Abstract
Yersinia pestis, the causative agent of bubonic and pneumonic plagues, has undergone detailed study at the molecular level. To further investigate the genomic diversity among this group and to help characterize lineages of the plague organism that have no sequenced members, we present here the genomes of two isolates of the “classical” antiqua biovar, strains Antiqua and Nepal516. The genomes of Antiqua and Nepal516 are 4.7 Mb and 4.5 Mb and encode 4,138 and 3,956 open reading frames, respectively. Though both strains belong to one of the three classical biovars, they represent separate lineages defined by recent phylogenetic studies. We compare all five currently sequenced Y. pestis genomes and the corresponding features in Yersinia pseudotuberculosis. There are strain-specific rearrangements, insertions, deletions, single nucleotide polymorphisms, and a unique distribution of insertion sequences. We found 453 single nucleotide polymorphisms in protein-coding regions, which were used to assess the evolutionary relationships of these Y. pestis strains. Gene reduction analysis revealed that the gene deletion processes are under selective pressure, and many of the inactivations are probably related to the organism's interaction with its host environment. The results presented here clearly demonstrate the differences between the two biovar antiqua lineages and support the notion that grouping Y. pestis strains based strictly on the classical definition of biovars (predicated upon two biochemical assays) does not accurately reflect the phylogenetic relationships within this species. A comparison of four virulent Y. pestis strains with the human-avirulent strain 91001 provides further insight into the genetic basis of virulence to humans.
Plague is a zoonotic disease, endemic throughout the world, and highly infectious in humans. The causative agent, Yersinia pestis, primarily infects a wide range of rodents and is transmitted via flea vectors. Throughout history, plague has ravaged human populations in three major pandemic waves: Justinian's plague (AD 541 to 767), which started in Africa and spread to the Mediterranean; the Black Death of 1346 to the early 19th century, which may have originated in central Asia and spread from the Caspian Sea to Europe; and modern plague (since 1894), which began in southwest China and spread globally via marine shipping routes from Hong Kong. Although human disease is rare, Y. pestis is dangerous and highly infectious and thus has been identified as having potential for use in bioterrorism or as a biological weapon.
It was shown that Y. pestis recently diverged from Yersinia pseudotuberculosis, an enteropathogen, and likely comprises a clonal lineage (1, 3, 37, 40). Y. pestis strains have historically been classified according to their ability to utilize glycerol and reduce nitrate and have been grouped into three main subtypes or biovars: antiqua, medievalis, and orientalis. Isolates from the orientalis biovar have worldwide distribution due to spreading via steamship beginning 100 years ago. In contrast, isolates of the antiqua and medievalis biovars are generally limited to localized regions containing long-term plague foci from enzootic rodent hosts in Africa and central Asia. It has been argued that each of the biovars was associated with one of the plague pandemics (14, 20, 34), and recent studies have tried to provide direct evidence of whether Y. pestis was associated with any of the historical pandemics (15, 44). DNA sequences from ancient human remains dispute the assertion that different biovars were responsible for each of the last three pandemics and suggest that instead, orientalis-like Y. pestis may have been involved in all three (15). This suggestion remains highly controversial.
Isolates from the biovar antiqua have been thought to represent a more ancestral branch of the plague pathogen, primarily due to their association with long-established plague foci as well as sharing an additional set of genetic regions with Y. pseudotuberculosis and “nonclassical” (e.g., the microtus biovar) subspecies of Y. pestis. Our previous work using suppression-subtractive hybridization demonstrated a pattern of difference fragments (DFR profiles), including a 15,603-bp segment of chromosomal DNA that was shared by Y. pseudotuberculosis and a portion of both the “nonclassical” subspecies of Y. pestis and the “classical” biovar antiqua (38). There are currently three completed genome sequences for Y. pestis, one each from the orientalis, medievalis, and “nonclassical” microtus biovars. To get a better understanding of the detailed genetic changes in a pathogen that is adapting to an intracellular lifestyle, we have sequenced two isolates from the classical antiqua biovar. Strain Antiqua is fully virulent and possesses a DFR profile closest to that of Y. pseudotuberculosis. A pgm derivative of the virulent strain Nepal516 was found to have a different DFR profile and is believed to represent a different lineage of this biovar. A comparison with the genome sequence of the previously sequenced Y. pestis strains as well as that of Y. pseudotuberculosis gave further insight into the loci required for adaptation to an intracellular pathogenic lifestyle. Additional insight into the acquisition of virulence to humans was obtained by the comparison to the human-avirulent isolate 91001.
MATERIALS AND METHODS
Bacterial strains.
Y. pestis Nepal516 was isolated from a human infection in Nepal (possibly from a 1967 outbreak of pneumonic plague), while strain Antiqua was isolated from a human infection in Africa (Republic of Congo in 1965). Both have been biochemically characterized to belong to the antiqua biovar and carry the three previously described “virulence” plasmids found in most classical isolates of Y. pestis. Both strains have been previously used in a variety of studies (4, 21, 27, 36, 37, 45). The wild-type Antiqua strain and a pgm version of the Nepal516 strain were available and used in this genome-sequencing project. The Nepal516 strain lacks the ∼100-kb pgm region, including the high-pathogenicity island, the pesticin/yersiniabactin complex, and the hemin storage locus that are normally located between two parallel IS100 insertion sequence (IS) elements (5, 8, 18, 22, 29, 35, 42).
Construction, sequencing, and assembly.
Genomic DNA was isolated from Y. pestis strains Antiqua and Nepal516.
The two genomes were sequenced using the whole-genome shotgun method as previously described (9). Briefly, 3-kb- and 8-kb-sized, randomly sheared DNA fragments were isolated and cloned into pUC18 and pMCL200, respectively, for amplification in Escherichia coli. A larger fosmid library was constructed containing approximately 40-kb inserts of sheared genomic DNA cloned into the pCC1Fos cloning vector. Double-ended plasmid-sequencing reactions were performed from all three libraries at the Department of Energy Joint Genome Institute (JGI) using ABI 3730xl DNA analyzers and MegaBACE 4500 genetic analyzers as described at the JGI website, http://www.jgi.doe.gov/.
Approximately 110,556 and 113,541 sequences were assembled for Antiqua and Nepal516, respectively, producing an average of 11-fold coverage across the genomes. The processing of sequence traces, base calling, and assessment of data quality were performed with PHRED and PHRAP (P. Green, University of Washington, Seattle, WA), respectively. Assembled sequences were visualized with CONSED. The initial assemblies consisted of 154 and 113 contigs (≥20 reads per contig). Gaps in the sequence were primarily closed by resolving the many repetitive regions found within the genome. The remaining gaps were closed by primer walking on gap-spanning library clones or PCR products from genomic DNA. True physical gaps were closed by combinatorial (multiplex) PCR. Sequence finishing and polishing added roughly 300 reads, and assessment of final assembly quality was completed as described previously (9).
For the genome of Nepal516, the ∼70-kb pCD plasmid was underrepresented and was not completed as part of the sequencing project. Nepal516 is known to contain the pCD plasmid (S. Bearden, personal communication). The existence of the pCD plasmid was verified by PCR in our laboratory (data not shown). We believe that the failure to obtain a sufficient quantity of pCD DNA for sequencing is due to particular laboratory conditions and has no biological implication on the sequences of the chromosome and pMT and pPCP plasmids.
Sequence analysis and annotation.
Automated gene modeling was completed by combining results from Critica, Generation, and Glimmer modeling packages and comparing the translations to GenBank's nonredundant database using the basic local alignment search tool for proteins (BLASTP). The protein set was also searched against the KEGG Genes, InterPro, TIGRFams, PROSITE, and Clusters of Orthologous Groups of Proteins (COGs) databases to further assess function. Manual functional assignments were assessed on an individual gene-by-gene basis as needed. Sequence alignment and protein domain search tools (BLAST, CLUSTALW, Pfam, etc.) were applied in various stages of comparison. CO92 gene nomenclature is used in this work when possible; other nomenclature is mentioned and used when no CO92 ortholog is available.
Single nucleotide polymorphism analysis.
Yersinia genomes are known to harbor extensive rearrangements as well as a large number of insertion sequence elements and other duplicated regions (10, 13, 33, 41). These repeats and insertion elements were excluded from consideration in single nucleotide polymorphism (SNP) analysis. Genome-wide SNP discovery was achieved by whole-genome alignments using the software package Mummer3 (28) and by subsequent orthologous gene alignments. For coding regions, pairwise reciprocal BLASTP analyses were performed with the five sets of Y. pestis proteins. An ortholog pair was defined as reciprocal best top hits using a cutoff of 95% sequence identity. If an ortholog was not found in any one of the five genomes, the proteins were removed from further analysis. The sequences of the orthologous genes were used to find SNPs using Mummer3. Whole-genome comparisons were also carried out using Mummer3. SNPs were selected from regions not covered by the ortholog alignment method described above. Synonymous and nonsynonymous sites were calculated as follows: for every position in the genome, whether it was located in an intergenic or coding region was assessed; if it was in a coding region (excluding coding regions from insertion elements and other repetitive elements) and the nucleotide substitution resulted in no change in amino acid sequence, it was classified as a potential synonymous SNP site; otherwise, it was regarded as a potential nonsynonymous site.
Comparative analysis of gene deletions in Y. pestis genomes.
We analyzed the loss-of-function patterns in all Y. pestis genomes, focusing on the presence and absence of protein functions. Some deletions, such as a tufB deletion in Nepal516 (described below), were not included because of gene duplication. Complete datasets of proteins for each Y. pestis genome were downloaded from published reports or from the final annotations of the newly sequenced genomes. Transposases and enzymes related to insertion elements were removed. The final protein data sets for deletion analysis were 3,723, 3,909, 3,896, 3,769, 3,777, and 3,867 proteins for Y. pestis strains CO92, KIM, Antiqua, Nepal516, and 91001 and Y. pseudotuberculosis IP32953, respectively. Pairwise alignments of proteins of all five Y. pestis genomes were accomplished by BLASTP. A protein function was deemed absent if there was no top hit greater than 95% identity or at least 75% of the query sequence. This analysis focused on the presence or absence of protein and functional representation; therefore, if the protein had a closely related paralog or was duplicated in the genome, it was considered present. Due to the differences in annotation (particularly with smaller gene calls), we applied a cutoff criterion to remove all small proteins since these were more frequently found to be differentially annotated across the Y. pestis genomes. With a size filter of 75 amino acids, we are certain to have missed a small number of real proteins that are smaller than 75 amino acids, such as the 61-amino-acid carbon storage regulator crsA (YPO3304). The final set of proteins found to be absent in at least one genome was manually inspected with the aid of the multiple sequence alignment tool CLUSTALW and the nucleotide sequence alignment tool BLASTN. If the deletion was comprised solely of repetitive units, the protein was removed from this analysis because the deletion mechanism may be different in those cases and may revert frequently. Additionally, these final sets of proteins were inspected in multiple genome alignments to distinguish annotation differences versus true differences in the genomes. A similar set of criteria was employed to see if homologs of these proteins exist in the Y. pseudotuberculosis IP32953 genome.
Nucleotide sequence accession numbers and locus tag prefixes.
The annotated sequences of the complete genomes of Y. pestis strains Antiqua and Nepal516 are available at GenBank/EMBL/DDBJ and are as follows: for strain Nepal516, CP000305 (chromosome), CP000306 (pMT), and CP000307 (pPCP); for strain Antiqua, CP000308 (chromosome), CP000309 (pMT), CP000310 (pPCP), and CP000311 (pCD). The prefixes YPA and YPN are used for locus tags (gene identifier prefixes) in strains antiqua and Nepal516, respectively. When referring to specific genes throughout the text, we use the CO92 gene numbers (prefixed with YPO) where possible, unless the gene does not exist in CO92 (or if it is clearer to use a different prefix); then the locus tags for a different genome are used and mentioned.
RESULTS
Genome overviews.
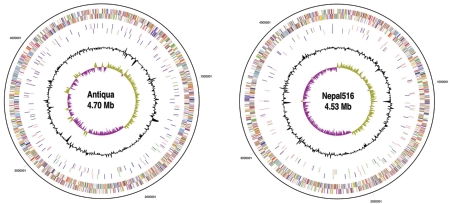
The genomes of Y. pestis strains Antiqua and Nepal516 each consist of a single circular chromosome and the three virulence plasmids, pMT, pCD, and pPCP, which are associated with most classical Y. pestis strains. Here we report all replicons except the pCD plasmid of Nepal516 (see Materials and Methods). The salient genomic features of each genome are detailed in Table 1, while gross chromosomal comparisons of strains are summarized in Fig. 1. Although the global characteristics of the five genomes are quite similar, a number of strain-specific insertions, deletions, rearrangements, and SNPs were identified, along with a unique distribution of IS elements (see Tables 2, 3, and 4 and the supplemental material).
TABLE 1.
General genome features for Yersinia pestis strains Antiqua and Nepal516
| Characteristic | Value of characteristic for:
|
|
|---|---|---|
| Antiqua | Nepal516 | |
| Chromosome size (bp) | 4,702,289 | 4,534,590 |
| G+C content (%) | 47.70 | 47.58 |
| Coding sequences | 4,138 | 3,956 |
| Average gene length (bp) | 953 | 958 |
| Coding density (%) | 83.8 | 83.6 |
| 16S-23S-5S rRNAs | 7 | 7 |
| Transfer RNAs | 68 | 72 |
| pMT size (bp) | 96,471 | 100,918 |
| G+C content | 50.24 | 50.16 |
| Coding sequences | 99 | 104 |
| Average gene length (bp) | 832 | 820 |
| Coding density (%) | 85.3 | 84.5 |
| pCD size (bp) | 70,299 | —a |
| G+C content | 44.83 | |
| Coding sequences | 89 | |
| Average gene length (bp) | 601 | |
| Coding density (%) | 76.1 | |
| pPCP size (bp) | 10,777 | 10,778 |
| G+C content | 45.44 | 45.44 |
| Coding sequences | 9 | 9 |
| Average gene length (bp) | 573 | 573 |
| Coding density (%) | 47.8 | 47.8 |
pCD of Nepal516 was not completed in this study; see Materials and Methods.
FIG. 1.
Circular representation of the strain Antiqua (A) and strain Nepal516 (B) chromosomes. The different rings represent (from outer to inner) all genes color-coded by functional category (rings 1 and 2), IS elements (IS100, IS285, IS1541, IS1661) (rings 3 and 4), deviation from average G+C content (ring 5), and GC skew (ring 6).
TABLE 2.
| Chromosome characteristic | No. of IS elements for:
|
||||
|---|---|---|---|---|---|
| Antiqua | Nepal516 | 91001 | KIM | CO92 | |
| Total IS elements | |||||
| IS100 | 75 | 32 | 30 | 34 | 44 |
| IS285 | 24 | 25 | 23 | 19 | 21 |
| IS1541 | 67 | 64 | 47 | 55 | 65 |
| IS1661 | 10 | 8 | 8 | 8 | 8 |
| Unique IS elementsc | |||||
| IS100 | 39 | 4 | 15 | 6 | 13 |
| IS285 | 2 | 4 | 11 | 1 | 1 |
| IS1541 | 5 | 4 | 0 | 0 | 8 |
| IS1661 | 1 | 0 | 0 | 0 | 0 |
Molecular groupings for the strains were 1.ANT, 2.ANT, 0.PE4, 2.MED, and 1.ORI for Antiqua, Nepal516, 91001, KIM, and CO92, respectively.
Genome sizes for the strains were 4,702,289, 4,534,595, 4,595,065, 4,600,755, and 4,653,728 bp for Antiqua, Nepal516, 91001, KIM, and CO92, respectively.
Some IS elements were shared between expected partners, such as three IS100 and one IS285 shared between Antiqua and CO92, as well as one IS100, one IS1661 and one IS1541 shared between Nepal516 and KIM. However, five exceptions were observed: one IS100 shared between 91001, Antiqua, and CO92 but not present in Nepal516 or KIM; one IS100 in 91001, Antiqua, CO92, and KIM but not in Nepal516; one IS285 in 91001, CO92, KIM, and Nepal516 but not in Antiqua; one IS1541 in CO92 and in Y. pseudotuberculosis IP32053; and one IS1541 in Antiqua and in Y. pseudotuberculosis IP32053.
TABLE 3.
Genome-specific inactivation of genes
| Strain(s) with deletions specific to the genome(s)a | No. of proteins inactivated |
|---|---|
| CO92 | 4 |
| KIM | 20 |
| Antiqua | 41 |
| Nepal516 | 13 |
| 91001 | 69 |
| CO92, KIM | 0 |
| CO92, Antiqua | 11 |
| CO92, Nepal516 | 0 |
| CO92, 91001 | 0 |
| KIM, Antiqua | 2 |
| KIM, Nepal516 | 16 |
| KIM, 91001 | 0 |
| Antiqua, Nepal516 | 0 |
| Antiqua, 91001 | 8 |
| Nepal516, 91001 | 1 |
For rows with two strains, the data indicate inactivations in the same CDS of both strains.
TABLE 4.
Prophage-like fragments specific to virulent Y. pestis strains
| Gene |
Y. pestisb
|
Y. pseudotuberculosis IP32953b | COG functional classc | Product | |||
|---|---|---|---|---|---|---|---|
| CO92 | KIM | Antiqua | Nepal | ||||
| YPO2095 | + | + | − | + | − | − | Hypothetical protein |
| YPO2096 | + | +a | − | +a | − | − | Hypothetical protein |
| YPO2097 | + | +a | − | +a | − | − | Hypothetical protein |
| YPO2098 | + | + | − | + | − | R | Putative phage lysozyme |
| YPO2099 | + | + | − | + | − | − | Putative prophage endopeptidase |
| YPO2100 | + | + | − | + | − | S | Phage regulatory protein |
| YPO2101 | + | + | − | + | − | − | Hypothetical protein |
| YPO2102 | + | + | − | + | − | − | Hypothetical protein |
| YPO2104 | + | + | − | + | + | L | Transposase for the IS285 insertion element |
| YPO2108 | + | + | + | + | − | − | Hypothetical protein |
| YPO2109 | + | + | + | + | − | − | Hypothetical protein |
| YPO2110 | + | + | + | + | − | − | Hypothetical protein |
| YPO2111 | + | + | + | + | − | − | Hypothetical protein |
| YPO2112 | + | + | + | + | − | − | Hypothetical protein |
| YPO2113 | + | + | + | + | − | − | Hypothetical protein |
| YPO2114 | + | + | + | + | − | − | Hypothetical protein |
| YPO2115 | + | + | + | + | − | − | Hypothetical protein |
| YPO2116 | + | + | + | + | − | − | Hypothetical protein |
| YPO2117 | + | + | + | + | − | − | Hypothetical protein |
| YPO2118 | + | +a | +a | +a | − | − | Hypothetical protein |
| YPO2119 | + | + | + | + | − | S | Putative phage tail protein |
| YPO2120 | + | + | + | + | − | S | Hypothetical protein |
| YPO2122 | + | + | + | + | − | S | Hypothetical protein |
| YPO2123 | + | + | + | + | − | R | Putative phage minor tail protein |
| YPO2124 | + | + | + | + | − | − | Hypothetical protein |
| YPO2125 | + | + | + | + | − | − | Putative phage regulatory protein |
| YPO2126 | + | + | + | + | − | K | Hypothetical protein |
| YPO2127 | + | + | + | + | − | − | Putative phage-related membrane protein |
| YPO2128 | + | +a | +a | +a | − | − | Putative phage-related lipoprotein |
| YPO2129 | + | + | + | + | − | S | Putative phage tail assembly protein |
| YPO2130 | + | +a | +a | +a | − | − | Hypothetical protein |
| YPO2131 | + | + | + | + | − | S | Putative phage host specificity protein |
| YPO2132 | + | + | + | + | − | − | Hypothetical protein |
| YPO2133 | + | + | + | + | − | − | Hypothetical protein |
| YPO2134 | + | + | + | + | − | − | Putative phage tail fiber assembly protein |
| YPO2135 | + | +a | + | + | − | − | Hypothetical protein |
| YPO2487 | + | +a | +a | +a | +a | − | Putative membrane protein |
| YPO2488 | + | + | + | + | + | − | Hypothetical protein |
| YPO2489 | + | + | + | + | + | S | Hypothetical protein |
The protein sequence was not found in the genome; however, the DNA fragment did exist in the intergenic region.
+, present; −, deleted.
COGs: K, transcription; L, DNA replication or repair; R, general function prediction only; S, function unknown; −, no COG assignment.
Strain-specific synonymous SNPs and nonsynonymous SNPs.
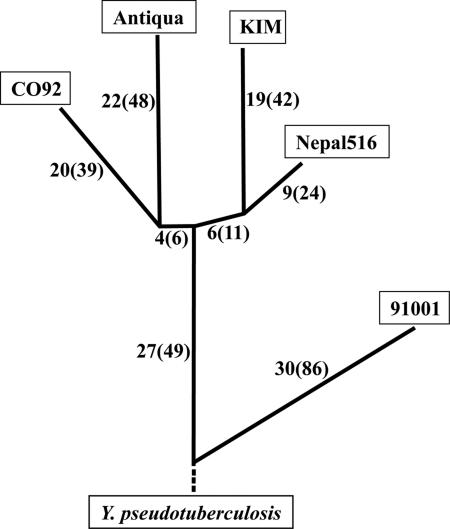
The numbers of synonymous SNPs (sSNPs) and nonsynonymous SNPs (nsSNPs) specific to one or to two genomes are shown in Fig. 2. There are 57 sSNPs (135 nsSNPs) specific to strain 91001 relative to all other Y. pestis strains. While 27 of these sSNPs (49 nsSNPs) are shared with the ancestral Y. pseudotuberculosis IP32953, the remaining 30 sSNPs (and 86 nsSNPs) differ with respect to Y. pseudotuberculosis IP32953, indicating that they likely arose in 91001 since its lineage diverged from the remaining Y. pestis-sequenced isolates. Likewise, the 27 sSNPs (and 49 nsSNPs) specific to 91001 (and identical to Y. pseudotuberculosis) are mutations predicted to have arisen in the other Y. pestis lineage which gave rise to the remaining sequenced strains (Fig. 2). No SNPs (sSNPs or nsSNPs) are found to be specific to strain pairs Antiqua and KIM, Antiqua and Nepal516, CO92 and KIM, or CO92 and Nepal516. However, four sSNPs (and six nsSNPs) are found specifically in CO92 and Antiqua (i.e., CO92 and Antiqua share the same SNP state, while all other Y. pestis strains have a different SNP state) and six sSNPs (and 11 nsSNPs) are specific to KIM and Nepal516. Taken together, these data suggest a separation of these four strains into two distinct branches, where Antiqua and CO92 belong to one branch and KIM and Nepal516 occupy the other (Fig. 2). These two sets of sSNP and nsSNP mutations have accumulated in the short period of time after the KIM/Nepal516 and CO92/Antiqua lineages diverged but before each lineage further split into two (Fig. 2). Thus, this analysis strongly supports the notion that, although strains Antiqua and Nepal516 are grouped into the same biovar (antiqua), they represent distinct lineages.
FIG. 2.
Phylogenetic ordering of Yersinia by SNP analysis. The number of sSNPs and the number of nsSNPs (in parentheses) are illustrated at the corresponding positions.
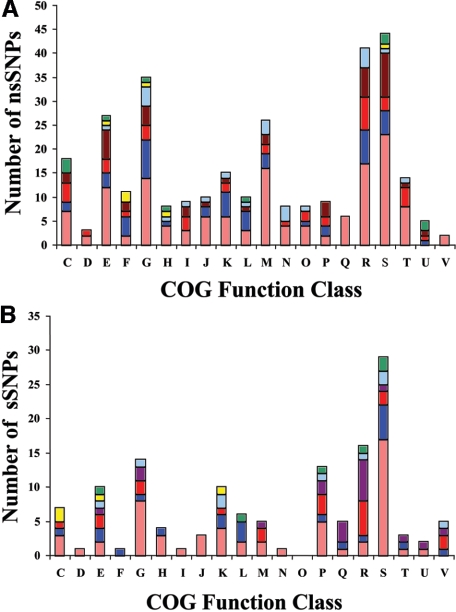
The genes harboring sSNPs (cumulatively for all the Y. pestis strains) can be distributed into functional gene categories (based on COGs) as shown in Fig. 3. Nonsynonymous SNPs are found distributed in 20 COG categories (Fig. 3A), while synonymous SNPs were distributed in 19 COG categories (Fig. 3B). We investigated whether SNPs belonging to the various branches in Fig. 2 are biased towards any functional categories, but no unique or biased distribution patterns were found. All strain-specific mutations share approximately 1/3 of the COG categories. Although some strain-specific SNPs are found in functional categories not represented in any other strain (sSNP, three categories are unique to 91001 and one category is unique to Antiqua; nsSNP, two categories are unique to 91001) (Fig. 3), the number of SNPs is too small to determine whether these were random events. No sSNP versus nsSNP bias was readily apparent (the average nsSNP/sSNP ratio is approximately 2.9) except for the large proportion of nsSNPs versus sSNPs in the cell wall/membrane biogenesis COG of 91001, with a ratio of 9:2.
FIG. 3.
Functional distribution of genes bearing SNPs. The numbers of genome-specific nsSNPs (A) and sSNPs (B) were grouped into COG functional classes. These were subcategorized based on what genome(s) they were found in (light orange, 91001; red, CO92; dark red, KIM; blue, Antiqua; light blue, Nepal516; green, KIM and Nepal516; and yellow, Antiqua and CO92). C, energy production; D, cell division; E, amino acid metabolism; F, nucleotide metabolism; G, carbohydrate metabolism; H, coenzyme metabolism; I, lipid metabolism; J, translation; K, transcription; L, DNA replication or repair; M, cell wall/membrane biogenesis; N, cell motility; O, posttranslational modification; P, inorganic ion metabolism; Q, secondary metabolite biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown; T, signal transduction; U, intracellular trafficking and secretion; V, defense mechanism.
The gene sequences and gene organization in the plasmids are highly conserved. There is only one synonymous SNP in the plasminogen activator protease and five SNPs (including small deletions) in noncoding regions of all pPCP plasmids. There are 8 and 17 SNPs in the predicted gene sets of all pCD and pMT plasmids, respectively. The majority of these SNPs are in hypothetical proteins and are distributed more or less randomly across strains.
IS elements and genome rearrangements.
As determined by several groups already (10, 13, 33, 46), IS elements have expanded tremendously in Y. pestis since its divergence from Y. pseudotuberculosis and have served as the delimiters for recombination events that have led to genomic deletions and genome rearrangements. Due to its presumed continued transposition activity in the wild, IS100 elements have been successfully used for typing and grouping strains (32). We thus performed a detailed analysis of the precise locations of IS100 and the other three major IS elements (IS1541, IS285, and IS1661) as well as investigated their contribution to the observed rearrangements.
Each sequenced strain of Y. pestis has a unique set of IS elements (Table 2) and a core IS set that is shared among them. It was previously observed that Y. pestis shares a set of 12 IS elements with Y. pseudotuberculosis (3 of each major IS element) that, based on identical locations of insertion, appear to have been acquired by Y. pseudotuberculosis before the evolution of Y. pestis (10). In addition to this set of 12, the core set of IS elements shared among all five sequenced Y. pestis strains is 45 IS1541, 15 IS100, 11 IS285, and 6 IS1661, indicative of elements present in the last common ancestor of all of these sequenced strains. Several more (two IS1541, one IS100, two IS285, and two IS1661) are predicted to be shared among all five Y. pestis strains, but in at least one strain, these IS elements have subsequently been involved in a deletion event between two IS copies (leaving only one behind) or have been lost as part of a larger deletion. Similarly, the four classical Y. pestis strains (Antiqua, Nepal516, KIM, and CO92) also share a subset of the remaining IS elements distinct from nonclassical strain 91001: five IS1541, five IS100, and three IS285 with four additional IS100 elements that have likely been lost in a strain-specific manner via deletion as described above. In addition, there are four IS elements shared between CO92 and Antiqua and three shared between KIM and Nepal516 (Table 2), supporting the SNP-based phylogeny shown in Fig. 2 as well as that depicted by Achtman and colleagues (2). Interestingly, a small number of IS elements were shared by unexpected groups of strains that disagree with the proposed phylogeny: one IS100 shared between 91001, Antiqua, and CO92; one IS100 shared between Antiqua and KIM; one IS100 in Antiqua, CO92, and KIM; one IS1541 in CO92 and Y. pseudotuberculosis IP32053; one IS285 in CO92, KIM, and Nepal516 but not Antiqua; and one IS1541 in Antiqua and Y. pseudotuberculosis IP32053.
While IS1541 was the most active IS element between the divergence of Y. pseudotuberculosis and the most recent common ancestor of the five Y. pestis strains, with 45 common IS1541 insertions among all sequenced isolates, IS100 has been the most active in more recent times, though not equally among the strains (Table 2). This suggests that insertion sequence activity may be punctuated and does not occur at a constant rate across different strains. With the exception of Nepal516, the number of unique IS100s is significantly more than that for any other IS element (Table 2), though the reason for this is unclear.
Despite their extensive sequence similarity, the Y. pestis genomes appear to be in a state of flux with respect to large genome rearrangements. Similar to previous observations, all breaks in colinearity between the Yersinia genomes occur at IS elements or other repeated sequences. Differences in the GC-skew patterns in Y. pestis genomes, including the many breaks observed in Antiqua (Fig. 1), are also the result of rearrangements between IS elements as previously observed (10, 33, 46). Similarly, IS elements have played a large role in deletion events observed in the Y. pestis genomes. For example, strains KIM and CO92 have undergone overlapping IS1541-mediated deletions of 32 kb (YP0966 to YP0994, using 91001 nomenclature) and 21.5 kb (YP0966 to YP0986, using 91001 nomenclature), respectively. These deletions accounted for the DFR patterns observed by suppression-subtractive hybridization (37). A similar strain-specific, IS-mediated deletion of a phage region is described further below. Additionally, an IS1661-mediated 13-kb deletion in both KIM and Nepal516 has removed a large cluster of flagellar genes (YPO0738 to YPO0754), including the flagellar RNA polymerase sigma factor and chemotaxis membrane proteins.
Functional reduction.
It has been postulated that the genomes of Y. pestis have undergone functional reduction as it made a transition from an orally and fecally spread pathogen causing gastroenteritis to a vector-borne pathogen causing a fatal, invasive, septicemic disease, where genes have been inactivated by various mechanisms, such as deletions/insertions, frameshifts, interruptions by insertion elements, and homologous or even nonhomologous recombination. We determined all the genes and associated functions that have been lost since Y. pestis emerged from Y. pseudotuberculosis (by comparing Y. pestis strains and identifying functional losses) to better understand Y. pestis diversity and evolution. We have identified a large number of strain-specific gene inactivations/deletions as well as some that are specific to only two of the five genomes (Table 3). Other than hypothetical proteins, there is one dominant category of proteins in these strain- or lineage-specific deletions: proteins contributing to the interaction of bacterium with its environment or host, including membrane proteins, membrane receptors, ABC transporters, flagellar proteins, and chemotaxis proteins.
The number of strain-specific lost functions was not equal among all strains. In part, this can be explained by several large deletions that effectively delete many genes (functions) in a single event. While strains 91001 and Antiqua had the greatest number of strain-specific function losses, with 69 and 41, respectively, it is interesting to note that CO92 was found to have the fewest compared to those of other strains. Though strains Antiqua and 91001 share several gene inactivations (8 proteins), these are the result of independent mutations (homoplasy), while those lost in CO92 and Antiqua (12 proteins), as well as those lost in KIM and Nepal516 (16 proteins), share lineage-specific, inherited mutations (function losses) that support the phylogenetic tree.
Of the 69 functional losses specific to 91001, 24 are hypothetical proteins, 7 are membrane proteins, 7 are phage-related proteins, 5 are regulatory proteins, and 3 are transporters. Some of these 91001-specific losses of function may be related to its human-avirulent phenotype. Twenty-one of the 69 belong to a single IS285-mediated deletion event (YPO2108 to YPO2134).
Ten of the 41 Antiqua-specific lost functions were the results of two deletion events. Several inactivated or deleted genes were predicted to be directly or indirectly involved in interactions with environment. For example, YPO2367, a glutathione S-transferase, is missing from the Antiqua genome. The glutathione S-transferase family of enzymes routinely responds to oxidative stress or detoxification, which can be encountered during entry into phagocytic cells (12). Since bacteria usually have multiple glutathione S-transferases (CO92 has at least four based on the annotation), losing one may not result in a distinct phenotype. Interestingly, there were several cases where similar functional inactivations/deletions that affect different genes that may have overlapping functions were observed in two strains. For example, a potassium efflux pump (YPO3129) was inactivated in Antiqua and a Na+/H+ antiporter (YPO2142) was inactivated in KIM. The role of the Na+/H+ antiporter is sodium extrusion (24), and the Na+/H+ antiporter and the potassium efflux pump are both involved in pH homeostasis. Since both Antiqua and KIM are geographically limited to localized regions, one possible explanation for the differential inactivations is local adaptation to selectively maintain one of the two similar functions. Alternate possibilities are that neither gene is required, that the differences observed in these sequenced strains were random events, and that both genes are not required and are on their way to being lost from the Y. pestis gene pool. Whether the similarity of the deletion profiles mentioned above reflects adaptations to their environmental niches or convergent evolution remains to be investigated.
Of the 20 KIM-specific inactivated genes, 7 are concentrated in one deletion and the inactivation via nonsense mutation of YPO3038 (NapA, a periplasmic nitrate reductase) is proposed to be one of the causes of the nitrate-negative phenotype of the medievalis biovar. Nepal516 has 13 specific lost functions, including those of two transporters (YPO2835 and YPO1350), the chromosomally encoded type III secretion system protein YPO0266, and two classical virulence factors (YPO2291, a putative virulence factor, and YPO0599, a hemolysin/adhesin mentioned further below). Some of the deletions in Nepal516 have been previously demonstrated experimentally by microarray hybridizations (21). There are only four lost functions (pseudogenes YPO1087 and YPO3679, along with y1377 and y2928, using KIM nomenclature) that are CO92-specific losses, yet all are putative proteins without clear functional predictions.
All of the genetic regions identified in the genomes of Antiqua or Nepal516 were present in at least one of the previously sequenced Y. pestis or Y. pseudotuberculosis strains. Though there were a number of Antiqua and Nepal516 genes or domains of genes that were found to differ significantly from the other strains, many of these differences consisted of various numbers of degenerate tandem repeat elements within the coding sequences (CDSs) of surface proteins, such as in the invasin YPO3944 described further below, and were not interpreted as losses of function. Surprisingly, our analysis revealed only one example of a strain-specific unique genetic region with no similar DNA sequence in any of the other four sequenced strains or the Y. pseudotuberculosis genome. This was a single-stranded DNA prophage found inserted in CO92 (YPO2271 to YPO2281). This region has been found by PCR in all tested biovar orientalis strains as well as a few African strains of the biovar antiqua (10, 19).
Differences in putative virulence factors.
Most characterized and putative classical virulence factors are identical throughout all five Y. pestis strains, including those found on the virulence plasmids, such as the pPCP-located plasminogen activator Pla required for successful subcutaneous infection (11) and the pMT-encoded murine toxin (Ymt) (23) and F1 capsular protein (16) (important for Y. pestis life cycle and vector-mammal transmission). Similarly, loci on the chromosome are also nearly identical between strains; the RTX-like toxin gene YPO0947, the attachment invasion locus ail (YPO2905), and two ail-like genes (YPO1860 to YPO2190) are virtually identical among Y. pestis strains and with Y. pseudotuberculosis as well. Interestingly, a fourth ail-like gene (YPO2506) has been deleted from the Antiqua genome.
Other loci, known to differ between Y. pestis and Y. pseudotuberculosis, were also investigated. The Y. pestis invasin YPO1793 is interrupted by an IS1541 in all strains, while putative adhesin YPO1562 is interrupted by an IS285 in all Y. pestis strains except for 91001, which harbors a nonsense mutation instead. Both are intact in Y. pseudotuberculosis. Similarly, RTX transporter YPO2250 and the TccC-family insecticidal toxin YPO2312 are frameshift pseudogenes in all Y. pestis strains but appear intact in Y. pseudotuberculosis.
A second TccC insecticidal toxin homolog, YPO2380, has been deleted in only Y. pestis KIM. Two additional TccC toxins are found in tandem in all Y. pestis strains (YPO3674 to YPO3673), while only a single copy is found in Y. pseudotuberculosis. In Y. pestis, a second family of insecticidal toxins is found upstream of these two TccC homologs and consists of a complex of three genes (YPO3681, YPO3679, and YPO3678). While these are present and highly similar in amino acid sequence in Y. pseudotuberculosis, YPO3681 is inactivated by a frameshift in only Antiqua and YPO3679 harbors a frameshift in only CO92.
One Y. pestis hemolysin/adhesin, YPO0599, located within a possible pathogenicity island (YPO0641a-YPO0590) is different from that of Y. pseudotuberculosis at the C terminus. Several additional CDSs that appear to be “modules” or adhesin fragments that share high similarity to portions of the C terminus of this CDS can be seen downstream in Y. pseudotuberculosis. Different “modules” are found downstream of the Y. pestis gene. Interestingly Nepal516 is missing a large section of the C-terminal portion of this protein, likely due to a recombination between one of these modules and the corresponding section in the Nepal516 gene. The remainder of this pathogenicity island is highly similar between Y. pestis strains and Y. pseudotuberculosis. A similar module-recombination scenario is envisioned to have resulted in a modified C terminus of hemolysin/adhesin YPO2490 in 91001 compared to that of the other Y. pestis strains and Y. pseudotuberculosis. A different mechanism, the expansion or contraction of degenerate repeat units within the putative invasin YPO3944, has resulted in different-sized invasins in Y. pseudotuberculosis (5,623 amino acids [aa]) and the various Y. pestis strains (91001, 3,108 aa; Nepal516, 4,270 aa; and KIM, CO92, and Antiqua, 3,013 aa). Further study is required to understand any phenotypic effect these two classes of differences may have.
Loss of functional TufB in Nepal516.
The genomes of Y. pestis and several other organisms have two copies of the elongation factor Tu (EF-Tu). Due to their highly conserved function and ubiquitous distribution, elongation factors are considered a valuable phylogenetic marker and have been used in evolution studies of Enterococcus (26), Lactobacillus (43), and other eubacteria (39). Interestingly, in Y. pestis, the two copies of this gene are not as conserved as the two genes of Escherichia coli are, and yet the conservation within each copy (among Y. pestis strains) is maintained. In E. coli, tufA and tufB gene products differ by only a single amino acid (7) and exhibit identical physical, chemical, and catalytic properties (17). We found that all five Y. pestis genomes and Y. pseudotuberculosis have two copies of tuf genes (tufA and tufB), and the general operon structure is similar to that of E. coli; however, the sequence identity between the Y. pestis tufA and tufB is considerably lower. There are 17 amino acid differences (4%) and a total of 138 nucleotide changes (11.7%) in addition to a large C-terminal deletion in tufB of Nepal516. This result is unexpected, since previous studies show that duplicate tuf genes within a genome differ on average by 0.7% in nucleotide sequence (30). Whether the two copies of the tuf genes in Y. pestis have different origins requires further investigation. All six Yersinia TufA proteins are 100% identical, and all Y. pestis tufA genes have identical nucleotide sequences, while Y. pseudotuberculosis has a single synonymous SNP (G to A). Although the tufB gene products differ from those of tufA, a similar conservation is evident. We have found, however, that the TufB of Nepal516 harbors a large C-terminal deletion which affects 57 aa (or 67%) of the GTP-EF-Tu-D3 domain, which is involved in binding charged tRNAs and EF-TUs (6). This deletion is likely to cause loss of functionality, and thus we believe that TufB is not functional in Nepal516. It is not known whether the two copies are expressed under different conditions or have slightly different functions or kinetic properties; however, this deletion leads us to infer that Y. pestis requires only one functional copy for its life cycle.
Comparison to the human-avirulent strain 91001.
A previous comparison between strain 91001 and human virulent strains CO92 and KIM revealed a number of differences, including a 33-kb prophage-like sequence (YPO2096 to YPO2135) that was absent in 91001 but intact in both CO92 and KIM, and suggested that this difference may have contributed to this strain's lack of virulence in human (41). This entire region was also absent in Y. pseudotuberculosis (10), consistent with this claim. Our analysis shows that this phage-like region is intact in Nepal516 but partially deleted in Antiqua (Table 4). While the deletion in 91001 can be attributed to recombination between two parallel IS100 elements, the smaller deletion in Antiqua (from YPO2087 to YPO2106) is likely due to excision between two IS285 sequences (see the supplemental material). Since Antiqua is a fully virulent strain, the region deleted in Antiqua would not seem to contribute to human pathogenicity; however, the remaining portion of the prophage region deleted from 91001 may indeed contain genes that are important for human virulence.
Previous comparisons with CO92 and KIM also revealed a list of 91001-specific pseudogenes that may be related to Y. pestis pathogenicity and host range (41). Our gene reduction analysis included two additional virulent strains and confirmed the presence or absence of orthologs in Y. pseudotuberculosis based on our cutoffs (see Materials and Methods). While all of these 91001-specific pseudogenes were also intact in Antiqua (see the supplemental material), only one (YPO2258) was also found to be inactivated in Nepal516, suggesting that this gene has no impact on the avirulent property of 91001 in humans. Among the 91001-specific pseudogenes, there are only four that are also absent in Y. pseudotuberculosis, YPO0733 (flagellar hook-associated protein), YPO0737 (flagellin), YPO0962 (hypothetical protein), and YPO3110 (putative O-unit flippase). In addition, the nsSNP mutations that contribute to changes in single amino acids in many proteins may affect 91001 or other Y. pestis strain virulence in subtle ways. For example, nsSNPs with a possible role in virulence were found in some genes (e.g., the antiqua ail gene YPO2905 carries an nsSNP, as does the 91001 RTX toxin gene YPO0947) but the significance of these substitutions is not known.
DISCUSSION
This work presents the complete genome sequences for the two previously unsequenced Y. pestis major lineages (both designated biovar antiqua using classical nomenclature). Phylogenetic relationships were elucidated clearly with the distribution of synonymous SNPs (Fig. 2). Since synonymous mutations do not affect protein functions (unlike nsSNPs or some IS elements), their accumulation is not under selective pressure, making this the least biased method for inferring evolutionary relationships. The distribution of sSNPs convincingly demonstrates that a single biovar, antiqua, is an inaccurate phylogenetic representation, supporting previous claims that categorize biovar antiqua strains into two groups (2, 10). Using terminology proposed previously (2), lineage 1.ANT (African strain Antiqua) is closely related to orientalis strain CO92, while 2.ANT (Asian strain Nepal516) is more closely related to medievalis strain KIM. These four classical isolates fall on a branch separate from the nonclassical, human-avirulent Chinese strain 91001. This analysis also revealed a relatively rapid divergence of the four distinctive lineages from two ancestral lines for the classical Y. pestis strains. Although it is only possible to make very crude estimations of the age of descent for these four lineages, the numbers of sSNPs are consistent with all of the lineages being present within the last 1,500 years of the three great pandemics (calculation not shown).
A comparison of all five Y. pestis sequences reveals extensive DNA sequence rearrangement, widespread gene reduction, and strain-specific IS elements as well as SNPs. It was previously reported that Y. pestis strains differ greatly in genome synteny and that repeated sequences most often were found at the borders of rearrangements (10, 13). Indeed, most rearrangements occur at IS elements and, regardless of which genomes were chosen for two-way comparisons, we identified numbers of rearrangements similar to those previously observed between Y. pestis strains (13) and even between Y. pestis and Y. pseudotuberculosis (10) (data not shown). The question remains whether these observed rearrangements have any effect on transcription or whether they have an overall destabilizing influence on the genome.
The distribution pattern of IS elements in the sequenced strains generally supports the SNP-derived phylogeny, with several IS elements shared across all classical strains except 91001 as well as IS elements found in only the CO92/Antiqua or KIM/Nepal516 pair of strains. Only a few (five in total) IS elements found to be shared by two or more strains did not conform to the predicted phylogeny (footnoted in Table 2); similar observations have been reported previously (2). Our analyses suggest that a small number of IS elements may have been precisely excised from their insertion locations, that identical insertion events have occurred in two different strains/lineages, or that there may be some limited horizontal transfers between Y. pestis strains that have resulted in mobilizing IS elements from one strain to a different strain/lineage (or, alternatively, removing an IS element by introduction of the wild-type sequence). One example is an aminotransferase (YPO3250) that is disrupted by an IS100 in all sequenced Y. pestis strains except Nepal516, which instead has the wild-type gene and no trace of an IS100. These data also suggest that certain IS elements may not be useful for typing or grouping strains and may explain certain discrepancies in phylogenetic groupings using different methods.
Interestingly, the entire complement of IS1541 (and almost all IS1661) elements in strain 91001 was acquired by the ancestor of all Y. pestis strains. In contrast, since 91001 diverged from the other strains, it has acquired a number of strain-specific IS100 and IS285 elements, supporting the idea of actively integrating IS elements within the genome of Y. pestis. With the exception of Nepal516, IS100 appears to have been more active (greater number of new transposition events) than other IS elements, but the reason for this is unknown.
Functional reduction analysis also generally agrees with the SNP-based phylogenetic tree (Fig. 3) as well as with a more limited study that identified the loss of gene regions across a panel of Y. pestis isolates using a CO92 gene-specific microarray (21). Similar to the IS and SNP data, the four classical strains appear to share an evolutionary path distinct from that of strain 91001 based on functional reduction, and the KIM/Nepal516 and CO92/Antiqua pairs also exhibit a larger number of shared function loss. The exceptions are the result of independent mutations: two shared losses between KIM and Antiqua, one shared loss between Nepal516 and 91001, eight shared losses between Antiqua and 91001, and 16 shared losses between CO92 and KIM (Table 3 and see the supplemental material). The two shared function losses between KIM and Antiqua are a putative siderophore biosynthetic enzyme and a putative membrane protein. The predicted functions suggest that both proteins could be involved in interactions with the environment; therefore, these losses may reflect adaptations to the Y. pestis microenvironment. Similarly, the single functional loss shared between Nepal516 and 91001 is the arabinose operon regulatory protein. Although the observed shared loss of function between Antiqua and 91001 contained several genes, they are exclusively in the prophage region described above and it is the result of independent deletion events. The shared losses between CO92 and KIM were possibly from a single deletion event.
Strain 91001 has the highest number of strain-specific losses of function, with a total of 69. Interestingly, all but four of the 91001-specific pseudogenes have homologs with >90% identity in Y. pseudotuberculosis, suggesting that 91001 lost those genes, while other virulent Y. pestis strains retained them. It is possible that these genes may be involved in human virulence and/or fitness in the human host. Some inactivated proteins, such as hemolysin (YPO2045), sulfatase and sulfatase modifier protein (YPO3046 and YPO3047), UDP-glycosyltransferase (YPO1985), and O-unit flippase-like protein (YPO3110), may be related to pathogenicity (see the supplemental material). Hemolysin is a toxin that forms transmembrane channels and is involved in heme utilization and adhesion. The precise function of the sulfatase operon (YPO3046 and YPO3047) in Y. pestis is not known; however, these enzymes belong to a family of proteins that hydrolyze various sulfate esters or catalyze sulfur insertions. In mammalian cells, the oligosaccharide moieties on glycoproteins, glycolipids, and proteoglycans are frequently modified with sulfate. Sulfatase from pathogenic bacteria has been shown to interact with mucin (47), and a previous study suggested that mucin-sulfatase activity in Burkholderia cepacia and Pseudomonas aeruginosa may contribute to their associations with airway infection in cystic fibrosis patients by possibly facilitating bacterial colonization (25). Thus, the deletion of the sulfatase and sulfatase modifier protein in strain 91001 may have contributed to its human-avirulent phenotype. Finally, the O-unit flippase is involved in translocating a polysaccharide unit across the membrane while UDP-glycosyltransferase (YPO1985) is typically involved in O-antigen biosynthesis. Since Y. pestis is known to lack O antigen, the actual functions of YPO3110 and YPO1985 may not directly involve O antigen but perhaps other surface polysaccharides.
Antiqua also had a high number of strain-specific losses, even after discounting the deletion events which involved several genes (41 and 31, respectively). Interestingly, we found a correlation with the observed higher IS100 transposition activity in Antiqua, with 13 of the 31 inactivations due to IS100 interruptions. The profile of Antiqua-specific loss of function contains a significant number of proteins which interact with environment, such as glutathione S-reductase, chemotaxis protein, porin C protein, potassium efflux pump, insecticidal toxin, flagellar motor switch protein, and six membrane proteins without specific known functions. A possible explanation for this may be that the genome has been adapting to the niche the Antiqua organism occupies.
Discounting those genes lost in a single deletion event, the numbers of KIM-specific (14) and Nepal516-specific (13) functional loss are similar. Surprisingly, only three CO92-specific losses of function were identified. It is possible that there was a selective advantage for the orientalis biovar to maintain a greater repertoire of genes and to maintain flexibility and be able to adapt quickly to a new host(s). The worldwide distribution of this group and the small number of CO92-specific putative gene inactivations are consistent with this theory. A 31-amino-acid deletion in YPO3937 (473 amino acids) confers the glycerol-negative phenotype of biovar orientalis (33); however, since the deletion was below the cutoff threshold, it was not included in our study as a loss of function. Unique to strain CO92 are a hypothetical protein (YPO2469), a hemolysin activator protein (YPO3720), and a prophage that do not exist or have been inactivated in the other sequenced Y. pestis strains or Y. pseudotuberculosis. These genes may again have been retained by CO92 to maintain its ability to interact with a more variable environment.
Unexpectedly we found that Nepal516 has many exceptions relative to the other sequenced Y. pestis strains, including the apparent loss of function of TufB, the number of Nepal516-specific SNPs much smaller than the numbers specific for other strains (Fig. 2), and the fact that IS100 has not been as active in Nepal516 as in the other strains (Table 2). Since both nsSNPs and sSNPs are equally affected, it is unlikely that this is due to selective pressure (which should have a neutral effect on sSNPs) but rather the mutation rate is responsible, suggesting the rate of mutation or evolution is slower in Nepal516. The reason for this is not known; however, a possible explanation may be that this phenomenon is driven by fewer rounds of bacterial division with a relatively cooler local environment and hibernation of the host(s) that fostered fewer opportunities for transmission.
Despite the observed differences between different strains of Y. pestis, the sequenced genomes reveal a highly conserved chromosomal backbone reminiscent of what is observed in Bacillus anthracis (31). Within the five genomes of Y. pestis compared here, a single region present in strain CO92 was found to be unique (not shared with another Y. pestis genome), though independent studies have shown that this region, which encodes phage genes, is present in most, if not all, 1.ORI strains as well as some 1.ANT strains (10, 19). We thus believe that most of the genomic sequence shared among the “classical” Y. pestis isolates is represented within this data set, though other sequences of nonclassical isolates may harbor novel genomic regions not revealed in these analyses.
Conclusions.
The two completed genomes presented here, from the previously unrepresented antiqua biovar, have provided important references for SNP discovery and for the study of insertion element distribution, genome rearrangement, and reductive evolution in Y. pestis. Comparisons of the four virulent “classical” strains to the human-avirulent strain 91001 have also provided further insight into Y. pestis human virulence. With sSNPs as the preferred method for elucidating phylogenetic relationships, strains Nepal516 and Antiqua were convincingly placed in two clearly separate branches, with one branch shared by strains KIM (medievalis) and Nepal516 and the other shared by strains CO92 (orientalis) and Antiqua. While IS element distributions and function loss across the strains generally agreed with such a phylogenetic representation, certain exceptions were found and are thought to be the result of a lack of selective pressure in the Y. pestis strain-inhabited niche, of possible horizontal gene exchange between Y. pestis strains, or of homoplasy in the reductive processes. Though there is some evidence of convergent evolution, whether this is the primary mechanism underlying the observed discrepancies remains to be investigated. The Y. pestis genome is a clear example of one actively undergoing reductive evolution, as its lifestyle has altered from an enteropathogen to an intracellular pathogen. The genome has slowly accumulated inactivations and deletions that result in loss of function, which, for the virulent strains (all strains except 91001), have little effect on pathogenicity. The differences between these strains and the human-avirulent 91001 provide an ideal starting point for future experiments to elucidate the mechanisms involved in Yersinia pathogenicity.
Supplementary Material
Acknowledgments
We thank Matt Van Ert, Ryan Easterday, Aubree Hinckley, and Maria Shin for technical assistance.
This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Berkeley National Laboratory, under contract no. DE-AC02-05CH11231 and Lawrence Livermore National Laboratory under contract no. W-7405-Eng-48. This work was supported by an Intelligence Technology Innovation Center grant.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Achtman, M. 2004. Age, descent and genetic diversity within Yersinia pestis, p. 432. In E. Carniel and B. J. Hinnebusch (ed.), Yersinia: molecular and cellular biology. Horizon Bioscience, Norwich, United Kingdom.
- 2.Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 101:17837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adair, D. M., P. L. Worsham, K. K. Hill, A. M. Klevytska, P. J. Jackson, A. M. Friedlander, and P. Keim. 2000. Diversity in a variable-number tandem repeat from Yersinia pestis. J. Clin. Microbiol. 38:1516-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bearden, S. W., and R. D. Perry. 1999. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32:403-414. [DOI] [PubMed] [Google Scholar]
- 6.Berchtold, H., L. Reshetnikova, C. O. A. Reiser, N. K. Schirmer, M. Sprinzl, and R. Hilgenfeld. 1993. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365:126-132. [DOI] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 8.Buchrieser, C., C. Rusniok, L. Frangeul, E. Couve, A. Billault, F. Kunst, E. Carniel, and P. Glaser. 1999. The 102-kb pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun. 67:4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chain, P., J. Lamerdin, F. Larimer, W. Regala, V. Lao, M. Land, L. Hauser, A. Hooper, M. Klotz, J. Norton, L. Sayavedra-Soto, D. Arciero, N. Hommes, M. Whittaker, and D. Arp. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chain, P. S. G., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowan, C., H. A. Jones, Y. H. Kaya, R. D. Perry, and S. C. Straley. 2000. Invasion of epithelial cells by Yersinia pestis: evidence for a Y. pestis-specific invasin. Infect. Immun. 68:4523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Groote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devignat, R. 1951. Varietes de l'espece Pasteurella pestis: nouvelle hypothese. Bull. W. H. O. 4:247-263. [PMC free article] [PubMed] [Google Scholar]
- 15.Drancourt, M., V. Roux, L. V. Dang, L. Tran-Hung, D. Castex, V. Chenal-Francisque, H. Ogata, P. Fournier, E. Crubézy, and D. Raoult. 2004. Genotyping, orientalis-like Yersinia pestis, and plague pandemics. Emerg. Infect. Dis. 10:1585-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du, Y., R. Rosqvist, and A. Forsberg. 2002. Role of fraction 1 antigen of Yersinia pestis in inhibition of phagocytosis. Infect. Immun. 70:1453-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furano, A. V. 1977. The elongation factor Tu coded by the tufA gene of Escherichia coli K-12 is almost identical to that coded by the tufB gene. J. Biol. Chem. 252:2154-2157. [PubMed] [Google Scholar]
- 18.Gehring, A. M., E. DeMoll, J. D. Fetherston, I. Mori, G. Mayhew, F. R. Blattner, C. T. Walsh, and R. D. Perry. 1998. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5:573-586. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez, M. D., C. A. Lichtensteiger, R. Caughlan, and E. R. Vimr. 2002. Conserved filamentous prophage in Escherichia coli O18:K1:H7 and Yersinia pestis biovar orientalis. J. Bacteriol. 184:6050-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guiyoule, A., F. Grimont, I. Iteman, P. Grimont, M. Lefevre, and E. Carniel. 1994. Plague pandemics investigated by ribotyping of Yersinia pestis strains. J. Clin. Microbiol. 32:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. F. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 23.Hinnebusch, B. J., A. E. Rudolph, P. Cherepanov, J. E. Dixon, T. G. Schwan, and A. Forsberg. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733-735. [DOI] [PubMed] [Google Scholar]
- 24.Ito, M., A. Guffanti, J. Zemsky, D. Ivey, and T. Krulwich. 1997. Role of the nhaC-encoded Na+/H+ antiporter of alkaliphilic Bacillus firmus OF4. J. Bacteriol. 179:3851-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jansen, H. J., C. A. Hart, J. M. Rhodes, J. R. Saunders, and J. W. Smalley. 1999. A novel mucin-sulphatase activity found in Burkholderia cepacia and Pseudomonas aeruginosa. J. Med. Microbiol. 48:551-557. [DOI] [PubMed] [Google Scholar]
- 26.Ke, D., M. Boissinot, A. Huletsky, F. J. Picard, J. Frenette, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Evidence for horizontal gene transfer in evolution of elongation factor Tu in enterococci. J. Bacteriol. 182:6913-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klevytska, A. M., L. B. Price, J. M. Schupp, P. L. Worsham, J. Wong, and P. Keim. 2001. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J. Clin. Microbiol. 39:3179-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurtz, S., A. Phillippy, A. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kutyrev, V. V., A. A. Filippov, O. S. Oparina, and O. A. Protsenko. 1992. Analysis of Yersinia pestis chromosomal determinants Pgm super+ and Pst supers associated with virulence. Microb. Pathog. 12:177-186. [DOI] [PubMed] [Google Scholar]
- 30.Lathe, I., C. Warren, and P. Bork. 2001. Evolution of tuf genes: ancient duplication, differential loss and gene conversion. FEBS Lett. 502:113-116. [DOI] [PubMed] [Google Scholar]
- 31.Medini, D., C. Donati, H. Tettelin, V. Masignani, and R. Rappuoli. 2005. The microbial pan-genome. Curr. Opin. Genet. Dev. 15:589-594. [DOI] [PubMed] [Google Scholar]
- 32.Motin, V. L., A. M. Georgescu, J. M. Elliott, P. Hu, P. L. Worsham, L. L. Ott, T. R. Slezak, B. A. Sokhansanj, W. M. Regala, R. R. Brubaker, and E. Garcia. 2002. Genetic variability of Yersinia pestis isolates as predicted by PCR-based IS100 genotyping and analysis of structural genes encoding glycerol-3-phosphate dehydrogenase (glpD). J. Bacteriol. 184:1019-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. F. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 34.Perry, R., and J. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry, R. D., A. G. Bobrov, O. Kirillina, H. A. Jones, L. Pedersen, J. Abney, and J. D. Fetherston. 2004. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J. Bacteriol. 186:1638-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prentice, M. B., K. D. James, J. Parkhill, S. G. Baker, K. Stevens, M. N. Simmonds, K. L. Mungall, C. Churcher, P. C. F. Oyston, R. W. Titball, B. W. Wren, J. Wain, D. Pickard, T. T. Hien, J. J. Farrar, and G. Dougan. 2001. Yersinia pestis pFra shows biovar-specific differences and recent common ancestry with a Salmonella enterica serovar Typhi plasmid. J. Bacteriol. 183:2586-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radnedge, L., P. G. Agron, P. L. Worsham, and G. L. Andersen. 2002. Genome plasticity in Yersinia pestis. Microbiology 148:1687-1698. [DOI] [PubMed] [Google Scholar]
- 38.Radnedge, L., S. Gamez-Chin, P. M. McCready, P. L. Worsham, and G. L. Andersen. 2001. Identification of nucleotide sequences for the specific and rapid detection of Yersinia pestis. Appl. Environ. Microbiol. 67:3759-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sela, S., D. Yogev, S. Razin, and H. Bercovier. 1989. Duplication of the tuf gene: a new insight into the phylogeny of eubacteria. J. Bacteriol. 171:581-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skurnik, M., J. A. Bengoechea, and K. Granfors (ed.). 2003. Advances in experimental medicine and biology, vol. 529. The genus Yersinia: entering the functional genomic era. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 41.Song, Y., Z. Tong, J. Wang, L. Wang, Z. Guo, Y. Han, J. Zhang, D. Pei, D. Zhou, H. Qin, X. Pang, Y. Han, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, F. Chen, S. Li, C. Ye, Z. Du, W. Lin, J. Wang, J. Yu, H. Yang, J. Wang, P. Huang, and R. Yang. 2004. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11:179-197. [DOI] [PubMed] [Google Scholar]
- 42.Une, T., and R. R. Brubaker. 1984. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect. Immun. 43:895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ventura, M., C. Canchaya, V. Meylan, T. R. Klaenhammer, and R. Zink. 2003. Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium species and their direct application for species identification. Appl. Environ. Microbiol. 69:6908-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiechmann, I., and G. Grupe. 2005. Detection of Yersinia pestis DNA in two early medieval skeletal finds from Aschheim (Upper Bavaria, 6th century A.D.). Am. J. Phys. Anthropol. 126:48-55. [DOI] [PubMed] [Google Scholar]
- 45.Wilmoth, B., M. Chu, and T. Quan. 1996. Identification of Yersinia pestis by BBL crystal enteric/nonfermenter identification system. J. Clin. Microbiol. 34:2829-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wren, B. W. 2003. The yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 1:55-64. [DOI] [PubMed] [Google Scholar]
- 47.Wright, D. P., C. G. Knight, S. G. Parkar, D. L. Christie, and A. M. Roberton. 2000. Cloning of a mucin-desulfating sulfatase gene from Prevotella strain RS2 and its expression using a Bacteroides recombinant system. J. Bacteriol. 182:3002-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.