNA in the cell (13, 19, 25, 26). The extra, plasmid-borne rrn transcripts (either intact or with partial deletions) could affect ribosomal protein synthesis and ribosome assembly, which in turn could disturb the balance of cell growth, leading ultimately to a reduced growth rate. However, another simple explanation is that when RNAP was diverted from the nucleoid into the cytoplasm in the presence of a plasmid-borne rrnB gene, the growth rate of a culture in rich medium was reduced dramatically. The complete correlation between the ability of a plasmid-borne rrn operon (either intact or with a partial deletion) to recruit RNAP into the cytoplasm and the ability to cause a reduced growth rate is striking (Table 2). This result is consistent with the notion that RNAP is limiting in the cell. Due to recruitment of RNAP into the cytoplasmic rrn operons, not only is the synthesis of chromosomal rrn operons reduced, as manifested by the decreased number of transcription foci in the nucleoid, but also the expression of other crucial genes that are sensitive to the concentration of RNAP is probably decreased. These combined effects are probably responsible for the reduced growth rate, the formation of small colonies, and the strong selective pressure to mutate the extrachromosomal element. The proposition that the amount of RNAP is limiting in E. coli and consequently its distribution and redistribution in the genome is sensitive to changing physiological conditions has been proposed to be an important element in global gene regulation, including the stringent (nutrient starvation) response and the carbon source limiting response (1, 16, 29).
Acknowledgments
We thank Tim Durfee, Don Court, Masayasu Nomura, and Catherine Squires for their helpful comments on the manuscript.
This research was supported by the Intramural Research Program of the NIH National Cancer Institute Center for Cancer Research.
REFERENCES
- 1.Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305:689-702. [DOI] [PubMed] [Google Scholar]
- 2.Berg, K. L., C. Squires, and C. L. Squires. 1989. Ribosomal RNA operon anti-termination. Function of leader and spacer region box B-box A sequences and their conservation in diverse micro-organisms. J. Mol. Biol. 209:345-358. [DOI] [PubMed] [Google Scholar]
- 3.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 4.Bremer, H., and P. Dennis. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p. 1553. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 5.Burgess, R. R., B. Erickson, D. Gentry, M. Gribskov, D. Hager, and S. Lesley. 1987. Bacterial RNA polymerase subunits and genes, p. 3-15. In W. S. Reznikoff (ed.), RNA polymerase and the regulation of transcription. Elsevier Science Publishing, New York, NY.
- 6.Cabrera, J. E., and D. J. Jin. 2003. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol. Microbiol. 50:1493-1505. [DOI] [PubMed] [Google Scholar]
- 7.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 8.Cook, P. R. 1999. The organization of replication and transcription. Science 284:1790-1795. [DOI] [PubMed] [Google Scholar]
- 9.Eliasson, A., R. Bernander, S. Dasgupta, K. Nordstrom, H. Niki, and S. Hiraga. 1992. Direct visualization of plasmid DNA in bacterial cells. Subcellular localization of plasmids containing the oriC region of the Escherichia coli chromosome, with or without the sopABC partitioning system. Mol. Microbiol. 6:165-170.1545702 [Google Scholar]
- 10.French, S. L., and O. L. Miller, Jr. 1989. Transcription mapping of the Escherichia coli chromosome by electron microscopy. J. Bacteriol. 171:4207-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon, G. S., D. Sitnikov, C. D. Webb, A. Teleman, A. Straight, R. Losick, A. W. Murray, and A. Wright. 1997. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90:1113-1121. [DOI] [PubMed] [Google Scholar]
- 12.Gourse, R. L., H. A. de Boer, and M. Nomura. 1986. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell 44:197-205. [DOI] [PubMed] [Google Scholar]
- 13.Heinrich, T., C. Condon, T. Pfeiffer, and R. K. Hartmann. 1995. Point mutations in the leader boxA of a plasmid-encoded Escherichia coli rrnB operon cause defective antitermination in vivo. J. Bacteriol. 177:3793-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinks-Robertson, S., R. L. Gourse, and M. Nomura. 1983. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell 33:865-876. [DOI] [PubMed] [Google Scholar]
- 15.Levin, P. A., I. G. Kurtser, and A. D. Grossman. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 96:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, M., T. Durfee, J. E. Cabrera, K. Zhao, D. J. Jin, and F. R. Blattner. 2005. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J. Biol. Chem. 280:15921-15927. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 18.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Mori, H., C. Dammel, E. Becker, K. Triman, and H. F. Noller. 1990. Single base alterations upstream of the E. coli 16S rRNA coding region result in temperature-sensitive 16S rRNA expression. Biochim. Biophys. Acta 1050:323-327. [DOI] [PubMed] [Google Scholar]
- 20.Niki, H., and S. Hiraga. 1999. Subcellular localization of plasmids containing the oriC region of the Escherichia coli chromosome, with or without the sopABC partitioning system. Mol. Microbiol. 34:498-503. [DOI] [PubMed] [Google Scholar]
- 21.Nomura, M., R. Gourse, and G. Baughman. 1984. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 53:75-117. [DOI] [PubMed] [Google Scholar]
- 22.Pogliano, J., T. Q. Ho, Z. Zhong, and D. R. Helinski. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sentenac, A. 1985. Eukaryotic RNA polymerases. Crit. Rev. Biochem. 18:31-90. [DOI] [PubMed] [Google Scholar]
- 24.Siemering, K. R., R. Golbik, R. Sever, and J. Haseloff. 1996. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 6:1653-1663. [DOI] [PubMed] [Google Scholar]
- 25.Stevenson, B. S., and T. M. Schmidt. 1998. Growth rate-dependent accumulation of RNA from plasmid-borne rRNA operons in Escherichia coli. J. Bacteriol. 180:1970-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theissen, G., S. E. Behrens, and R. Wagner. 1990. Functional importance of the Escherichia coli ribosomal RNA leader box A sequence for post-transcriptional events. Mol. Microbiol. 4:1667-1678. [DOI] [PubMed] [Google Scholar]
- 27.Voulgaris, J., S. French, R. L. Gourse, C. Squires, and C. L. Squires. 1999. Increased rrn gene dosage causes intermittent transcription of rRNA in Escherichia coli. J. Bacteriol. 181:4170-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou, Y. N., and D. J. Jin. 1998. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2908-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]