Abstract
Saccharophagus degradans strain 2-40 is a representative of an emerging group of marine complex polysaccharide (CP)-degrading bacteria. It is unique in its metabolic versatility, being able to degrade at least 10 distinct CPs from diverse algal, plant and invertebrate sources. The S. degradans genome has been sequenced to completion, and more than 180 open reading frames have been identified that encode carbohydrases. Over half of these are likely to act on plant cell wall polymers. In fact, there appears to be a full array of enzymes that degrade and metabolize plant cell walls. Genomic and proteomic analyses reveal 13 cellulose depolymerases complemented by seven accessory enzymes, including two cellodextrinases, three cellobiases, a cellodextrin phosphorylase, and a cellobiose phosphorylase. Most of these enzymes exhibit modular architecture, and some contain novel combinations of catalytic and/or substrate binding modules. This is exemplified by endoglucanase Cel5A, which has three internal family 6 carbohydrate binding modules (CBM6) and two catalytic modules from family five of glycosyl hydrolases (GH5) and by Cel6A, a nonreducing-end cellobiohydrolase from family GH6 with tandem CBM2s. This is the first report of a complete and functional cellulase system in a marine bacterium with a sequenced genome.
The plant cell wall is both chemically and structurally complex, containing cellulose, hemicellulose (including xylans, arabinans, and mannans), pectin, and lignin (6, 9, 55). Cellulose microfibrils are the structural backbone of the plant cell wall, forming amorphous and crystalline regions. Cellulose degradation requires multiple enzymes acting in concert to accommodate this structural heterogeneity (6, 35, 53). Such multienzyme cellulase systems contain an array of catalytic modules, substrate-binding pockets, and/or active sites (55). Complicating matters further, the cellulose microfibrils are embedded in a matrix of hemicelluloses (including xylans, arabinans, and mannans), pectins (galacturonans and galactans), and β-1,3 and β-1,4 glucans. Hemicellulose polymers are often substituted with arabinose, galactose, and/or xylose residues, yielding arabinoxylans, galactomannans, and xyloglucans, among others (32, 35, 53, 55). The complexity and sheer number of different glycosidic bonds present in noncellulosic cell wall polysaccharides require specific enzyme systems which often rival cellulase systems in enzyme count and complexity. Because of its heterogeneity and complexity, plant cell wall degradation often requires a consortia of microorganisms (34, 53). However, there are also cellulolytic organisms that have enzyme systems that act on hemicellulose components, e.g., certain members of the genus Clostridium (1, 59).
The microbial degradation of plant cell walls, particularly the cellulose and hemicellulose components, is not as well characterized in the oceans as it is in terrestrial systems. Among the cellulolytic organisms chosen for genome sequencing, there are few marine representatives (http://microbialgenome.org/organisms.shtml). On the other hand, the cellulase systems of a number of soil and ruminant bacteria have been the subject of considerable study (4, 37). Very recently, however, several related marine bacterial genera that can degrade cellulose have been discovered (2, 14, 20). One species is Saccharophagus degradans strain 2-40T (formerly “Microbulbifer degradans” strain 2-40). S. degradans is a pleomorphic, gram-negative, aerobic, motile γ-proteobacterium (16) that was isolated from decaying salt marsh cord grass Spartina alterniflora in the Chesapeake Bay watershed (2). It has extraordinary metabolic versatility, demonstrated to degrade at least 10 different complex polysaccharides (CP) (17). S. degradans represents a third genus in the newly emerging Microbulbifer-Teredinibacter-Saccharophagus group of marine carbohydrate degraders (16). Of the 20 isolates in this group, only the genome of S. degradans has been sequenced.
S. degradans can grow using Avicel, filter paper, and plant matter as the sole carbon and energy sources, wherein growth is concurrent with visible degradation of the substrates (52). Not surprisingly, annotations of the S. degradans genome reveal approximately 180 open reading frames (ORFs) related to a wide variety of carbohydrate-active enzymes. Roughly half of these ORFs appear to be dedicated to metabolizing common structural polysaccharides, including cellulose, which are found in green plants and/or algae (43). There are 112 ORFs that contain catalytic and/or carbohydrate-binding modules (CBM) with predicted specificity for plant-derived polysaccharides, 20 of which contain a CBM but do not have a predicted catalytic module. Although the function of these “CBM-only” proteins is uncertain, it is noteworthy that the genome of S. degradans contains more of them than any other organism that has been studied thus far.
Although plant cell walls often require consortia of microorganisms to degrade them, S. degradans does this in pure culture (52). The genomic analyses corroborate this observation. In addition to complete cellulase and xylanase systems, the genome contains at least five other multienzyme systems which appear to be involved in plant wall degradation: arabinanases, mannanases, β-1,3-glucanases, and pectinases. Each of these systems contain multiple depolymerases with diverse catalytic and substrate-binding modules.
Based upon the recently finished genome sequence, we present here genomic, proteomic, and functional analyses of the S. degradans cellulase system—the first in a marine proteobacterium. The present study focuses on the architecture of the catalytic and substrate-binding modules and their contribution to cellulose degradation.
MATERIALS AND METHODS
Genomic analyses.
The finished assembly of the genome sequence of S. degradans, obtained in conjunction with the U.S. Department of Energy's Joint Genome Institute (JGI) comprises 5.06 Mb in a single, circular chromosome. Automated annotations by the computational genomics division of the Oak Ridge National Laboratory (ORNL) are available on the World Wide Web at http://genome.ornl.gov/microbial/mdeg/.
In an extension of the ORNL annotation, all of the ORFs were analyzed on the CAZy ModO (Carbohydrase Active enZyme Modular Organization) server at AFMB-CNRS (11). Catalytic and substrate-binding modules were identified and named according to the CAZy nomenclature scheme (25). The known characteristics of the GH and CBM families that are commonly involved in cellulose metabolism and also occur in the genome of S. degradans are summarized in Table 1.
TABLE 1.
Properties of studied examples from the GH and CBM families occurring within the predicted cellulases, CBM proteins, and glycanases of uncertain specificity of S. degradans
| Modulea | Demonstrated function(s) of examples found in the literature | Anomeric reaction conformationb | Referencec |
|---|---|---|---|
| GH1 | β-Glucosidase, β-galactosidase, β-mannosidase, others | r | |
| GH3 | β-1,4-Glucosidase, β-1,4-xylosidase, β-1,3-glucosidase, others | r | |
| GH5 | β-1,4-Endoglucanase, β-1,4-endoxylanase, β-1,4-endomannanase, licheninase, others | r | |
| GH6 | Nonreducing end cellobiohydrolase, β-1,4-endoglucanase | i | |
| GH9 | β-1,4-Endoglucanase, cellobiohydrolase, β-glucosidase | i | |
| GH16 | β-1,3-Endoglucanase, β-1,3(4)-endoglucanase, licheninase, β-agarase, others | r | |
| GH26 | β-1,4-Endomannanase, β-1,3-xylanase | r | |
| GH28 | α-1,4-Galacturonanase (polygalacturonase), exo-polygalacturonase | i | |
| GH30 | β-1,6-Endoglucanase, glucosylceramidase | r | |
| GH43 | β-1,4-Xylosidase, α-arabinofuranosidase, α-endoarabinanase, β-1,4-endoxylanase | i | |
| GH94 | Cellobiose phosphorylase, cellodextrin phosphorylase, chitobiose phosphorylase | i | |
| CBM2a | Binds crystalline cellulose | - | 49 |
| CBM6 | Binds glycan chains: cellulose, xylan, β-glucans | - | 8 |
| CBM10 | Binds crystalline substrates: cellulose, mannan | - | |
| CBM13 | Lectin-like binding of mono- and oligosaccharides, xylan | - | 38 |
| CBM16 | Found in alginases, chitinases, and mannanases; binding unknown | - | |
| CBM32 | Lectin-like binding of small, soluble oligosaccharides, dp 1-3 | - |
GH, glycosyl hydrolase; CBM, carbohydrate-binding module. Numbers refer to families according to CAZy (http://www.cazy.org).
r, retaining; i, inverting; −, none.
Data are from the CAZy database (http://www.cazy.org) unless otherwise indicated.
Identified carbohydrase ORFs were also analyzed for lipoprotein signature sequences (lipoboxes) by the DOLOP Web server (Database of LipOProteins; http://www.mrc-lmb.cam.ac.uk/genomes/dolop/) and the Technical University of Denmark's CBS LipoP 1.0 server (http://www.cbs.dtu.dk/services/LipoP), which uses hidden Markov models (30) to identify candidate lipoproteins.
Bacterial growth media and conditions.
Escherichia coli strains were grown according to standard protocols. S. degradans strain 2-40 (ATCC 43961T) was grown at 27°C on half-strength marine agar (18.7 g of Difco Marine Broth 2216/liter amended with 1.5% agar) or in minimal broth medium consisting of 23 g of Instant Ocean Sea salts (Aquarium Systems, Mentor, OH), 1 g of yeast extract, 50 mM Tris buffer (pH 7.4), and 0.05% (wt/vol) NH4Cl/liter. Minimal medium was supplemented with 0.2% (wt/vol) Avicel PH-101 (Fluka, Bruchs, Switzerland), barley β-glucan (medium viscosity; Megazyme, Bray, Ireland), laminarin (from Laminaria digitata; Sigma, St. Louis, MO), or xylan (Birchwood; Sigma).
Cloning and expression of S. degradans proteins in E. coli.
ORF sequences were used to design primers (Table 2), permitting in-frame cloning into pETBlue2 (Novagen, Madison, WI). PCRs included the primers (Invitrogen, Frederick, MD) and S. degradans genomic template under standard conditions for tailed primers and Proof Pro Polymerase (Continental Lab Products, San Diego, CA). The pETBlue2 constructs were transformed into E coli DH5α, screened, and transformed into the E. coli BL21(DE3)pLysS (Tuners; Novagen). The production of an appropriate-sized, IPTG (isopropyl-β-d-thiogalactopyranoside)-induced, His-tagged protein was confirmed by Western blotting with anti-HisTag monoclonal antibody (Novagen).
TABLE 2.
Primers used in this study
| Primer | Restriction site | Nucleotide sequencea |
|---|---|---|
| rCel5A-F | EcoRV | AGTCTTGATATCCATAACCGCCATGCAAAGCACT |
| rCel5A-R | XhoI | CCGCTCGAGGTTGAACATGCTGGGTGGTTCT |
| rGly3D-F | BamHI | CGGGATCCGAAGGCTACTGCGCAAACGAAT |
| rGly3D-R | AscI | AGGCGCGCCAGAACCAGGCCTGTGCCATTTA |
| rGly5K-F | AscI | AGGCGCGCCCGCAATCACACCACAATACCAA |
| rGly5K-R | ClaI | CCATCGATGACGTTATTAGCGCTGCCATTT |
| rGly5M-F | BamHI | CGGGATCCGGCACTGTTAACAAGCGCCAAGT |
| rGly5M-R | AscI | AGGCGCGCCGCCTCGGTTTGCCATGTTTGCCA |
| rGly9C-F | AscI | AGGCGCGCCAAACCGCGCCCATCAAATTAGC |
| rGly9C-R | ClaI | CCATCGATCTGATTAAATGGCGGTGAAGGCG |
| rGly43M-F | BamHI | CGGGATCCGACCGTAGCGCTAGACACAACAA |
| rGly43M-R | AscI | AGGCGCGCCCCTTCGGCGAACAGTTCACTTACA |
| rCbm2A-F | EcoRV | AGTCTTGATATCCAGTATGGGGACGTTGTCGTCT |
| rCbm2A-R | XhoI | CCGCTCGAGTGGCACCGTGGTGCTAATAAC |
| rCbm2B-F | AscI | AGGCGCGCCTTTCAGCGCTCTCGCTAGGTTT |
| rCbm2B-R | ClaI | CCATCGATTACACCAACGCCCACGGAATAA |
| rCbm2C-F | AscI | AGGCGCGCCCTATTTCGCTTGGTGCAATGA |
| rCbm2C-R | ClaI | CCATCGATCTTATCCTTCCCACCAGCATTC |
Underlining delineates the restriction sites introduced to permit in-frame cloning into the pETBlue2 vector.
Production and purification of recombinant proteins.
Expression cultures were induced by treatment with 1 mM IPTG for 4 h at 37°C or for 16 h at 20°C. Cell pellets were frozen overnight at −20°C, thawed, and lysed in urea buffer (8 M Urea, 100 mM NaH2PO4, 25 mM Tris [pH 8.0]) at 4 ml/g (wet weight). Cell lysates were column affinity purified on nickel-NTA resin (QIAGEN, Valencia, CA). Renaturation was done on the column at 4°C by using 25 mM Tris (pH 7.4), 500 mM NaCl, and 20% glycerol, with urea in decreasing concentrations of 6 to 1 M in 1 M steps. The samples were eluted in 1 M urea refolding buffer amended to 250 mM imidazole. Millipore (Billerica, MA) Centricon and/or Microcon devices were used to concentrate the recombinant protein fractions and exchange them into storage buffer (20 mM Tris [pH 7.4], 10 mM NaCl, 10% glycerol). Aliquots were held at −80°C for later use. Final protein concentrations ranged from 500 to 2,200 μg/ml. Where needed, samples were adjusted to 500 μg/ml prior to further analyses.
Zymograms.
Gels were prepared as miniformat sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (33) with substrate incorporated directly into the separating gel at final concentrations of 0.1% for HE-cellulose (medium viscosity; Fluka) and 0.15% for barley β-glucan (medium viscosity; Megazyme). Each lane contained 5 μg of recombinant protein or 15 μg of filtered supernatant from S. degradans cultures grown in Avicel, barley β-glucan, xylan (Birchwood; Sigma), or laminarin (Laminaria digitata; Sigma). Gels were incubated in 80 ml of refolding buffer (20 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer [pH 6.8], 2.5% Triton X-100, 2 mM dithiothreitol, 2.5 mM CaCl2) for 1 h at room temperature and then held overnight in fresh refolding buffer at 4°C. The gels were transferred to PIPES buffer, incubated 4 h at 37°C, and stained in 0.25% Congo red.
Nelson-Somogyi reducing-sugar assays.
Expressed proteins were assayed by the Nelson-Somogyi reducing sugar method adapted to 96-well microwell plates (21). The substrates included Avicel, carboxymethyl cellulose (CMC), phosphoric acid swollen cellulose (PASC) prepared as described by Wood (56), and xylan dissolved at 1% or barley β-glucan and laminarin at 0.5% in 20 mM PIPES (pH 6.8). Barley β-glucan, laminarin, and xylan assays were incubated for 2 h at 37°C; Avicel, CMC, and PASC assays were incubated for 36 h at 37°C. Assays were done in triplicate, with protein concentrations measured in triplicate using the bicinchoninic acid protein assay (50) (BCA Assay; Pierce Biotechnology, Rockford, IL). The enzymatic activity was calculated, with one unit defined as 1 μmol of reducing sugar released/min and reported as the specific activity in mU/mg of protein.
Exoglycosidase activity assays: pNP derivatives.
Purified proteins were assayed using 5 mM para-nitrophenol (pNP) derivatives of α-l-arabinofuranoside, α-l-arabinopyranoside, β-l-arabinopyranoside, β-d-cellobioside, α-d-glucopyranoside, β-d-glucopyranoside, α-d-xylopyranoside, and β-d-xylopyranoside in 20 mM PIPES (pH 6.8). The A405 was compared to a p-nitrophenol standard curve and is reported as specific activities in mU/mg of protein, with one unit (U) defined as 1 μmol of pNp/min. All pNp conjugates were obtained from Sigma.
Cellulose binding assays.
Aliquots of Cbm2A:His6, Cbm2B:His6, and Cbm2C:His6 were adjusted to 500 μg/ml in storage buffer (20 mM Tris [pH 7.4], 10 mM NaCl, 10% glycerol), and 500-μg/ml solutions of the control proteins, pepsin (37 kDa), porcine lipase (50 kDa), and ovalbumin (45 kDa) were prepared in the same buffer. A 20-μl drop of 10% (wt/vol) Avicel in Milli-Q water was added to 75 μl of 500 μg/ml of bovine serum albumin (BSA) in 20 mM PIPES buffer (pH 6.8) and intermittently vortexed for 15 min at 25°C. The Avicel pellet was washed twice in 400 μl of PIPES, resuspended in 20 μl of the test or control protein solution, and intermittently vortexed 3 min at 25°C. The pellets were washed four times for 10 min each time in 2 M urea at 56°C and then analyzed by SDS-PAGE. Staining with Coomassie blue revealed proteins remaining bound to the washed Avicel, and specific binding attributed to CBM modules was defined as the persistence of the test protein under conditions that removed all three negative control proteins.
MS and proteomic analyses.
Supernatants of Avicel-, CMC-, and xylan-grown cultures were concentrated to ∼25-fold in Centricon or Microcon devices (Millipore). Protein concentrations were determined by the BCA protein assay (Pierce). Samples were denatured and reduced in 8 M urea-10 mM dithiothreitol-100 mM Tris (pH 8.5) for 2 h at 37°C, after which they were alkylated in 50 mM iodoacetate and then exchanged into 50 mM Tris-1 mM CaCl2 (pH 8.5). The samples were digested overnight at 37°C with proteomics-grade trypsin (Promega). Digestions were analyzed by reversed-phase high-pressure liquid chromatography (RPHPLC)-tandem mass spectrometry (MS/MS) at the University of Maryland at College Park College of Life Sciences CORE Mass Spectrometry facility using a Waters 2960 HPLC linked to a Finnagin LCQ tandem mass spectrometer. Alternatively, to identify specific proteins from SDS-PAGE gels of Avicel-grown supernatants, regions containing 120-, 100-, and 75-kDa proteins were excised from an 8% gel and sent to Stanford University's mass spectrometry facility (http://mass-spec.stanford.edu), where they were processed similarly and analyzed by using an ESI-Quadrupole-Time-of-Flight (Micromass Q-Tof) mass spectrometer. All peptide fragment masses were analyzed by the peptide analysis packages SEQUEST and MASCOT (15, 39) and compared to amino acid sequence translations of all ORFs in the S. degradans draft genome and to the nonredundant Mass Spectrometry Database (ftp://ftp.ncbi.nih.gov/repository/MSDB/msdb.nam). Peptide identity matches were evaluated by using the accepted thresholds of statistical significance specific to each program.
RESULTS
Overview of the plant wall-degrading enzymes.
Since S. degradans degrades plant matter in monoculture, it is expected to have a broad spectrum of carbohydrases. The CAZy ModO database identifies 77 ORF coding for predicted S. degradans plant wall degradases, including apparent xylanase, arabinoxylanase, β-mannanase, β-1,3-glucanase, and pectinase systems comprised of multiple depolymerases and accessory enzymes for the removal of substituents and the processing of oligomers to monomers. We focus here on the 13 cellulose depolymerases and 7 associated accessory enzymes from six different glycoside hydrolase (GH) families (Table 3). Most of these enzymes contain one or more CBM.
TABLE 3.
Predicted cellulases and accessory enzymes of S. degradans
| Namea | Sde no.c | Predicted functiond | Module(s)d | No. of amino acidsg | MM (kDa)g | MSh |
|---|---|---|---|---|---|---|
| Cel5Ab | 3003 | Endo-1,4-β-glucanase (EC 3.2.1.4) | GH5/CBM6/CBM6/CBM6/GH5 | 1,167 | 127.2 | |
| Cel5B | 2490 | Endo-1,4-β-glucanase | LPB/PSL(47)/CBM6/GH5 | 566 | 60.8 | |
| Cel5C | 0325 | Endo-1,4-β-glucanase | LPB/PSL(47)/GH5 | 451 | 49.1 | |
| Cel5D | 2636 | Endo-1,4-β-glucanase | CBM2/PSL(58)/CBM10/PSL(36)/GH5 | 621 | 65.9 | |
| Cel5E | 2929 | Endo-1,4-β-glucanase | CBM6/CBM6/GH5 | 673 | 72.6 | |
| Cel5F | 1572 | Endo-1,4-β-glucanase | GH5 | 365 | 42.0 | |
| Cel5G | 3239 | Endo-1,4-β-glucanase | GH5/PSL(21)/CBM6/PSL(32)/Y95 | 638 | 67.9 | |
| Cel5H | 3237 | Endo-1,4-β-glucanase | GH5/PSL(32)/CBM6/EPR(16) | 630 | 66.9 | av |
| Cel5I | 3420 | Endo-1,4-β-glucanase | CBM2/PSL(33)/CBM10/PSL(58)/GH5 | 725 | 77.2 | av |
| Cel5J | 2494 | Endo-1,4-β-glucanase | GH5/CBM6/CBM6 | 610 | 65.2 | |
| Cel6A | 2272 | Cellobiohydrolase (EC 3.2.1.91)e | CBM2/PSL(43)/CBM2/PSL(85)/GH6 | 791 | 81.9 | |
| Cel9A | 0636 | Endo-1,4-β-glucanase | GH9 | 578 | 62.7 | |
| Cel9B | 0649 | Endo-1,4-β-glucanase | GH9/PSL(54)/CBM10/PSL(50)/CBM2 | 867 | 89.5 | av |
| Ced3A | 2497 | Cellodextrinase (EC 3.2.1.74) | LPB/GH3/PLP | 1,072 | 116.0 | av, cm, xn |
| Ced3B | 0245 | Cellodextrinase | LPB/GH3 | 862 | 92.9 | xn |
| Bgl1A | 3603 | Cellobiase (EC 3.2.1.21) | GH1f | 461 | 52.8 | |
| Bgl1B | 1394 | Cellobiase | GH1f | 444 | 49.8 | |
| Bgl3C | 2674 | Cellobiase | LPB/GH3/UNK(511) | 866 | 95.4 | av, cm, xn |
| Cep94A | 1318 | Cellobiose phosphorylase (EC 2.4.1.20) | GH94f | 811 | 91.7 | |
| Cep94B | 0906 | Cellodextrin phosphorylase (EC 2.4.1.49) | GH94f | 788 | 88.7 | cm |
Cel, cellulase; Ced, cellodextrinase; Bgl, β-glucosidase; Cep, cellobiose/cellodextrin phosphorylase.
cel5A was cloned, expressed as Cel5A:His6, and assayed for activity.
ORF number with the prefix “Sde,” for S. degradans as assigned in 15 June 2005 genome assembly (http://genome.ornl.gov/microbial/mdeg/15jun05/mdeg.html).
Predictions of function and module determination by CAZy ModO at AFMB-CNRS. Module abbreviations: CBM, carbohydrate-binding module; GH, glycosyl hydrolase (numbers refer to families according to CAZy [http://www.cazy.org]); EPR, glutamic acid-proline-rich region; LPB, lipobox signature sequence; PLP, phospholipase-like domain; PSL, polyserine linker; UNK, unknown function. Numbers in parentheses indicate the length of a region in amino acid residues.
Cel6A is predicted to act on the nonreducing end of cellulose chains.
No type II secretion signal sequence.
Molecular mass (MM) and amino acid count calculated by the protParam tool at http://us.expasy.org/tools/protparam.html using DOE/JGI gene model amino acid sequence translations including any predicted signal peptide.
MS, protein identified by MS/MS in concentrated supernatant from cultures grown in Avicel (av), CMC (cm), or xylan (xn).
In addition to the 77 easily annotated plant-degrading ORFs, there are another 35 with poorly conserved CBM and/or GH modules from families commonly involved in plant CP binding and/or degradation that require further analyses. Nineteen contain a CBM but have no identifiable carbohydrase module; one of these includes a zinc-dependent protease module, suggesting a role as an accessory protease. The rest of these sequences lack any recognizable catalytic modules, and we have designated them “CBM proteins” and given them the prefix “Cbm” (Table 4). There are also 16 genes (Table 5) with GH modules of uncertain function or identity. We termed these “glycanases of uncertain specificity” and identify them with the acronym “Gly” pending an authoritative determination of their substrate. Several of these proteins of uncertain specificity were subjected to functional analyses.
TABLE 4.
Selected CBM proteins
| Namea | Sde no.c | Module(s)d | No. of amino acidse | MM (kDa)e | MSf |
|---|---|---|---|---|---|
| Cbm2Ab | 1182 | CBM2/EPR(48)/UNK(191) | 371 | 38.0 | |
| Cbm2Bb | 1183 | CBM2/UNK(914) | 1,042 | 112.1 | av |
| Cbm2Cb | 0182 | CBM2/PSL(58)/Y94/PSL(25)/UNK(577) | 933 | 97.5 | xn |
| Cbm2D-cbm10A | 0650 | CBM2/PSL(43)/CBM10/PSL(101)/UNK(231) | 558 | 56.3 | |
| Cbm2E | 0569 | CBM2/PSL(18)/PSL(18)/UNK(471) | 781 | 85.0 | |
| Cbm2F | 2939 | CBM2/PSL(33)/PSL(17)/UNK(544) | 787 | 84.8 | |
| Cbm6A | 0242 | LPB/PSL(28)/CBM6/UNK(476) | 681 | 72.5 | |
| Cbm6B | 0216 | CBM6/CBM6/UNK(217) | 630 | 67.2 | |
| Cbm6C | 3892 | CBM6/UNK(216) | 550 | 59.8 | |
| Cbm6D | 3260 | CBM6/UNK(262) | 500 | 54.6 | |
| Cbm6E | 1445 | LPB/UNK(311)/CBM6 | 473 | 53.0 | |
| Cbm6F | 3927 | UNK(311)/CBM6 | 465 | 50.4 | |
| Cbm6G-Cbm16B | 0112 | UNK(620)/CBM6/CBM16 | 1,024 | 111.4 | |
| Cbm6H-Cbm32F | 2795 | UNK(514)CBM6/CBM32 | 912 | 97.5 | |
| Cbm16A-Cbm32E | 3272 | CBM16/PTR(16)/CBM32/PTR(22)/UNK(232) | 552 | 58.5 | |
| Cbm32A | 0478 | CBM32/CBM32/UNK(251)/COG3488 | 1,028 | 111.9 | xn |
| Cbm32B | 3845 | UNK(109)/CBM32/CBM32/CBM32 | 557 | 59.5 | |
| Cbm32C | 1503 | ZDP/CBM32/FN3/FN3/CBM32 | 674 | 73.6 | |
| Cbm32D | 3709 | CBM32/FCL/UNK(521) | 818 | 88.8 |
Cbm, carbohydrate-binding module.
ORF was cloned, expressed as a HisTag fusion protein, and assayed for cellulose binding and enzyme activity.
ORF number having the prefix “Sde,” for Saccharophagus degradans as assigned in the 15 June 2005 genome assembly (http://genome.ornl.gov/microbial/mdeg/15jun05/mdeg.html).
Predictions of function and module determination by CAZy ModO at AFMB-CNRS. Module abbreviations: CBM, carbohydrate-binding module; COG3488, thiol-oxidoreductase like domain; EPR, glutamic acid-proline-rich region; FCL, similar to fucose-binding lectins in eel serum proteins; FN3, fibronectin type 3 module; LPB, lipobox signature sequence; PSL, polyserine linker; PTR, proline-threonine repeat; UNK, unknown function; Y94, novel domain identified in this protein and two xylanases by CAZyme analysis; ZDP, zinc-dependent protease domain.
Molecular mass (MM) and amino acid count calculated by the protParam tool at http://us.expasy.org/tools/protparam.html using DOE/JGI gene model amino acid sequence translations including any predicted signal peptide.
Protein identified by MS/MS in concentrated supernatant from cultures grown in Avicel (av) or xylan (xn).
TABLE 5.
Glycanases of uncertain substrate specificity
| Namea | Sde no.c | Predicted functiond | Module(s)d | No. of amino acidse | MM (kDa)e | MSf |
|---|---|---|---|---|---|---|
| Gly3Db | 0475 | β-Glycosidase (EC 3.2.1.-) | CBM32/CBM32/CBM32/GH3/CBM32 | 1,581 | 173.0 | xn |
| Gly5Kb | 2993 | Endo-(1,3 or 1,4)-β-glucanase (EC 3.2.1.4/6) | GH5/CBM6/CBM6/CBM13 | 863 | 94.6 | |
| Gly5L | 2996 | Endo-(1,3 or 1,4)-β-glucanase | LQAC/GH5/CBM6 | 853 | 93.3 | |
| Gly5Mb | 3023 | Endo-(1,3 or 1,4)-β-glucanase | LPB/EPR(34)/GH5/FN3/CBM6 | 869 | 94.9 | |
| Gly5R | 1121 | β-Glycosidase or mannanase (EC 3.2.1.25) | GH5 | 457 | 50.9 | |
| Gly5S | 2285 | β-Glycosidase | GH5 | 383 | 42.8 | xn |
| Gly9Cb | 0558 | β-Glycosidase | LPB/GH9 | 665 | 73.2 | |
| Gly16H | 2878 | β-Glycosidase | GH16/CBM4 | 968 | 107.2 | |
| Gly26B | 3022 | β-Glycosidase | GH26/CBM13/CBM13 | 644 | 71.6 | |
| Gly30A | 2992 | β-Glycosidase | LPB/GH30/CBM6/CBM6/CBM13 | 982 | 107.4 | |
| Gly30B | 2994 | β-Glycosidase | GH30 | 481 | 52.8 | |
| Gly43Mb | 3317 | β-Glycosidase | LPB/GH43/LGL/LGL | 1,174 | 127.2 |
Poorly conserved modules from GH families which contain plant-wall depolymerases are designated by the generic prefix “Gly” pending identification of substrate specificity.
ORF was cloned, expressed as a HisTag fusion protein, and assayed for enzyme activity.
Gene number having the prefix “Sde” for Saccharophagus degradans as assigned in the 15 June 2005 genome assembly (http://genome.ornl.gov/microbial/mdeg/15jun05/mdeg.html).
Predictions of function and module determination by CAZy ModO at AFMB-CNRS. Module abbreviations: CBM, carbohydrate-binding module; EPR, glutamic acid-proline-rich region; FN3, fibronectin type 3 module; LPB, lipobox signature sequence; LQAC, putative alternate lipobox sequence with atypical second amino acid.
Molecular mass (MM) and amino acid count calculated by the protParam tool at http://us.expasy.org/tools/protparam.html using DOE/JGI gene model amino acid sequence translations including any predicted signal peptide.
Protein identified by MS/MS in concentrated supernatant from cultures grown in Avicel (av) or xylan (xn).
An unusually high percentage of S. degradans carbohydrases contain lipobox sequences, which are normally acylated and anchored to the outer membrane so that the enzymes function on the cell surface. In fact, DOLOP analysis reveals that of the 112 predicted or potential plant wall carbohydrases, 32 (28.5%) contain consensus lipobox sequences: [LVI][ASTVI][GAS][C]. Five of these are cellulases and cellulase accessory enzymes (Table 3), five are glycanases of unknown specificity (Table 5), and two are “CBM only” proteins (Table 4). There are also two genes (Gly5L and Man5Q) with lipobox-like sequences that carry an atypical amino acid in the second position (LQAC); these two were identified as probable lipoproteins by LipoP 1.0.
Cellulases and related accessories.
Of the 13 predicted S. degradans cellulases, 12 annotate as β-1,4-endoglucanases and one as a cellobiohydrolase that acts on the nonreducing ends of cellulose chains (Table 3). The seven “cellulase accessory enzymes” process cellulose oligosaccharides and include two cellodextrinases, three cellobiases, a cellodextrin phosphorylase, and a cellobiose phosphorylase. All of these proteins include an N-terminal type II secretion signal sequence except for two of the cellobiases and both phosphorylases. Two endoglucanases, Cel5B and Cel5C, the cellodextrinases Ced3A and Ced3B, and cellobiase Bgl3C contain lipobox motifs. Eight of the thirteen cellulose depolymerases contain one or more extended regions comprised mainly of serine residues (Table 3). These “polyserine linkers” (PSL) are similar to those in certain carbohydrases of Cellvibrio japonicus (19, 22) and Clostridium thermocellum (62), where they are believed to function as flexible spacers or linkers. Such sequences occur in an unusually high number of S. degradans carbohydrases (26).
The activities of several predicted cellulases were confirmed by using zymograms. Analysis of Avicel-grown cultures in these zymograms revealed a number of protein bands with activity against HE-cellulose and barley β-glucan (Fig. 1), showing that there are numerous endoglucanases that can be induced to degrade cellulose. However, the number of predicted endoglucanases with molecular masses between 60 and 100 kDa made specific bands of activity difficult to correlate with genomically identified enzymes.
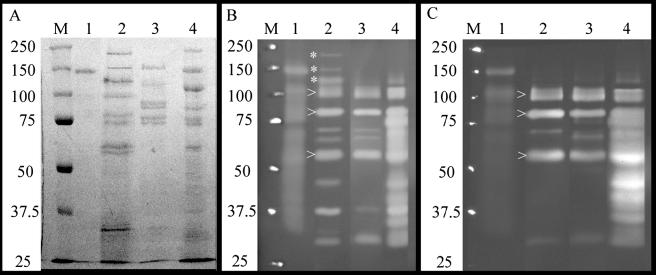
FIG. 1.
Zymograms of Cel5A:His6- and CP-grown stationary-phase 2-40 culture supernatants. (A) SDS-8% PAGE gel stained with Coomassie blue. Lanes: M, Bio-Rad Precision Plus molecular weight markers (Bio-Rad, Hercules, CA); 1, ∼5 μg of Cel5A:His6; 2, ∼15 μg of laminarin-grown culture supernatant; 3, ∼15 μg of Barley β-glucan-grown culture supernatant; 4, ∼15 μg of Avicel-grown culture supernatant. (B) SDS-8% PAGE zymogram containing 0.15% barley β-glucan (medium viscosity; Megazyme, Bray, Ireland). Lanes are as defined in panel A. (C) SDS-8% PAGE zymogram containing 0.1% HE-cellulose (medium viscosity; Fluka). Lanes are as described in panel A. Note the high-molecular-mass bands (✽) in panel B, lane 2, showing three large β-1,3-endoglucanases produced in response to laminarin but not in response to cellulose or barley β-glucan. Also note the appearance of three conserved bands (arrows) in lanes 2 to 4 of panels B and C, suggesting that as many as three β-1,4-endoglucanases function as common members of laminarin, mixed glucan, and cellulose degradative pathways.
To identify individual carbohydrases, culture supernatants were analyzed by RPHPLC-MS/MS. Three cellulases and four cellulase accessory enzymes were repeatedly detected in cellulose- and/or xylan-grown culture supernatants (Table 3). The cellulases were Cel5H, Cel5I, and Cel9B, and the accessory enzymes included the cellodextrinases, Ced3A and Ced3B, as well as the cellobiase, Bgl3C. Also detected was the cellodextrin phosphorylase, Cep94B, which lacks a secretion signal sequence. The detection of Cep94B in CMC-grown culture supernatants indicated lysis of some of these stationary-phase cells.
Enzymatic and binding domain analysis.
The cellulases of S. degradans are unlike those of other sequenced cellulolytic bacteria and contain a fascinating array of modules. Compared to other cellulolytic prokaryotes with sequenced genomes, S. degradans has an unusually high number of enzymes from glycoside hydrolase family 5 (Table 6). Twelve of the predicted cellulases annotate as endoglucanases, ten of which have GH5 modules, and the other two have GH9 modules (Table 3). There is also a GH6 nonreducing end cellobiohydrolase. Unlike two well-studied bacterial models of cellulose degradation, Thermobifida fusca and Clostridium thermocellum (29), the genome of S. degradans does not appear to encode a GH48 reducing-end cellobiohydrolase (Table 6).
TABLE 6.
Distribution of candidate cellulases and accessory cellodextrinases, β-glucosidases, and phosphorylases in the genomes of completely sequenced cellulolytic bacteria
| Familya | Subfamilyb | Distribution (no.)
|
||||
|---|---|---|---|---|---|---|
| Saccharophagus degradans 2-40c | Cytophaga hutchinsoniic | Thermobifida fuscac | Clostridium thermocellumc | Thermotoga maritimad | ||
| GH5 | 16 | 5 | 2 | 7 | 2 | |
| GH6 | Endo | 1 | ||||
| GH6 | Exo | 1 | 1 | |||
| GH8 | 4 | 1 | ||||
| GH9 | 3 | 7 | 2 | 14 | ||
| GH12 | 2 | |||||
| GH48 | 1 | 2 | ||||
| GH74 | 1 | 1 | 1 | 1 | ||
| GH1 (β-glucosidases) | 2 | 1 | 2 | 2 | 1 | |
| GH3 (β-glucosidases) | 5 | 5 | 1 | 1 | 3 | |
| GH94 (phosphorylases) | 2 | 2 | 1 | |||
Predictions of function and module determination by CAZy ModO at AFMB-CNRS.
GH family 6 includes endoglucanases and exo-acting cellobiohydrolases.
Data obtained by BLAST analysis on the NCBI microbial genomes server (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi).
Data from the CAZy database (http://www.cazy.org).
Most S. degradans cellulases contain one or more CBM belonging to families 2, 6, and 10 (Table 3). Sequence analysis shows that all of the CBM2 modules in the genome are from subfamily 2a, indicating that they bind crystalline cellulose (7). Cellobiohydrolase Cel6A contains two such CBM2 modules, making it the first known GH6 protein with dual CBM2 modules. Three endoglucanases, Cel5D, Cel5I, and Cel9B, also have CBM that are predicted to bind crystalline cellulose, each enzyme containing one CBM2a and one CBM10 (Table 3). Six other endoglucanases contain one or more CBM6 modules, and only three lack a CBM entirely (Table 3). The cellodextrinases, cellobiases, and phosphorylases lack CBM, in accordance with the soluble nature of their substrates. The substrate binding-specificities of the CBM are considered important in assessing the binding sites, and thereby the function, of the cellulases (see Discussion). For example, although the GH5 module of Cel5D is a predicted endoglucanase, the CBM2 and CBM10 both bind crystalline cellulose. Thus, Cel5D is thought to bind and act on or near crystalline regions (7).
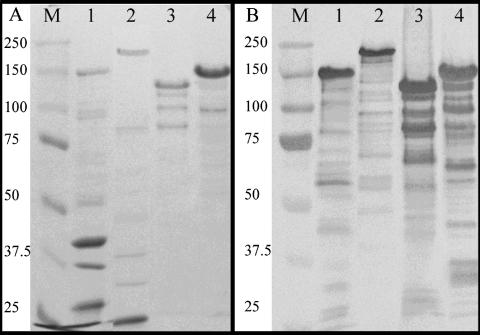
Cel5A was studied further because of its intriguing features. In addition to its three CBM6s, Cel5A contains dual GH5 modules, both of which annotate as endoglucanases although their sequences are dissimilar (37% identity and 54% similarity in amino acid sequence; 60.9% identical nucleotide sequence). Cel5A was cloned, expressed as Cel5A:His6 (Fig. 2), and assayed for activity. Zymograms indicated that Cel5A:His6 was active on HE-cellulose and barley β-glucan (Fig. 1). Reducing sugar assays revealed that it had approximately 10- and 20-fold more activity versus barley β-glucan than versus CMC and PASC, respectively (Table 7). No activity was detected against Avicel, xylan, or laminarin. The activity of Cel5A:His6 against pNP-β-cellobioside and the absence of activity against pNP-β-glucopyranoside suggest that the enzyme does not cleave oligo-cellodextrins of less than three residues and corroborates the results from reducing-sugar assays. Thus, it is concluded that Cel5A is a β-1,3(4) endoglucanase with secondary activity on amorphous cellulose chains.
FIG. 2.
Expression of His-tagged recombinant proteins. (A) SDS-8% PAGE gel stained with Coomassie blue. Lanes: M, Bio-Rad Precision Plus molecular weight markers; 1, Gly43M:His6; 2, Gly3D:His6; 3, Gly5M:His6; 4, Cel5A:His6. (B) Western blot of SDS-8% PAGE gel prepared identically to that in panel A probed with anti-His6 antibody (Novagen). Lanes are as described in panel A.
TABLE 7.
Activity assays of cloned enzymes
| Substratea | Activity (mU mg protein) (SD)
|
|
|---|---|---|
| Cel5A:His6b | Gly5K:His6c | |
| Barley β-glucan | 33.1 (15.0) | ND |
| CMC | 3.4 (1.1) | ND |
| PASC | 1.9 (1.0) | ND |
| pNP-cellobiose | 12.4 (0.5) | ND |
| pNP-α-arabinopyranoside | ND | 1.1 (0.04) |
| pNP-α-arabinofuranoside | ND | 1.1 (0.2) |
| pNP-β-xylopyranoside | ND | 0.8 (0.03) |
| pNP-β-glucopyranoside | ND | 1.0 (0.04) |
CMC, carboxymethylcellulose; PASC, phosphoric acid swollen cellulose; pNP, para-nitrophenol. No activity was detected against Avicel, laminarin, xylan, pNP-β-arabinopyranoside, pNP-α-glucopyranoside, pNP-α-xylopyranoside.
Activity is expressed in milliunits/mg protein as glucose reducing equivalents, determined by Nelson-Somogyi reducing sugar assay (20). ND, not detected. The assays were run in triplicate.
Activity is expressed as milliunits/mg protein as nitrophenol released from p-NP derivative. ND, not detected. The assays were run in triplicate.
CBM proteins.
Analysis of the proteins that incorporate a CBM, but not an annotated carbohydrase domain (CBM proteins), reveals that they all have a type II secretion signal, and most have extended regions of unknown sequence large enough to accommodate binding and/or catalytic modules. Many of these CBM proteins contain unusual sequence features. In Cbm2C, Cbm2D-Cbm10A, Cbm2E, Cbm2F, and Cbm6A, a number of binding modules are separated from the rest of the protein by PSLs (Table 4). Other proteins with unusual features include Cbm2A, with a glutamate-proline repeat (EPR), and Cbm2B containing a 914-amino-acid region that has as yet no homolog in the nonredundant database of the National Center for Biotechnology Information. Cbm2C and two predicted xylanases contain a novel module, designated Y94, which is thus far unique to S. degradans. RPHPLC-MS/MS analyses detected Cbm2C xylan-grown culture supernatants and Cbm2B in Avicel-grown cultures.
Cbm6A and Cbm6E have lipobox motifs. Two other CBM6 proteins include an unusual second binding module; Cbm6G-Cbm16B and Cbm6H-Cbm32F contain a family 16 and family 32 CBM, respectively. There are four other ORFs with CBM32s, three of which have multiple copies of the binding module. Cbm32A was detected by MS/MS in xylan-grown supernatant and contains tandem CBM32s, followed by a 251-amino-acid unknown region and a thiol oxidoreductase-like module (Table 4). Cbm32D has an N-terminal CBM32, followed by a novel module, designated FCL, which has homology to fucose-binding lectins that are found in serum proteins of the Japanese eel, Anguilla japonica. An INTERPRO database search (http://www.ebi.ac.uk/interpro/displayIproEntry?ac =IPR006585) did not identify any prokaryotic examples of this module except for S. degradans.
In order to assess whether the cloned CBM proteins were successfully renatured after expression and purification and to verify the predicted binding of their CBM2a modules, a novel cellulose-binding assay was developed in which Avicel was preincubated with BSA, blocking nonspecific protein binding. Cbm2A:His6, Cbm2B:His6, and Cbm2C:His6 were subjected to this Avicel-binding assay, examined in zymograms, and checked for enzyme activity in reducing-sugar assays. All three of these fusion proteins specifically bound to Avicel (Fig. 3A and B), and none exhibited enzymatic activity in zymograms or reducing-sugar assays.
FIG. 3.
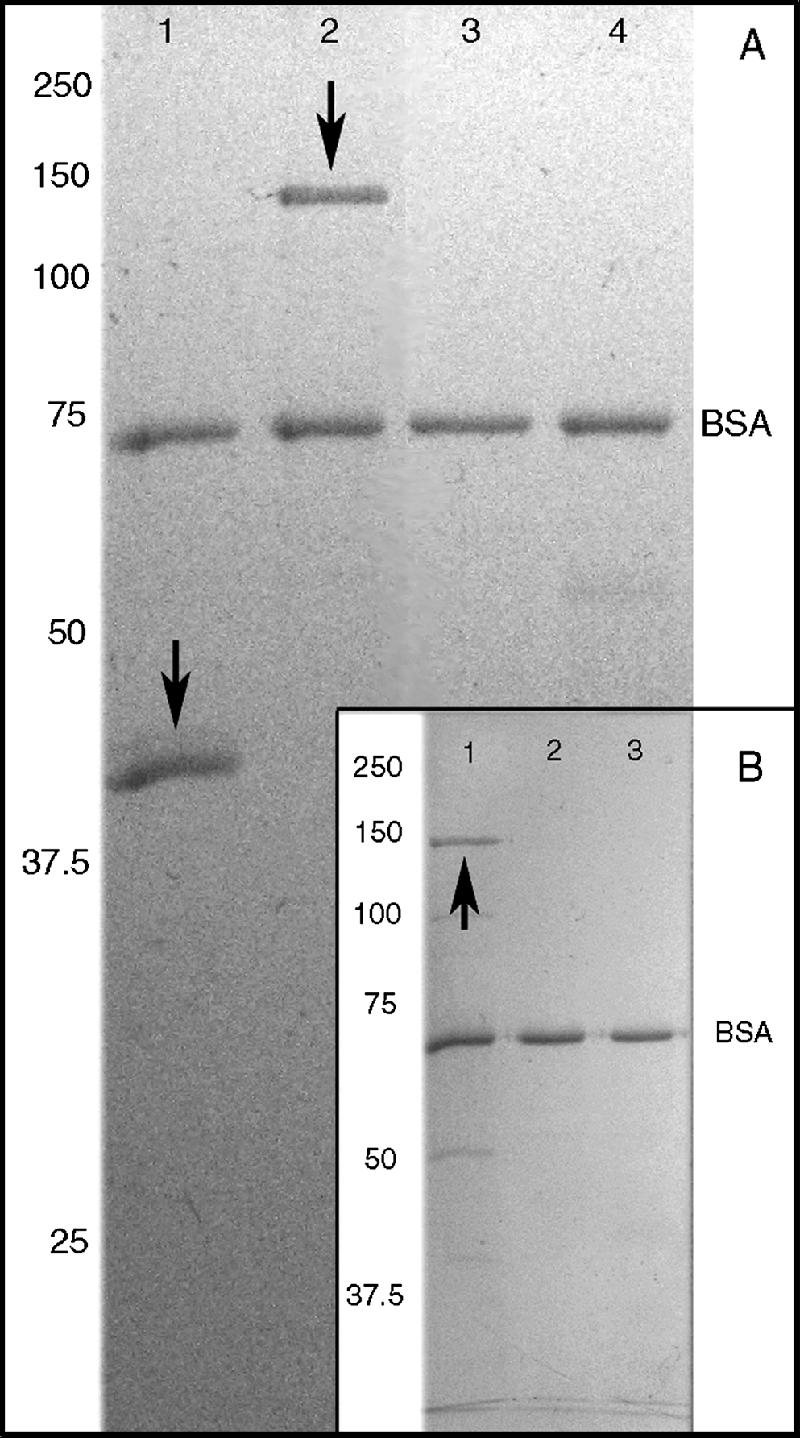
Cbm2-containing proteins specifically bind to Avicel. (A) SDS-11% PAGE gel. Sample proteins were incubated with BSA-treated Avicel as described in Materials and Methods. Arrows indicate test protein bands. Lanes: 1, Cbm2A:His6; 2, Cbm2B:His6; 3, pepsin. Porcine lipase and ovalbumin controls are not shown. (B) SDS-8% PAGE gel prepared as in panel A. Lanes: 1, rCbm2C; 2, pepsin; 3, porcine lipase. The ovalbumin control is not shown. For reasons yet to be determined a number of cloned 2-40 proteins, including Cbm2B and Cbm2C, consistently run higher in SDS-PAGE than their predicted molecular masses.
Glycanases of uncertain function.
The glycanases for which precise substrate specificity cannot be predicted by sequence analyses (glycanases) belong to five different well-characterized GH families (Table 5), although many deviate from the typical consensus sequence. For example, the catalytic modules of Gly5K, Gly5L, and Gly5M are categorized as possible β-1,3 or β-1,4 endoglucanases but are not well enough conserved to permit more specific prediction of their substrate specificity. Similarly, Gly5R is distantly related to known β-mannanases, and the sequence of Gly9C is only 25% conserved with known GH9 cellulases.
There are also glycanases with modules from less-well-characterized families, such as Gly26B, Gly30A, and Gly30B. Approximately half of the glycanases contain one or more CBM from families 6, 13, or 32, suggesting that these enzymes bind to glycan chains or oligosaccharides. Although CBM families 6 and 13 are well characterized, relatively little is known about CBM family 32, quadruply represented in Gly3D. Unlike the cellulases and CBM proteins, none of the glycanases contain PSLs; however, there are other unusual motifs. Gly5M contains a 34-amino-acid glutamate-proline repeat (EPR), and Gly43M carries two modules (designated LGL; Table 5) with homology (BLASTP; 2e-12, 2e-18) to concanavalin A-like lectins (SCOP: d1epwa).
To determine the function of selected glycanases, Gly3D, Gly5K, Gly5M, Gly9C, and Gly43M were cloned, expressed, and assayed for activity. Of these, only Gly5K was active under assay conditions (Table 7). Western blots of the recombinant protein preparations showed that the observed molecular mass of the His-tagged band agreed with the predicted molecular mass of the target protein (Fig. 2B), and the proteins of interest appeared as a single predominant band, with the exception of Gly5M, which consistently appeared as a major band of ∼120 kDa with three minor bands of lower molecular mass. Although these, and other, minor bands visible in SDS-PAGE and/or Western blots were probably degradation products of the target protein, there may have been impurities present as well; in either event, these minor components appear unlikely to affect the functional analysis.
Amino acid sequence comparisons showed that the active sites of Gly5K and Gly5L were so similar to each other as to share common substrate specificity. Accordingly, Gly5K was the chosen representative for cloning and functional analyses. Gly3D:His6, Gly5M:His6, Gly9C:His6, and Gly43M:his6 were not active in the zymograms or activity assays, and their substrate specificities remain to be determined. However, Gly3D was detected by MS/MS in xylan-grown supernatants, implying a role in plant wall degradation. Gly5K:His6 was moderately active against pNP-conjugated α-l-arabinofuranoside, α-l-arabinopyranoside, β-d-glucopyranoside, and β-d-xylopyranoside (Table 7), suggesting that Gly5K may be active against oligomers of arabinoxylan.
DISCUSSION
Enzyme complement and domain analysis.
The degradation of CP requires consortia of enzymes with diverse and complementary activities. Genomic and functional analyses reveal that S. degradans has 10 distinct CP-degrading systems (17) and more carbohydrases and accessory proteins than any marine bacterium studied thus far. To our knowledge, S. degradans is the first marine bacterium shown to have complete systems for metabolizing every major polysaccharide component of the plant cell wall, including cellulose, common hemicelluloses (xylans, arabinans, glucans, and β-mannans), and pectin. This is particularly remarkable in the context of its other complete systems which metabolize other polysaccharides commonly found in the marine environment: agar, alginate, and chitin (17, 28).
It is also noteworthy that the genome of S. degradans contains more predicted cellulases than many other cellulolytic organisms, including the cellulolytic fungus Trichoderma reesei, which has seven cellulases (two cellobiohydrolases and five endoglucanases (18). Examples of cellulase systems from genomically sequenced bacteria include the soil organisms Cytophaga hutchinsonii, Clostridum thermocellum, and Thermobifida fusca and the marine bacterium Thermotoga maritima (Table 6). Like Thermobifida fusca and Clostridium thermocellum, S. degradans has a nonreducing end cellobiohydrolase (53, 57). However, unlike these organisms, S. degradans does not have a GH48 reducing-end cellobiohydrolase. Furthermore, by sequence analysis, it is uncertain whether S. degradans has any processive endoglucanases, such as the cellotetraolases Cel9R and Cel9I of C. thermocellum (60, 61) or Cel48F of C. cellulolyticum (41, 42). BLASTP analysis of the S. degradans genome, using the GH9 sequences of Ct-Cel9I and Ct-Cel9R, revealed that Sd-Cel9A and Sd-Cel9B are not strongly conserved with the C. thermocellum tetraolases (∼25% identity, ∼38% similarity in amino acid sequence). However, because the genome contains so many predicted enzymes from well-studied GH families with sequences that are not conserved with characterized examples (Table 5), it remains possible that S. degradans has processive endoglucanases which have novel sequences. It is also possible, although less likely, that such activity may reside within one or more cellulases with conserved sequence (Table 3).
Studies of organisms such as T. fusca and T. reesei have strongly suggested that the synergistic action of endoglucanases and reducing-end and nonreducing-end cellobiohydrolases (the so-called endo-exo synergy) is essential for efficient cellulose utilization (3, 55). This perceived requirement is often extended to cellulolytic microorganisms in general. However, accumulating evidence suggests that there may be other operative mechanisms, as well. For example, the gram-negative gliding soil bacterium, Cytophaga hutchinsonii, is regarded as an efficient cellulolytic bacterium (36) despite the absence of either a GH6 or GH48 cellobiohydrolase (Table 6). Furthermore, recent studies of Clostridium thermocellum reveal that it has at least two processive cellotetraolases (60, 61) and that, during growth on Avicel, cellotetraose is the major oligomer imported into the cytoplasm, where it is metabolized further via phosphorolytic cleavage (58). C. thermocellum is reported to have 23 ORFs that appear to participate in cellulose degradation (59), among which only four are proven or predicted cellobiohydrolases (59). Thus, it appears that in C. thermocellum the combined activities of the abundant endoglucanases (including processive cellotetraolases) produce the bulk of assimilated carbon during cellulose metabolism (58), despite the presence of reducing-end and nonreducing-end cellobiohydrolases. In such a system, the main function of the cellobiohydrolases may be to promote the breakdown of particularly recalcitrant crystalline regions rather than to produce cellobiose as a primary energy source. Interestingly, S. degradans has an abundance of endoglucanases and only one cellobiohydrolase. Taken together, the presence of cellodextrin and cellobiose phosphorylases (Table 6) suggests that S. degradans cellulose metabolism proceeds, at least in part, through the uptake of endoglucanase-derived oligo-cellodextrins (n > 2) and that phosphorolytic cleavage is also an operative mechanism.
Most S. degradans cellulase components appear to be freely secreted, as indicated by type II secretion signals. However, two cellulases, both of the cellodextrinases, and one cellobiase have lipobox sequences (Table 3), suggesting that they are anchored to the cell surface (13, 40, 48). Within the cellulase system, the lipoprotein enzymes are specific for single glycan chains and cello-oligomers. Thus, it appears that the extracellular enzymes that process intermediate-length glucan chains into oligo-cellodextrins and glucose are maintained on the cell surface, thereby minimizing substrate and enzyme loss through diffusion (6, 45, 53).
Many carbohydrases of S. degradans have features that are unique and others that are unusual among prokaryotes. In addition to well-predicted plant wall carbohydrases, there are numerous ORFs with binding and/or catalytic modules for which precise substrate specificity cannot be predicted due to insufficient relatedness to a well-characterized enzyme. It has long been noticed that the sequence-based families of glycoside hydrolases contain enzymes of various substrate specificities (23), making predictions of substrate specificity difficult for distantly related members (12, 24).
S. degradans encodes many such genes. Although GH5 is among the best-studied CAZyme families, the genome contains five GH5 modules for which substrate specificity cannot be predicted by sequence analysis. Of the two glycanases of unknown function with GH5 modules that were cloned and assayed in the present study, Gly5M:His6 was not active on any tested substrate, and Gly5K:His6 was moderately active on arabinoside, xyloside, and glucoside pNP conjugates, but not on laminarin, barley β-glucan, xylan, or three forms of cellulose. The findings suggest that the substrates of these GH5 glycanases, much like their sequences, differ from those of previously characterized GH5 cellulases. This is also the likely case for Gly9C and Gly43M—both of which were inactive in reducing-sugar and pNp assays. For many of these glycanases, follow-up studies, testing activities on cellulose and hemicellulose oligosaccharides (many of which are not commercially available), as well as synergy tests with enzymes with known reaction characteristics, will help to clarify their role in CP metabolism. Such studies may advance knowledge of even well-studied GH families.
Although it is possible that one or more of the expressed recombinant proteins failed to refold properly, evidence suggests that many of them retained or recovered functionality. For example, as predicted, the CBM proteins Cbm2A:His6, Cbm2B:His6, and Cbm2C:His6 exhibited specific binding to Avicel. Furthermore, Cel5A:His6 was active against amorphous cellulose and barley β-glucan. [It is possible, based upon their low sequence conservation, that the dual GH5 modules of Cel5A have different reaction specificities. In fact, this was found, in another carbohydrase system of S. degradans, namely, the dual GH18 modules of chitinase, ChiB (27).] Including the detected activities of Gly5K:His6, four of the nine expressed recombinant proteins functioned as genomically predicted and a fifth was active against arabinoside and xyloside pNp linkages. Therefore, it is probable that Gly3D:His6, Gly5M:His6, Gly9C:His6, and Gly43M:His6 were expressed in a functional state and that they act on untested substrates.
S. degradans encodes four distinct modules which might be novel CBM. These include the fucolectin-like module (FCL) in Cbm32D (Table 4) and the concanavalin A lectin-like modules of Gly43M (Table 5). Two other candidate CBM are the conserved modules designated Y94 and Y95. Y94 modules are about 130 amino acids in length and are found in Cbm2C, which was identified by MS in supernatant from S. degradans grown with xylan as the sole carbon source. Y94 modules also appear in two predicted xylanases which, together with the MS/MS data, suggests a possible xylan-binding role. Y95 modules are about 95 amino acids in length and occur in Cel5G (Table 3), as well as in two poorly conserved polysaccharide lyases.
Alternatively, it is possible that one or more of these unknown modules are involved in specific attachment of proteins to the bacterial cell surface as exemplified by the dockerin-cohesin interaction in C. thermocellum (6). Surface attachments in gram-negative bacteria are not solely mediated by lipobox sequences (5, 31, 51). Other modules that may be involved in surface attachment include fibronectin-like (FN3), found in Cbm32C (Table 4), and the glycanases Gly5M and Gly5R (Table 5). The function of FN3 modules is uncertain (47), and it remains possible that they participate in protein-cell surface interactions.
Functional analysis.
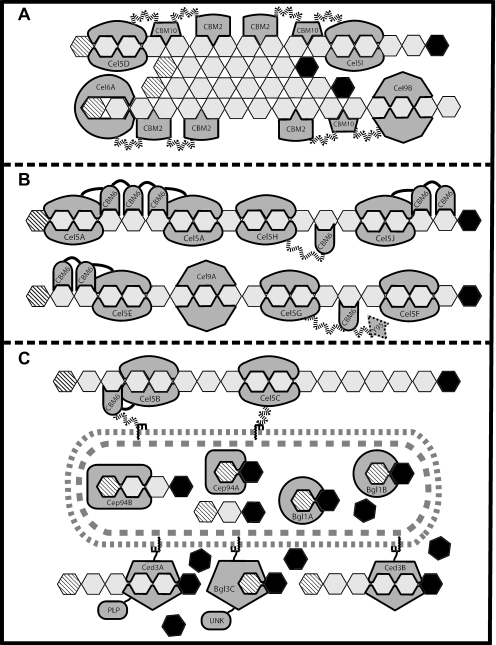
The general characteristics of the cellulase system of S. degradans can be inferred from the analysis of its proteome with the CAZy database tools. Annotations and analyses of the GH and CBM modules within an ORF allow prediction of its catalytic and binding activities. Furthermore, analyses of lipoprotein or secretion signal sequences, if present, suggest where the protein will perform its expected function. From this, the predicted location of each enzyme relative to the cell and its likely substrate are presented in Fig. 4. This model suggests the most probable location and activity of the well-conserved cellulases, based on functional predictions, and provides a testable framework for further study of cellulose metabolism in S. degradans.
FIG. 4.
Representation of cellulose degradation by S. degradans. The diagram shows the predicted cellulases of S. degradans participating in three distinct phases of cellulose degradation based on their expected location relative to the cell, as well as their enzymatic and substrate-binding specificities. In panel A, a region of crystalline cellulose located distal from the cell is attacked by the nonreducing-end cellobiohydrolase, Cel6A, and endoglucanases Cel5D, Cel5I, and Cel9B. Panel B shows the secreted cellulases which contain CBM6 modules (or lack a CBM) acting on freely accessible cellulose chains at an intermediate distance from the cell surface. In panel C, the lipoprotein-anchored components are shown processing oligo-cellodextrins. The endoglucanases Cel5B and Cel5C likely bind and degrade single cellulose chains at the cell surface, whereas the cellodextrinases cleave cellodextrins to generate glucose and (n-1) oligo-cellodextrins. Extracellular cellobiose is cleaved to glucose by Bgl3C. In addition, within the cell cytoplasm, the cellobiose and cellodextrin phosphorylases are shown bound to their substrates, as are the cytoplasmic cellobiases Bgl1A and Bgl1B. Internal glucose residues of cellulose chains are represented by open hexagons, while the nonreducing ends are indicated by diagonal lineed hexagons, with the reducing ends shown as solid filled hexagons. The bacterial inner and outer membranes are indicated by shaded dashed lines. Enzyme names (i.e., Cel9B) are indicated within catalytic module, while the CBM family type is indicated within the CBM. Wavy, dashed lines represent PSLs, and solid lines represent non-PSL regions. Lipoprotein anchors are shown embedded in the outer membrane with the lipid moiety represented by the zigzag line. Cel5H, Cel5I, Cel9B, Ced3A, Ced3B, Bgl3C, and Cep94B were detected by RPHPLC-MS/MS in cellulose- and/or xylan-grown culture supernatants.
Since the roles of the CBM-only proteins remain unclear, they were not included in Fig. 4 despite the detection of three of them in Avicel- or xylan-grown culture supernatants (Table 4) and the fact that Cbm2A, Cbm2B, and Cbm2C specifically bind to Avicel (Fig. 3). Perhaps one or more of the CBM proteins enhance cellulose degradation through a “disruption” mechanism similar to that recently shown for the nonhydrolytic, chitin-binding protein CBP21 of Serratia marcescens (54). Interestingly, and like S. marcescens, the S. degradans chitinase system also contains a CBM33 containing chitin-binding protein that is believed to assist in the degradation of chitin (28). To this end, certain CBM proteins of S. degradans may be bacterial equivalents of plant expansins (10) or the swollenin protein of T. reesei (44). These proteins also contain CBM and yet lack catalytic modules, and there is evidence that swollenins act to disrupt the surface of crystalline cellulose, thereby increasing the availability of cellulose chains for enzymatic attack (44).
In our model, crystalline cellulose is attacked by the freely secreted endoglucanases which bind insoluble cellulose, namely, Cel5D, Cel5I, and Cel9B (Fig. 4A). These endoglucanases appear to act on amorphous regions within insoluble cellulose, thereby generating shorter cellulose chains and increasing the availability of nonreducing ends for attack by cellobiohydrolase Cel6A. Although a reducing-end cellobiohydrolase has not been found by sequence analysis, the cellulase system of S. degradans is quite functional, permitting growth on Avicel and even filter paper as sole carbon sources (52). Therefore, it appears that its numerous endoglucanases (some of which may be processive) act synergistically to compensate for the absence of a reducing-end cellobiohydrolase. Alternatively, although less likely, there may be a reducing-end cellobiohydrolase that was not identified in the analyses of the recently closed genome.
Continuing with the cellulose degradation model, there are five freely secreted endoglucanases with CBM6s and two which do not have a CBM. Because of their CBM specificity (or lack of CBM) these enzymes are expected to act on more accessible forms of cellulose such as individual chains or extended amorphous regions (Fig. 4B). Six of these “secondary” endoglucanases are from GH family 5, and the other is a GH9. These enzymes cleave cellulose (and perhaps other glucans as well, e.g., 1,3-1,4-β-glucan) to shorter, random-length chains and cellodextrins. Although it is possible that one or more of the endoglucanases in this group may generate oligomers that are acted on by other endoglucanases, no such functional hierarchy can be inferred by sequence analysis.
Some of the cellulose chain fragments and cellodextrins generated by S. degradans enzymes are likely processed at the cell surface by the lipoproteins: endoglucanases Cel5B and Cel5C, cellodextrinases Ced3A and Ced3B, and the cellobiase Bgl3C (Fig. 4C). Cel5B and Cel5C hydrolyze individual cellulose chains to cellodextrins, which are cleaved to glucose and oligo-cellodextrins by Ced3A and Ced3B. Cellobiose may be hydrolyzed to glucose at the cell surface by Bgl3C and then imported to the cytoplasm. Alternatively, short cellodextrins and cellobiose may be imported into the cytoplasm for further metabolism by the cellobiases and/or phosphorylases (Fig. 4C).
The presence of the cellobiases and phosphorylases suggests that, in addition to the surface-bound enzymes, there are also two distinct cytoplasmic pathways for oligo-cellodextrins metabolism, both of which rely on energy-dependent import of cellobiose and/or cellodextrins. In the hydrolytic pathway, cellobiose is cleaved to glucose by either of the two cellobiases, Bgl1A and Bgl1B. In the phosphorolytic pathway, cellobiose and cellodextrins are cleaved into glucose-1-phosphate and glucose or cellodextrin (n-1) by the cellobiose or cellodextrin phosphorylase (Fig. 4C). Although it is impossible to predict the extent to which each pathway contributes to cellulose metabolism, the energy savings afforded by phosphorolytic cleavage may be advantageous under certain conditions (46, 58). The detection of cellobiase Bgl3C, cellodextrinase Ced3A, and the cellodextrin phosphorylase, Cep94B, by MS/MS in cultures grown in CMC as the sole carbon source suggests that surface-bound hydrolysis and intracellular phosphorolytic cleavage of oligo-cellodextrins may occur simultaneously in S. degradans.
Collectively, these data support the notion that S. degradans synthesizes a full complement of enzymes that degrade algal and green plant cell wall polymers. It is uniquely the first marine proteobacterium shown to do so in pure culture (52), having versatile arrays of enzymes with complementary activities. Until now reports on the degradation of algal and higher plant cell wall polymers in marine environments have been descriptive. The discovery and molecular biochemical characterization, beginning with S. degradans, of an emerging group of cellulolytic marine bacteria promises a more complete understanding of the marine carbon cycle. Further studies of the cellulases of S. degradans should reveal additional important similarities and differences between the cellulase systems of aerobic marine bacteria and anaerobic soil organisms such as the clostridia.
.
Acknowledgments
This study was supported by grants from the National Science Foundation, (DEB0109869), the U.S./Israel Bi-national Science Foundation, NOAA/Maryland Sea Grant, and the Maryland Technology Development Company (TEDCO).
We thank the U.S. Department of Energy Joint Genome Institute for determining the genome sequence of S. degradans and Oak Ridge National Laboratories for contributions to the annotation of the draft genome sequence. L.E.T. was supported by a Maryland Sea Grant Fellowship.
REFERENCES
- 1.Adelsberger, H., C. Hertel, E. Glawischnig, V. V. Zverlov, and W. H. Schwarz. 2004. Enzyme system of Clostridium stercorarium for hydrolysis of arabinoxylan: reconstitution of the in vivo system from recombinant enzymes. Microbiology 150:2257-2266. [DOI] [PubMed] [Google Scholar]
- 2.Andrykovitch, G., and I. Marx. 1988. Isolation of a new polysaccharide-digesting bacterium from a salt marsh. Appl. Environ. Microbiol. 54:3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr, B. K., Y. L. Hsieh, B. Ganem, and D. B. Wilson. 1996. Identification of two functionally different classes of exocellulases. Biochemistry 35:586-592. [DOI] [PubMed] [Google Scholar]
- 4.Bayer, E. A., H. Chanzy, R. Lamed, and Y. Shoham. 1998. Cellulose, cellulases, and cellulosomes. Curr. Opin. Struct. Biol. 8:548-557. [DOI] [PubMed] [Google Scholar]
- 5.Bayer, E. A., and R. Lamed. 1986. Ultrastructure of the cell surface cellulosome of Clostridium thermocellum and its interaction with cellulose. J. Bacteriol. 167:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beguin, P., and J. P. Aubert. 1994. The biological degradation of cellulose. FEMS Microbiol. Rev. 13:25-58. [DOI] [PubMed] [Google Scholar]
- 7.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boraston, A. B., V. Notenboom, R. A. Warren, D. G. Kilburn, D. R. Rose, and G. Davies. 2003. Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and “galactose-binding” domains. J. Mol. Biol. 327:659-669. [DOI] [PubMed] [Google Scholar]
- 9.Carpita, N. C. 1996. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47:445-476. [DOI] [PubMed] [Google Scholar]
- 10.Cosgrove, D. J., L. C. Li, H.-T. Cho, S. Hoffmann-Benning, R. C. Moore, and D. Blecker. 2002. The growing world of expansins. Plant Cell Physiol. 43:1436-1444. [DOI] [PubMed] [Google Scholar]
- 11.Coutinho, P. M., and B. Henrissat. 1999. The modular structure of cellulases and other carbohydrate-active enzymes: an integrated database approach, p. 15-23. In K. Ohmiya, K. Hayashi, K. Sakka, C. Kobayashi, S. Karita, and T. Kimura (ed.), Genetics, biochemistry, and ecology of cellulose degradation. Uni Publishers, Tokyo, Japan.
- 12.Coutinho, P. M., M. Stam, E. Blanc, and B. Henrissat. 2003. Why are there so many carbohydrate-active enzyme-related genes in plants? Trends Plant Sci. 8:563-565. [DOI] [PubMed] [Google Scholar]
- 13.d'Enfert, C., A. Ryter, and A. P. Pugsley. 1987. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 6:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Distel, D. L., W. Morrill, N. MacLaren-Toussaint, D. Franks, and J. Waterbury. 2002. Teredinibacter turnerae gen. nov., sp. nov., a dinitrogen-fixing, cellulolytic, endosymbiotic gamma-proteobacterium isolated from the gills of wood-boring molluscs (Bivalvia: Teredinidae). Int. J. Syst. Evol. Microbiol. 52:2261-2269. [DOI] [PubMed] [Google Scholar]
- 15.Ducret, A., I. Van-Oostveen, J. E. Eng, J. R. Yates III, and R. Aebersold. 1998. High throughput protein characterization by automated reverse-phase chromatography/electrospray tandem mass spectrometry. Protein Sci. 7:706-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekborg, N. A., J. M. Gonzalez, M. B. Howard, L. E. Taylor, S. W. Hutcheson, and R. M. Weiner. 2005. Saccharophagus degradans gen. nov., sp. nov., a versatile marine degrader of complex polysaccharides. Int. J. Syst. Evol. Microbiol. 55:1545-1549. [DOI] [PubMed] [Google Scholar]
- 17.Ensor, L., S. K. Stotz, and R. M. Weiner. 1999. Expression of multiple insoluble complex polysaccharide degrading enzyme systems by a marine bacterium. J. Ind. Microbiol. Biotechnol. 23:123-126. [DOI] [PubMed] [Google Scholar]
- 18.Foreman, P. K., D. Brown, L. Dankmeyer, R. Dean, S. Diener, N. S. Dunn-Coleman, F. Goedegebuur, T. D. Houfek, G. J. England, A. S. Kelley, H. J. Meerman, T. Mitchell, C. Mitchinson, H. A. Olivares, P. J. M. Teunissen, J. Yao, and M. Ward. 2003. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J. Biol. Chem. 278:31988-31997. [DOI] [PubMed] [Google Scholar]
- 19.Gilkes, N. R., B. Henrissat, D. G. Kilburn, R. C. Miller, Jr., and R. A. Warren. 1991. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol. Rev. 55:303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez, J., and R. M. Weiner. 2000. Phylogenetic characterization of marine bacterium strain 2-40, a degrader of complex polysaccharides. Int. J. Syst. Evol. Microbiol. 50:831-834. [DOI] [PubMed] [Google Scholar]
- 21.Green, F., III, C. A. Clausen, and T. L. Highley. 1989. Adaptation of the Nelson-Somogyi reducing-sugar assay to a microassay using microtiter plates. Anal. Biochem. 182:197-199. [DOI] [PubMed] [Google Scholar]
- 22.Hazlewood, G. P., and H. J. Gilbert. 1998. Structure and function analysis of Pseudomonas plant cell wall hydrolases. Prog. Nucleic Acid. Res. Mol. Biol. 61:211-241. [DOI] [PubMed] [Google Scholar]
- 23.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280((Pt. 2):309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henrissat, B., P. M. Coutinho, E. Deleury, and G. J. Davies. 2003. Sequence families and modular organisation of carbohydrate-active enzymes, p. 15-34. In A. Svendsen (ed.), Enzyme functionality: design, engineering, and screening. Marcel Dekker, New York, N.Y.
- 25.Henrissat, B., T. T. Teeri, and R. A. Warren. 1998. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett. 425:352-354. [DOI] [PubMed] [Google Scholar]
- 26.Howard, M. B., N. A. Ekborg, L. E. Taylor, S. W. Hutcheson, and R. M. Weiner. 2004. Identification and analysis of polyserine linker domains in prokaryotic proteins with emphasis on the marine bacterium Microbulbifer degradans. Protein Sci. 13:1422-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard, M. B., N. A. Ekborg, L. E. Taylor, R. M. Weiner, and S. W. Hutcheson. 2004. Chitinase B of “Microbulbifer degradans” 2-40 contains two catalytic domains with different chitinolytic activities. J. Bacteriol. 186:1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard, M. B., N. A. Ekborg, L. E. Taylor, R. M. Weiner, and S. W. Hutcheson. 2003. Genomic analysis and initial characterization of the chitinolytic system of Microbulbifer degradans strain 2-40. J. Bacteriol. 185:3352-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irwin, D. C., S. Zhang, and D. B. Wilson. 2000. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 267:4988-4997. [DOI] [PubMed] [Google Scholar]
- 30.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung, H. C., J. M. Lebeault, and J. G. Pan. 1998. Surface display of Zymomonas mobilis levansucrase by using the ice-nucleation protein of Pseudomonas syringae. Nat. Biotechnol. 16:576-580. [DOI] [PubMed] [Google Scholar]
- 32.Kosugi, A., K. Murashima, and R. H. Doi. 2002. Characterization of two noncellulosomal subunits, ArfA and BgaA, from Clostridium cellulovorans that cooperate with the cellulosome in plant cell wall degradation. J. Bacteriol. 184:6859-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature 277:680-685. [DOI] [PubMed] [Google Scholar]
- 34.Ljungdahl, L. G., and K. E. Eriksson. 1985. Ecology of microbial cellulose degradation. Adv. Microb. Ecol. 8:237-299. [Google Scholar]
- 35.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McBride, M. J., and S. A. Baker. 1996. Development of techniques to genetically manipulate members of the genera Cytophaga, Flavobacterium, Flexibacter, and Sporocytophaga. Appl. Environ. Microbiol. 62:3017-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrison, M., and J. Miron. 2000. Adhesion to cellulose by Ruminococcus albus: a combination of cellulosomes and Pil-proteins? FEMS Microbiol. Lett. 185:109-115. [DOI] [PubMed] [Google Scholar]
- 38.Notenboom, V., A. B. Boraston, S. J. Williams, D. G. Kilburn, and D. R. Rose. 2002. High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding. Biochemistry 41:4246-4254. [DOI] [PubMed] [Google Scholar]
- 39.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 40.Pugsley, A. P., C. Chapon, and M. Schwartz. 1986. Extracellular pullulanase of Klebsiella pneumoniae is a lipoprotein. J. Bacteriol. 166:1083-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reverbel-Leroy, C., A. Belaich, A. Bernadac, C. Gaudin, J. Belaich, and C. Tardif. 1996. Molecular study and overexpression of the Clostridium cellulolyticum celF cellulase gene in Escherichia coli. Microbiology 142:1013-1023. [DOI] [PubMed] [Google Scholar]
- 42.Reverbel-Leroy, C., S. Pages, A. Belaich, J. Belaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts, A. W., E. M. Roberts, and D. P. Delmer. 2002. Cellulose synthase (CesA) genes in the green alga Mesotaenium caldariorum. Eukaryot. Cell 1:847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saloheimo, M., M. Paloheimo, S. Hakola, J. Pere, B. Swanson, E. Nyyssonen, A. Bhatia, M. Ward, and M. Penttila. 2002. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 269:4202-4211. [DOI] [PubMed] [Google Scholar]
- 45.Salyers, A. A., A. Reeves, and J. D'Ella. 1996. Solving the problem of how to eat something as big as yourself: diverse bacterial strategies for degrading polysaccharides. J. Ind. Microbiol. 17:470-476. [Google Scholar]
- 46.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 47.Schwarz, W. H., V. V. Zverlov, and H. Bahl. 2004. Extracellular glycosyl hydrolases from clostridia. Adv. Appl. Microbiol. 56:215-261. [DOI] [PubMed] [Google Scholar]
- 48.Seydel, A., P. Gounon, and A. P. Pugsley. 1999. Testing the “+2 rule” for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34:810-821. [DOI] [PubMed] [Google Scholar]
- 49.Simpson, P. J., H. Xie, D. N. Bolam, H. J. Gilbert, and M. P. Williamson. 2000. The structural basis for the ligand specificity of family 2 carbohydrate-binding modules. J. Biol. Chem. 275:41137-41142. [DOI] [PubMed] [Google Scholar]
- 50.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 51.Tamm, L. K., A. Arora, and J. H. Kleinschmidt. 2001. Structure and assembly of beta-barrel membrane proteins. J. Biol. Chem. 276:32399-32402. [DOI] [PubMed] [Google Scholar]
- 52.Taylor, L. E. 2005. Degradation of plant cell wall polysaccharides by Saccharophagus degradans. Ph.D. thesis. University of Maryland, College Park.
- 53.Tomme, P., R. A. Warren, and N. R. Gilkes. 1995. Cellulose hydrolysis by bacteria and fungi. Adv. Microb. Physiol. 37:1-81. [DOI] [PubMed] [Google Scholar]
- 54.Vaaje-Kolstad, G., D. R. Houston, A. H. Riemen, V. G. Eijsink, and D. M. van Aalten. 2005. Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J. Biol. Chem. 280:11313-11319. [DOI] [PubMed] [Google Scholar]
- 55.Warren, R. A. 1996. Microbial hydrolysis of polysaccharides. Annu. Rev. Microbiol. 50:183-212. [DOI] [PubMed] [Google Scholar]
- 56.Wood, T. M. 1988. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol. 160:19-25. [Google Scholar]
- 57.Zhang, S., D. C. Irwin, and D. B. Wilson. 2000. Site-directed mutation of noncatalytic residues of Thermobifida fusca exocellulase Cel6B. Eur. J. Biochem. 267:3101-3115. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y.-H. P., and L. R. Lynd. 2005. Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation. Proc. Natl. Acad. Sci. USA 102:7321-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zverlov, V. V., J. Kellermann, and W. H. Schwarz. 2005. Functional subgenomics of Clostridium thermocellum cellulosomal genes: identification of the major catalytic components in the extracellular complex and detection of three new enzymes. Proteomics 5:3646-3653. [DOI] [PubMed] [Google Scholar]
- 60.Zverlov, V. V., N. Schantz, and W. H. Schwarz. 2005. A major new component in the cellulosome of Clostridium thermocellum is a processive endo-β-1,4-glucanase producing cellotetraose. FEMS Microbiol. Lett. 249:353-358. [DOI] [PubMed] [Google Scholar]
- 61.Zverlov, V. V., G. A. Velikodvorskaya, and W. H. Schwarz. 2003. Two new cellulosome components encoded downstream of celI in the genome of Clostridium thermocellum: the non-processive endoglucanase CelN and the possibly structural protein CseP. Microbiology 149:515-524. [DOI] [PubMed] [Google Scholar]
- 62.Zverlov, V. V., G. V. Velikodvorskaya, W. H. Schwarz, K. Bronnenmeier, J. Kellermann, and W. L. Staudenbauer. 1998. Multidomain structure and cellulosomal localization of the Clostridium thermocellum cellobiohydrolase CbhA. J. Bacteriol. 180:3091-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]