Abstract
The ability of Bacillus subtilis to form spores is a strategy for survival under unfavorable environmental conditions. It is equally crucial to break spore dormancy and return to vegetative growth at the appropriate time. Here we present data showing that the PrpE phosphatase is involved in the control of expression of genes coding for GerA receptors, which are necessary for l-alanine-induced spore germination. Moreover, PrpE is also involved in aspartic acid, glucose, fructose, and potassium (AGFK)-induced spore germination by controlling expression of genes coding for GerK receptors. In the absence of PrpE, the production of spores was essentially normal. However, l-alanine-induced spore germination and, to a lesser extent, the AGFK-induced pathway were abolished. In contrast, the germination pathway dependent on Ca2+-dipicolinate or dodecylamine remained intact. A protein phosphatase PrpE-green fluorescent protein fusion was localized to the prespore and to the dormant spore, consistent with a role in controlling expression of genes coding for GerA receptors. We propose that PrpE is an important element in a signal transduction pathway in Bacillus subtilis that controls the expression of genes coding for germination receptors.
Upon starvation for nutrients, cells of the gram-positive bacterium Bacillus subtilis differentiate into metabolically dormant spores that are well adapted to resist environmental damage (23, 26). Initiation of the sporulation process requires integration of information concerning the physiological state of the cell as well as the environmental conditions. This integration process is known to involve protein phosphorylation. Thus, at the level of sporulation initiation, information flows through a phosphorelay system that involves several histidine kinases and protein phosphatases (14, 32). All these proteins receive different signals from the interior and exterior of the cell. Processing of this information results in a phosphorylated form of the Spo0A protein, which acts as a major regulator of sporulation initiation (9, 32). Later, during asymmetrical cell division and formation of the prespore, the key role is played by the SpoIIA operon (37). This protein is necessary for successful division and continuation of the sporulation process by activation of the first forespore-specific sigma factor, the σF subunit of RNA polymerase (17). The resulting activation cascade of sigma subunits in the mother cell and forespore requires cross talk between these two compartments (11). Proteolysis is a key event in this process (16). Expression of germination receptors depends on σG, which becomes activated late in the prespore compartment (5, 8, 11). Modulation of expression of σG-dependent genes in the forespore is controlled by the SpoVT protein. Deletion of spoVT has variable effects on the expression of different genes, i.e., the level of the GerA receptor is elevated, while in contrast, that of SspB is reduced (2). Previous investigations regarding the mechanism of action of SpoVT suggest that it may require interaction with other, so far unknown proteins for proper action (7).
The mature spore is highly resistant to harsh, unfavorable conditions and can remain for long periods without loss of potential to return to vegetative growth (23, 34). Furthermore, the spore is able to monitor the environment continuously and to return to vegetative growth immediately when conditions become appropriate (21, 22, 31). This sensing ability and the consequent germination of spores are dependent on different pathways according to the conditions. Some pathways, for example, those triggered by exposure to l-alanine or an AGFK mixture (asparagine, glucose, fructose, potassium), depend on specific germination receptors encoded by three operons, gerA, gerB, and gerK (5, 6, 22, 38), although the proteins interacting with these receptors remain unknown (4). Similarly, mutants defective in these receptors that are able to sense d-alanine have been described (24). Expression of genes coding for germination receptors takes place only in the forespore and reaches a maximum about 4 to 6 h after initiation of sporulation (5, 8).
The gene encoding PrpE is located at 105 degrees of the B. subtilis genetic map between yjbO and yjbQ, transcribed in opposite directions. The location of the promoter driving expression of prpE is unknown. Previous studies of the PrpE protein showed the presence of motifs characteristic of PPP protein phosphatases and diadenosine-polyphosphate hydrolases. Indeed, purified PrpE displayed both activities (13). Analysis of the amino acid sequence of PrpE does not indicate any transmembrane region, so we conclude that it is a cytoplasmic protein. We now show that PrpE is implicated in the sporulation process; in particular, it is involved directly in germination of spores. Thus, we present evidence that the protein phosphatase PrpE is involved in controlling expression of the gerA and gerK operons and is part of a novel regulatory circuit, although any relationship between PrpE and σG and/or SpoVT remains unclear. In addition, the use of a PrpE-green fluorescent protein (GFP) fusion protein enabled us to detect this phosphatase in the forespore, with the fluorescent signal remaining detectable in the dormant spores.
MATERIALS AND METHODS
Construction of plasmids and strains.
The strains used in this study are listed in Table 1.
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype | Reference, source, or constructiona |
|---|---|---|
| 168 | trp | 1 |
| MM1325 | ΔprpE Spcr | 11 |
| BFA2853 | prpE::pMutin4 Eryr | Our collection |
| MM1453 | amyE::pSpac-prpE Catr | pKH18→168 |
| MM1502 | ΔprpE amyE::pSpac-prpE Catr Spcr | MM1325→MM1453 |
| MM1503 | prpE-gfp Catr | pKH21→168 |
| MO1615 | sigH::Kanr | From P. Stragier |
| 719 | trp2 pheA1 sigF::Kanr | From P. Stragier |
| RL1061 | sigE::Ermr | From R. Losick |
| 1446 | trp2 pheA1 sigG::Tetr | From P. Stragier |
| MM1504 | sigH::Kanr | MO1615→168 |
| MM1505 | trp2 pheA1 sigF::Kanr | 719→168 |
| MM1506 | trp2 pheA1 sigG::Tetr | 1446→168 |
| MM1601 | sigE::Ermr | RL1061→168 |
| MM1602 | sigH::KanrprpE-gfp Catr | MM1504→MM1503 |
| MM1507 | sigF::Kanr prpE-gfp Catr | MM1505→MM1503 |
| MM1508 | sigE::ErmrprpE-gfp Catr | MM1601→MM1503 |
| MM1509 | sigG::Tetr prpE-gfp Catr | MM1506→MM1503 |
| PS1245 | gerA-lacZ Catr | From P. Setlow |
| PS3441 | psspB-gerA MLSr | From P. Setlow |
| FB58 | psspB-gerB Spcr | From P. Setlow |
| PS3461 | psspB-gerK MLSr | From P. Setlow |
| MM1603 | gerA-lacZ Catr | PS1245→168 |
| MM1604 | psspB-gerA MLSr | PS3441→168 |
| MM1605 | gerA-lacZ ΔprpE Spcr Catr | MM1602→MM1325 |
| MM1606 | psspB-gerA amyE::pSpac-prpE MLSr Catr | MM1603→MM1453 |
| MM1607 | psspB-gerA ΔprpE MLSr Spcr | MM1603→MM1325 |
| MM1608 | psspB-gerB Spr | FB58→168 |
| MM1609 | psspB-gerK MLSr | PS3461→168 |
| MM1610 | psspB-gerB amyE::pSpac-prpE Spcr Catr | MM1608→MM1453 |
| MM1611 | psspB-gerB prpE::pMutin4 Eryr Spcr | BFA2853→MM1320 |
| MM1612 | psspB-gerK amyE::pSpac-prpE MLSr Catr | MM1609→MM1453 |
| MM1613 | psspB-gerK ΔprpE MLSr Spcr | MM1609→MM1325 |
| MM1615 | amyE::gerB-bgaB Catr | pKH26→168 |
| MM1616 | amyE::gerB-bgaB ΔprpE Catr Spcr | MM1615→MM1325 |
| MM1618 | amyE::gerK-bgaB Catr | pKH25→168 |
| MM1619 | amyE::gerK-bgaB ΔprpE Eryr Spcr | MM1618→MM1325 |
| MM1621 | sspB-lacZ Eryr | pKH24→168 |
| MM1622 | sspB-lacZ ΔprpE Eryr Spcr | MM1621→MM1325 |
| MM1624b | PprpE-gfp Eryr | pKH27→168 |
Strains were constructed by transformation of plasmid or chromosomal DNA from the strain to the left of the arrow into the strain to the right of the arrow.
Strain containing transcription fusion for gfp from the prpE promoter.
In order to construct a strain in which expression of prpE is under the control of an isopropyl-β-d-thiogalactoside (IPTG)-inducible promoter, prpE was PCR amplified from the chromosome with primers 18up and 18down, with addition of a SalI restriction site, and cloned into pDG66. Next, DNA of pKH18 was used for transformation of MM1325, selecting for chloramphenicol resistance, to generate strain MM1502. To express prpE-gfp at the normal prpE locus, a three-step cloning procedure was used. First, a distal fragment of prpE was PCR amplified from the chromosome with primers 19up-1 and 19down-1, with addition of BamHI and KpnI sites. The distal part of yjbO was also PCR amplified from chromosomal DNA with primers 19up-2 and 19down-2, with addition of EcoRI and KpnI sites. The PCR products were cloned together into the pUC19 cloning vector, generating pKH19. Second, the cat gene was PCR amplified (primers cat-up and cat-down), with addition of a BcuI restriction site from the pDL plasmid, and inserted into pMutinGFP+, generating pKH20. Third, the fragment containing the gfp+ and cat genes was PCR amplified (primers 20up and 20down) from pKH20 and cloned into the KpnI site in pKH19, generating pKH21. The DNA of pKH21, after linearization, was used for transformation of wild-type B. subtilis (strain 168), selecting for chloramphenicol resistance, to generate strain MM1503. The transcription fusion, expressing gfp from the prpE promoter (PprpE-gfp), was made by cloning a PCR fragment starting immediately downstream of the stop codon of prpE (primers 2557 and 2821) into Eco52I and KpnI sites in the pMutinGFP+ plasmid, generating pKH27, in which gfp depending on its own ribosome binding site is expressed from the prpE promoter. Next, the resulting plasmid was used for transformation of wild-type B. subtilis (strain 168), selecting for erythromycin resistance, to generate strain MM1624. Expression of GFP was confirmed by Western blotting with monoclonal antibodies against GFP (Roche).
For construction of strains expressing a thermostable variant of LacZ (i.e., BgaB), gerB-bgaB, or gerK-bgaB, a similar procedure was used. The amplified products from chromosomal fragments (primers 26up and 26down for gerB; primers 25up and 25down for gerK) were ligated into the pDL cloning vector, generating plasmids pKH26 and pKH25. DNA of such plasmids was used for transformation of 168, selecting for chloramphenicol resistance, to generate strains MM1615 (gerB-bgaB) and MM1618 (gerK-bgaB).
To obtain an IPTG-inducible copy of prpE at the α-amylase locus, prpE was PCR amplified from chromosomal DNA with addition of SalI sites and cloned downstream of the Pspac promoter into the pDR66 plasmid. The resulting plasmid was called pKH18.
The fragment of B. subtilis DNA containing the distal part of prpE and the proximal part of the yjbO gene was PCR amplified and cloned into pUC19 (35). The resulting plasmid was named pKH19. In parallel, the cat gene was PCR amplified from the pDL vector (36) and cloned into the BcuI site of the pMutinGFP+ plasmid (15), and the resulting plasmid was named pKH20. A fragment of pKH20 containing the gfp+ gene and cat was PCR amplified and cloned into the KpnI site of pKH19. The resulting plasmid, pKH21, encodes a C-terminal fusion of PrpE and GFP+ and in addition has cat as a marker, followed by the yjbO gene.
The sequences of all primers used are available on request.
Growth conditions.
B. subtilis strains were grown in Lurina-Bertani (LB), nutrient broth (NB), or Sterlini-Mandelstam (32) medium supplemented with antibiotics where appropriate.
Microscopy.
Strain MM1503 (prpE-gfp) was grown in Sterlini-Mandelstam sporulation medium for 24 h at 37°C. Five microliters of the culture was placed on a microscope slide and covered with a coverslip that had been treated for approximately 60 s with poly-l-lysine (Sigma). Cells were observed with a Zeiss Axioplan fluorescence microscope. Pictures were taken with the digital camera connected to the microscope, and final adjustments were made with Corel Photo-Paint software.
Sporulation efficiency test.
Bacteria were inoculated into sporulation medium (33) and grown with shaking at 37°C to an optical density at 570 nm (OD570) of 1. Cells were harvested by centrifugation and resuspended in the starvation sporulation medium (33), and incubation was continued for 24 h with shaking at 37°C. Then serial dilutions of the culture in 0.85% NaCl solution were performed, and bacteria were plated on NB medium. Solutions were heated to 80°C for 20 min in order to kill vegetative cells, and subsequently serial dilutions were plated. All plates were incubated overnight at 37°C. Colonies on plates were counted, and numbers of bacteria with spores (unheated) or spores only (heated) were calculated. The ratio between the second and first counts was called the sporulation efficiency. To simplify comparisons, the sporulation efficiency of the wild-type strain (under which conditions a nongerminating spore is counted as not produced) was taken as 100%.
Spore purification.
Forty-eight-hour cultures in nutrient broth were pelleted by centrifugation (10,000 × g, 10 min) and washed three times with 1/4 volume of cold water. The pellet was resuspended in 1/5 of the initial volume of cold MQ water and incubated overnight at 4°C. On subsequent days the suspension was centrifuged (20,000 × g, 20 min, 4°C). The pellet was resuspended in fresh cold MQ water. This procedure was repeated for 5 to 10 days. Purified spores were kept in water suspension at 4°C in the dark. Once per week the spores were centrifuged and suspended in fresh water to avoid spontaneous germination.
Protein isolation and Western blot analysis.
Bacteria were cultivated in nutrient broth medium with shaking at 37°C. At the indicated times, 5 OD units were centrifuged (14,000 × g, 5 min, 4°C). Bacterial pellets were suspended in 0.2 ml of lysis buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 10 mM β-mercaptoethanol). Then lysozyme was added to a final concentration of 100 μg/ml, and the suspension was incubated for 30 min on ice. Lysis was performed by adding 50 μl of a glass bead suspension with shaking in a Mini-Beadbeater for 30 s at maximal speed. Then the sample was chilled on ice for 2 min, and the breaking procedure was repeated eight times. The extract was centrifuged (15,000 rpm for 10 min at 4°C) in order to remove insoluble material, together with glass beads. Finally, protein concentrations in extracts were determined by the Bradford method. Proteins were isolated from the spores as described previously (3). Briefly, decoated spores were lyophilized, dry ruptured in a Mini-Beadbeater (Biospect Products), and pulsed with glass beads 18 times at maximal speed for 30 s each time, with a 30-s cooling period between pulses. The resulting dry powder was extracted with 200 μl of 50 mM HEPES (pH 7.5)-5 mM EDTA-1 mM phenylmethylsulfonyl fluoride for 30 min on ice, followed by a 5-min centrifugation at top speed in a microcentrifuge in the cold. The supernatant fluid (soluble fraction) was mixed with an equal volume of 2× sodium dodecyl sulfate (SDS) loading buffer and boiled for 3 min. The pellet (insoluble fraction) was suspended in 200 μl of 1× SDS loading buffer, incubated at 70°C for 15 min with occasional vortexing, boiled for 5 min, and spun in a microcentrifuge at room temperature. Samples, 20 μg per lane, were mixed with loading buffer and boiled for 3 min before being loaded onto an SDS-polyacrylamide gel. After electrophoresis, proteins were transferred to a nitrocellulose membrane and blocked by a 1-h incubation in 3% milk in Tris-buffered saline (TBS). Mouse monoclonal antibodies against GFP (Roche) were added at a 1:1,000 dilution and incubated for 1 h. After three washes in TBS (50 mM Tris-HCl [pH 7.5]-500 mM NaCl), alkaline phosphatase-conjugated goat secondary antibodies against mouse immunoglobulin G were added at a 1:20,000 dilution (Sigma) and incubated for 1 h, followed by three washes in TBS. The reaction was developed with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate according to the manufacturer's instructions.
Spore germination.
Spore germination measurements in the presence of l-alanine or AGFK solution were performed as follows. Spores were heat activated at 80°C for 10 min and diluted to an OD600 of 1 in 10 mM l-alanine and 10 mM Tris-HCl, pH 7.5 (for l-alanine-induced spore germination) or in 10 mM Tris-HCl at pH 7.5 with 3.3 mM l-asparagine, 5.6 mM d-glucose, 5.6 mM d-fructose, and 10 mM KCl (for AGFK-induced spore germination). Germination was then monitored by following the loss of absorbance of spore suspensions at 600 nm.
For analysis of spore germination in the presence of Ca2+-dipicolinate (DPA), spore suspensions (OD600, 1) were incubated for 1 h at 24°C in 60 mM CaCl2-DPA chelate in 20 mM Tris-HCl (pH 8.6), aliquots were diluted and spotted onto LB agar plates, and colonies were counted after incubation for 24 h at 37°C. In this assay, spores can germinate either with Ca2+-DPA during the 1-h incubation or with nutrients on the LB plates. Consequently, this assay is most informative when spores with poorly functional or nonfunctional nutrient receptors are tested.
For analysis of dodecylamine (DCA)-induced germination, spores were germinated without prior heat activation, since this is not required for the DCA method (29). Spore suspensions (OD600, 1) were incubated in the presence of 1 mM DCA in 20 mM phosphate buffer at pH 7.4 for 1 h. Next, serial dilutions were prepared, plated on LB, and incubated overnight at 37°C. The number of colonies and the germination efficiency were calculated.
Wet heat test.
Purified spores were resuspended in water to an OD570 of about 0.1. One hundred microliters of a spore suspension was placed in two PCR tubes; one was kept on ice, and the other was heated at 90°C for 10 min. Similar results were obtained at 100°C. Spores were then placed on ice and serial dilutions of spores prepared. Dilutions were plated on NB and incubated overnight at 37°C. The number of colonies and the ratio between heated and unheated samples were calculated.
Dry heat test and desiccation.
Purified spores were suspended in water to an OD570 of 0.1. Aliquots of suspensions of the spores were vacuum dried in a Speed-vac (Savant) for 60 min. The Eppendorf tubes were incubated at 80°C, 90°C, or 100°C for 10 min, tubes were cooled to room temperature, spores were resuspended in an initial volume of water, and serial dilutions were prepared. Plating and colony counting were performed as described above. For desiccation, test spores were rehydrated directly after a drying step. These experiments were repeated four times with different spore preparations, with a variation of ±6%. Repeated cycles of desiccation and rehydration slightly decreased the viability of spores, i.e., after the third cycle of desiccation-rehydration, we observed an average of 40% ± 5% viable spores.
Organic solvent resistance test.
Tests with water-miscible solvents, immiscible solvents, and organic acid were performed as follows. Purified spores were suspended in water to an OD570 of about 0.1; then 50 μl of solvent or acid was added to 0.45 ml of spore suspension, mixed vigorously with a Vortex mixer, and left at room temperature for 10 min. Next, serial dilutions were prepared. The percentage of survivors was calculated by comparing the number of colonies after overnight incubation at 37°C on NB plates.
Lysozyme test.
A 450-μl solution of lysozyme (250 μg/ml) was freshly prepared (water was used as a control), mixed with 50 μl of a spore suspension (OD570, about 0.1), and incubated for 10 min at 37°C; then serial dilutions were prepared and samples plated. The percentage of survivors was calculated by comparing the number of colonies after overnight incubation at 37°C on NB plates.
UV resistance test.
A 50-μl suspension of purified spores (OD570, 0.1) was distributed in a plastic petri dish and irradiated 60 s from 30 cm with the UV lamp (wavelength, 254 nm; 30 W), which had been switched on at least 10 min prior to use. Serial dilutions were performed, spores were plated, and the percentage of survivors was calculated as described above. Under UV resistance test conditions, about 10% of wild-type spores survived the test.
RT-PCR experiments.
Fifty nanograms of total RNA, isolated from the cells using the Total RNA kit (A & A Biotechnology), 3 h after resuspension of the cells in Sterlini-Mandelstam sporulation medium, was used. Prior to the real-time reverse transcription PCR (RT-PCR), the RNA was digested with RNase-free DNase I (Roche), and then the enzyme was thermally inactivated. Experiments were performed using the Brilliant SYBR Green QRT-PCR Master Mix kit (Stratagene) as recommended by the manufacturer. Products of reactions were analyzed for homogeneity by performing a melting curve after each PCR. The probes were designed as fragments (100 to 150 bp) complementary to the internal part of the respective open reading frames.
RESULTS
Localization of PrpE.
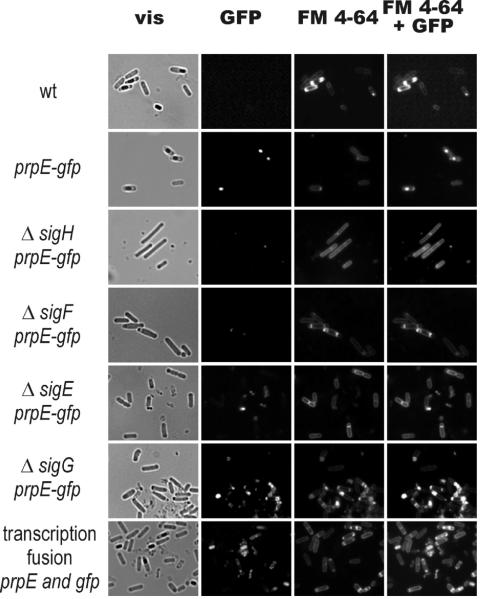
Bacteria with the prpE gene deleted displayed no detectable phenotype during either the exponential or the stationary phase in liquid media. Their responses to different stresses (salt, ethanol, temperature) also remained unchanged (data not shown). The amino acid sequence of PrpE lacks any signal sequence, so we hypothesized that this protein is localized in the cytoplasm. To verify this hypothesis, a gene fusion encoding PrpE with C-terminal GFP was constructed. The strain harboring the prpE-gfp fusion gene under the control of its own promoter was examined for GFP fluorescence. During the exponential-growth and early-stationary phases in LB medium, fluorescence was undetectable, but in stationary phase a GFP signal was detected by microscopy in forespores and mature spores (Fig. 1). GFP itself, when overproduced in B. subtilis cells as a control, was dispersed through the cells and was not detected in the spores (data not shown). The data suggested that localization of the PrpE phosphatase is mainly restricted to the forespore and that this enzyme is one of the components of the mature spore. The presence of the fusion protein in spores was confirmed by Western blot analysis using an antibody against GFP (data not shown).
FIG. 1.
Subcellular localization of PrpE-GFP. Cells of wild-type strain 168 (wt) and of strain 168 carrying the prpE-gfp fusion or a transcription fusion between prpE and gfp, together with the deletion of different sigma subunits, were induced to sporulate by growth in NB medium for 22 h at 37°C and were prepared for microscopy as described in Materials and Methods. Cells producing the PrpE-GFP fusion protein were observed in identical fields in visible light (vis), for fluorescence of GFP, for fluorescence of FM 4-64 dye, and for overlay fluorescence GFP and FM 4-64 dye. The localization of the fusion protein indicates that the PrpE phosphatase has reached a concentration sufficient for detection by fluorescence microscopy in the spores but not in the mother cell compartment. Spores produced by 168 analyzed under identical conditions showed extremely weak fluorescence at wavelengths used for GFP detection. However, this weak fluorescence is below the limit of detection of the camera installed in the microscope and thus is not visible.
Expression of prpE.
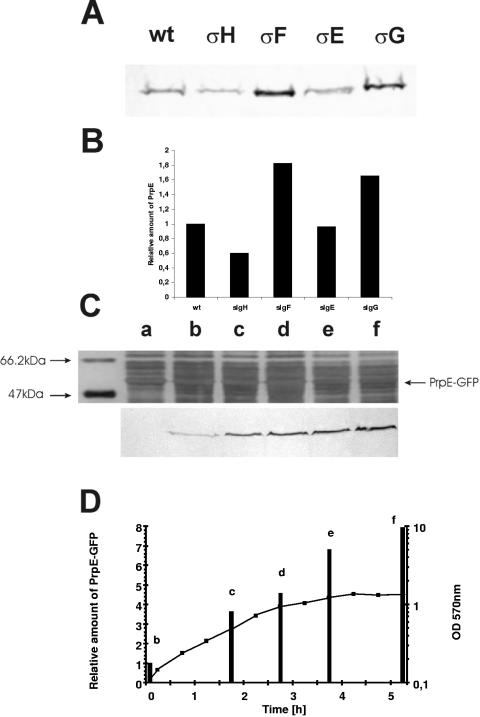
On the basis of microscope observations we wondered whether prpE is expressed during sporulation. In order to test this possibility, we constructed a series of deletion mutants in genes coding for different sigma subunits (σH, σF, σE, and σG, corresponding to strains MM1602, MM1507, MM1508, and MM1509, respectively) involved in the sporulation process. These mutants were analyzed for fluorescence of the PrpE-GFP fusion protein in stationary-phase cultures. No GFP fluorescence was observed in strains lacking the σH or σF subunit. A weak GFP signal was detected in the strain with σE deleted, and a stronger signal was detected in the σG mutant. To confirm these microscope observations, we performed Western blot analysis with monoclonal antibodies against GFP. However, in all the strains, the GFP fusion was found in cells from stationary phase, as shown in Fig. 2A, with relatively small differences (Fig. 2B) in amounts of PrpE between the mutants and the wild type. Thus, the lack of fluorescence in the ΔσF strain, for example, may be explained by the inability of such cells to complete asymmetric division and therefore to concentrate the fusion protein in the forespore compartment at a level sufficient to give a fluorescent focus. On the other hand, during exponential growth of the wild type (Fig. 2C) and all the mutants (data not shown), we detected small amounts of the PrpE-GFP fusion protein, which increased significantly as cells entered stationary phase (Fig. 2D). This suggests that expression of prpE normally depends on the housekeeping sigma factor, σA.
FIG. 2.
Expression of the prpE-gfp fusion gene. (A) Western blot analysis, with monoclonal antibodies against GFP (Bio-Rad), of the expression of prpE-gfp in the wild type (wt) and in strains with different sigma subunits deleted. Proteins were isolated from bacteria grown in NB to stationary phase (16 to 18 h) as described in Materials and Methods. (B) Bar graph of corresponding amounts of PrpE protein in the wild type and in sigma subunit deletion mutants. (C) Silver staining of proteins isolated from strain 168 and Western blot analysis of expression of prpE-gfp during the growth phase. Lane a, 168; lanes b to f, samples taken during the growth phase of strain MM1503 (prpE-gfp), analyzed by Western blotting with the anti-GFP antibody. The cells for protein extraction were taken with respect to OD, and identical amounts of protein (20 μg) were loaded in each lane. The antibody detected only a single band in the gel profile at a position corresponding to the expected molecular mass, about 54 kDa for the fusion. Note that lane a (strain 168 expressing no GFP fusion) gives no signal. (D) Growth curve of strain MM1503 (prpE-gfp) and relative amounts of PrpE-GFP proteins, indicated by bars.
When spores were induced for germination, the fluorescence signal remained detectable until the outgrowth phase, and finally the vegetative cells were found to lack a detectable signal when examined by fluorescence microscopy, although the fusion protein was still detectable by Western blotting (data not shown). These results could be interpreted to indicate that PrpE is specifically delivered to the forespore from the mother cell or is expressed in both compartments, with the possibility that it is rapidly degraded in the mother cell. In order to investigate the hypothesis that expression of the PrpE phosphatase may be enhanced in the forespore, we built a transcriptional fusion for gfp, expressing GFP from the prpE promoter (see Materials and Methods). Microscope observations of the strain showed that GFP was present in the forespore, apparently confirming that the PrpE promoter was active in the forespore compartment (Fig. 1). The presence of this GFP in the spores was also confirmed by Western blot analysis of proteins isolated from purified spores (data not shown).
Sporulation efficiency.
Localization of the PrpE-GFP fusion protein in the spore suggested that it may play a role in spore formation. To verify this hypothesis, we first analyzed the sporulation efficiency of a strain with the prpE gene deleted (MM1325) in comparison with the wild-type strain (168). The results showed that bacteria lacking PrpE grew normally (the growth rate and yield are indistinguishable from those of the wild-type strain) but produced a low number of colonies in comparison to the wild type after spores harvested from Sterlini-Mandelstam sporulation medium were plated onto LB agar (Table 2). Similar results were obtained on NB plates (data not shown). On the other hand, the sporulation efficiency of the strain carrying the prpE-gfp fusion replacing wild-type prpE (MM1503) remained almost unchanged, indicating that PrpE in the fusion was active and that probably the localization of PrpE-GFP in the forespore corresponds to that of the native PrpE.
TABLE 2.
Colony formation efficiency of prpE mutantsa
| Strain | IPTG concn (mM)
|
No. of spores | No. of colonies | Efficiency of colony formation (%) | |
|---|---|---|---|---|---|
| In medium | On plates | ||||
| 168 (wild-type) | 1.9 × 109 | 1.8 × 109 | 95 | ||
| MM1325 (ΔprpE) | 2.8 × 109 | 1.7 × 108 | 6 | ||
| MM1503 (prpE-gfp) | 5.3 × 107 | 4.7 × 107 | 89 | ||
| MM1502 (ΔprpE amyE::pSpac-prpE) | 1.2 × 109 | 4.0 × 108 | 33 | ||
| 0.5 | 1.3 × 109 | 5.5 × 108 | 42 | ||
| 0.5 | 1.6 × 109 | 1.3 × 109 | 80 | ||
| 0.5 | 0.5 | 8.0 × 109 | 5.8 × 109 | 73 | |
| 0.5 | 1 | 1.2 × 109 | 1.1 × 109 | 92 | |
Cells were induced to sporulate in Sterlini-Mandelstam sporulation medium, and spore suspensions were prepared as described in Materials and Methods. Spores were counted in a Thoma cell and plated on LB medium. After a 5-day incubation at 37°C, colonies were counted, and the efficiency of colony formation was calculated.
In order to determine whether PrpE is involved in germination rather than the production of spores, we analyzed directly the numbers of spores produced by the wild-type strain and the ΔprpE mutant by counting spores in a Thoma cell. The numbers of spores produced by the two strains grown in Sterlini-Mandelstam sporulation medium were essentially identical. However, the number of colonies obtained after plating of the spores on LB (or NB) was drastically reduced for the mutant strain (Fig. 3). This clearly indicated that prpE is involved in germination rather than spore production. In order to verify a germination defect, we employed the overlay germination test with 2,3,5-triphenyltetrazolinum chloride (Tzm) dye, which distinguishes colonies with germinating spores from those with nongerminating spores (10). The spores produced in bacterial colonies on agar plates were induced to germinate by the addition of fresh medium containing the Tzm dye. The results confirmed that spores lacking the PrpE phosphatase are defective in germination/early outgrowth (data not shown).
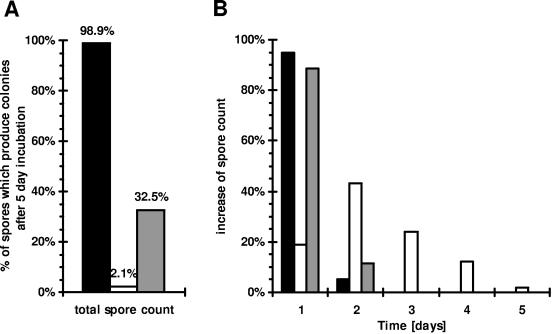
FIG. 3.
Formation of colonies from spores on the plates over time. Spores prepared from cells of strain 168 (solid bars), MM1325 (ΔprpE) (open bars), and MM1502 (ΔprpE amyE::pspac-prpE) (shaded bars) grown in NB medium were plated onto LB agar. The plates for strain MM1502 contained 0.5 mM IPTG. Plates were incubated at 37°C, and new colonies were counted each day. (A) Total spore count, total number of colonies arising on the plates as a percentage of the number of plated spores. (B) In order to show the distribution of colonies arising over time, the total spore count for each strain was taken as 100%.
We also analyzed over time the ability of spores produced by strain MM1325 (with prpE deleted) to germinate on LB plates in comparison to that of the wild type. We found that the majority of wild-type spores germinated rapidly in comparison to mutant spores (Fig. 3). After 24 h, about 94% of spores of the wild-type strain 168 produced colonies on the plates, but only 0.4% of MM1325 spores were able to do this. All colonies appeared within 2 days on plates with wild-type spores, while MM1325 spores continued to give rise to new colonies over 5 days. The delayed appearance of colonies on plates when spores produced by strain MM1325 were plated (Fig. 3B) indicates that the germination process was affected. Nevertheless, while 99% of wild-type spores finally produced colonies, only 2% of ΔprpE mutants produced colonies. In summary, all these results confirmed that PrpE is essential for germination.
Germination analysis.
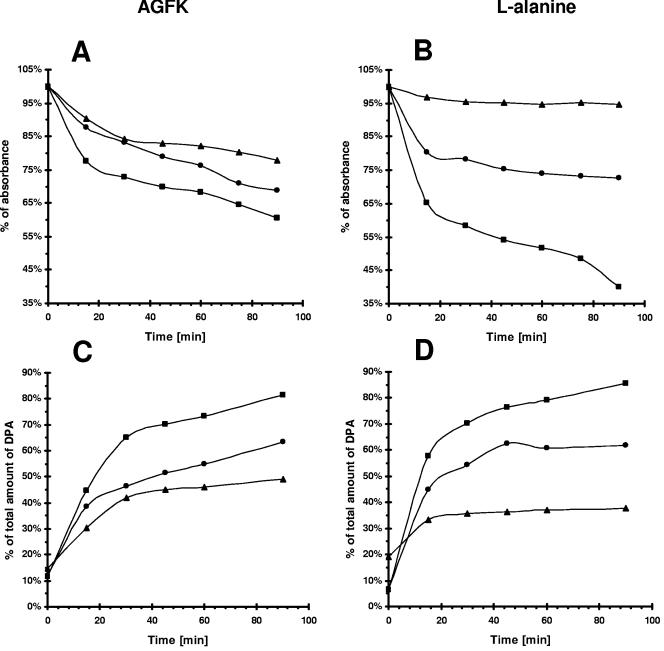
In the course of our investigation of germination of spores produced by strain MM1325 (ΔprpE) in Sterlini-Mandelstam medium, we observed that mutant spores produced about 20% more colonies on LB plates than in NB (data not shown). The major difference between LB and NB media is the presence of yeast extract in LB versus beef extract in NB. Yeast extract contains approximately 17.5% sugars versus 0.2% in beef extract. Consequently, we supposed that some spores produced by MM1325 might fail to germinate unless particular sugars or other signaling molecules were present in the medium. To explore this possibility, we analyzed the germination of spores suspended in an AGFK solution (see Material and Methods), again monitoring germination by measuring the A600 of spore suspensions. The mutant spores germinated much more slowly than the wild type (Fig. 4A). Moreover, the spores were gray under the microscope after 90 min of incubation in AFGK solution, indicating that spore hydration was not complete (data not shown). The germination of spores produced by the strain expressing the PrpE-GFP fusion protein was normal (data not shown). Consequently, we conclude that since the fusion protein restored wild-type germination, it must also be correctly localized within the sporulation cells.
FIG. 4.
Germination of spore suspensions in AGFK and l-alanine solutions. Spores prepared from cells of 168 (squares), MM1325 (ΔprpE) (triangles), and MM1502 (ΔprpE amyE::pspac-prpE) (circles) grown in NB medium were heat activated and subsequently incubated in 10 mM Tris-HCl (pH 7.5) with 3.3 mM l-asparaginate, 5.6 mM d-glucose, 5.6 mM d-fructose, and 10 mM KCl (A and C) or with 10 mM l-alanine (B and D). IPTG added to the germination solution alone has no effect on germination. (A and B) Germination was followed by measuring the A600 of the spore suspension. (C and D) Release of dipicolinic acid was followed by measuring the A280 of the spore suspension.
Normal spores can also be induced to germinate by suspension in 10 mM l-alanine (see Materials and Methods). However, spores produced by MM1325 (ΔprpE) were unable to respond to l-alanine and did not germinate (Fig. 4B). Microscopic examination of mutant spores after 90 min of incubation in l-alanine showed that the spores were still refractile (data not shown). However, when we tested the spores produced by strain MM1502 (ΔprpE amyE::pSpac-prpE) in the presence of 0.5 mM IPTG in sporulation medium, germination was now observed, confirming that PrpE is necessary for l-alanine-induced germination of B. subtilis spores (Fig. 4B). Measurements of DPA release from the spores produced by strain MM1325 (ΔprpE), induced for germination by incubation in an AGFK or l-alanine solution, showed a twofold reduction (Fig. 4C and D). This indicates that the germination process is affected in this strain. Partial release of DPA may suggest that germination is inhibited at this or a later step. These results indicated, therefore, that PrpE is a necessary element in a GerA signal transduction pathway, since GerA receptors are responsible for triggering germination in response to l-alanine (4).
However, since germination in AGFK solution, which involves GerB and GerK receptors (25), was also partially reduced in the ΔprpE strain, PrpE may also be involved in additional germination pathways. To test the possibility that the absence of PrpE affected receptor specificity in some way, we checked the ability of spores produced by the prpE deletion strain to germinate in the presence of d-alanine. Previously, studies have shown that mutations in gerBA or gerBB result in a germination receptor that is now able to respond to the presence of d-alanine by an unknown mechanism (24). We incubated spores produced by the wild-type and the ΔprpE strain in the presence of d-alanine (with or without glucose). However, both types of spores failed to germinate under such conditions (data not shown).
It has been reported that, in addition to nutrient-induced germination, a 1:1 chelate of Ca2+ and DPA can also break spore dormancy in B. subtilis, bypassing the known receptors (25, 27). Therefore, we tested whether Ca2+-DPA-induced germination was affected by the lack of PrpE phosphatase, using the procedure described previously by Igarashi and coworkers (12) (see also Materials and Methods). We did not observe any significant differences in germination of the wild-type or mutant spores (data not shown). This indicates that PrpE acts upstream or independently of this pathway (31).
Spore dormancy can also be broken by cationic surfactants such as DCA (28, 29). Previous detailed studies of DCA-induced germination showed that this pathway is distinct from the Ca2+-DPA germination pathway (31), is independent of nutrients, and acts downstream of the germination receptors. We analyzed DCA-induced germination of spores produced by the prpE deletion strain. As found with Ca2+-DPA, DCA-induced germination of spores lacking PrpE protein is indistinguishable from that of wild-type spores (data not shown). This indicates that PrpE also acts upstream of this pathway.
PrpE regulates expression of the gerA and gerK operons.
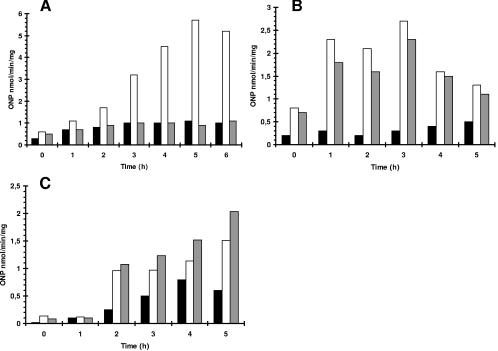
A possible explanation for a role for PrpE in germination might be that the phosphatase is involved in some way in the control of expression of some ger genes, the translation of these receptors, or the activities of the GerA and GerK proteins. In order to test this hypothesis, we prepared strains carrying a gerA-lacZ transcriptional fusion, expressed from the native promoter, together with deletion of prpE or overproduction of PrpE. Sporulation was induced in such strains by growth in Sterlini-Mandelstam sporulation medium, and β-galactosidase levels were measured. The results showed that deletion of prpE blocked β-galactosidase induction after sporulation initiation (Fig. 5A). This finding is in agreement with previous data showing that spores produced by the prpE deletion strain were unable to germinate in the presence of l-alanine, since GerA receptors are required for this pathway. Other investigations were performed with strains carrying gerB-lacZ and gerK-lacZ transcriptional fusions, using the thermostable form of β-galactosidase, BgaB from Bacillus stearothermophilus (36). With the gerB-lacZ and gerK-lacZ fusions, β-galactosidase levels were not affected in a prpE deletion strain (Fig. 5B and C). On the other hand, the expression of gerK was reduced by an elevated level of PrpE (Table 3). The results of this analysis clearly showed that PrpE is required in some way for the transcription of both the gerA and gerK genes.
FIG. 5.
Expression of genes coding for germination receptors in strains with prpE deleted, detected by using a β-galactosidase reporter. (A) Solid bars, 168; open bars, MM1604 (gerA-lacZ); shaded bars, MM1607 (gerA-lacZ ΔprpE). (B) Solid bars, 168; open bars, MM1615 (gerB-bgaB); shaded bars, MM1616 (gerB-bgaB ΔprpE). (C) Solid bars, 168; open bars, MM1618 (gerK-bgaB); shaded bars, MM1619 (gerK-bgaB ΔprpE). Bacteria were grown in Sterlini-Mandelstam medium at 37°C with shaking. At time zero, sporulation was initiated by replacing Sterlini-Mandelstam growth medium with sporulation medium. The bgaB gene is functionally equivalent to lacZ. ONP, o-nitrophenyl.
TABLE 3.
Effect of overexpression or deletion of prpE in cells overproducing Ger receptorsa
| Strain | Relative level of expressionb of:
|
Efficiency of colony formation (%)c | |||
|---|---|---|---|---|---|
| prpE | gerA | gerB | gerK | ||
| 168 (wild type) | N | N | N | N | 100 |
| MM1502 (ΔprpE amyE::pSpac-prpE) | O | N | N | N | 162 |
| MM1325 (ΔprpE) | D | N | N | N | 0.7 |
| BFA2853 (prpE::pMutin4) | I | N | N | N | 0.9 |
| MM1604 (psspB-gerA) | N | O | N | N | 0.9 |
| MM1606 (psspB-gerA amyE::pSpac-prpE) | O | O | N | N | 57 |
| MM1607 (psspB-gerA ΔprpE) | D | O | N | N | 35 |
| MM1608 (psspB-gerB) | N | N | O | N | 49 |
| MM1610 (psspB-gerB amyE::pSpac-prpE) | O | N | O | N | 66 |
| MM1611 (psspB-gerB prpE::pMutin4) | I | N | O | N | 55 |
| MM1609 (psspB-gerK) | N | N | N | O | 0.2 |
| MM1612 (psspB-gerK amyE::pSpac-prpE) | O | N | N | O | 43 |
| MM1613 (psspB-gerK ΔprpE) | D | N | N | O | 0.2 |
Bacteria were grown in nutrient broth for 24 h at 37°C. OD575 was measured; samples were heated at 80°C for 1 h and plated on NB plates. After overnight incubation at 37°C, colonies corresponding to viable spores were counted. IPTG at 1 mM was added for strains carrying pSpac fusions.
N, physiological level of expression; O, overproduction; D, deletion; I, inactivation.
The number of colonies arising from strain 168 was taken as 100% with respect to the OD.
Changes in the level of PrpE modify the effect of overproduction of GerA and GerK receptors.
Earlier investigations by Cabrera-Martinez and coworkers showed that overproduction of the GerA or GerK receptor, placed under the control of a PsspB promoter controlling production of a small acidic protein during sporulation (18), blocked the sporulation process; however, the molecular basis of this sporulation defect remains unknown (4). Whatever the mechanism of sporulation inhibition, we anticipated that if PrpE is implicated in expression of germination receptors, then changes in the level of prpE might suppress this sporulation defect. Surprisingly, both overproduction of PrpE and deletion of prpE in the strain overproducing GerA receptors (MM1606 and MM1607, respectively) showed dramatically increased production of viable spores on NB in comparison with the parental strains MM1604 (psspB-gerA) and MM1502 (ΔprppE amyE::pspac-prpE) (Table 3). Microscopic observation confirmed that both strains MM1606 and MM1607 produce many spores. Indeed, overproduction of PrpE largely restored sporulation when cells were grown on NB plates, to 57% of the wild-type level.
One may speculate that the production of high levels of GerA receptors somehow interferes with the function of GerB and GerK receptors or expression of their genes. In fact, deletion of prpE also largely suppressed the effect of GerA overproduction, restoring sporulation to 35% of the wild-type level. Similar results were observed when GerK receptors were overproduced together with prpE overexpression, resulting in restoration of spore production (Table 3). However, the results of overproduction of GerK combined with ΔprpE were more complicated; the number of viable spores produced remained very low with such a strain, i.e., deletion of prpE in this case did not “rescue” the production of spores. Since we showed that PrpE is required for normal expression of gerA, the deletion of prpE from the receptor-overproducing strain presumably counters this effect by reducing the level of the receptor. In the case of GerA, this reduction is sufficient to alleviate any effect on sporulation.
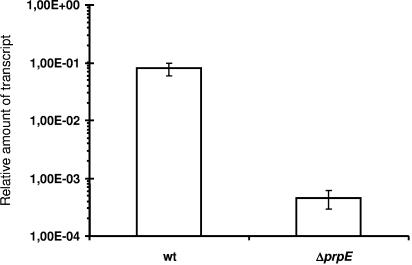
Notably, in order to fully explain the results of sporulation in cells overproducing gerA and gerK by changing the level of PrpE, it was necessary to predict that the sspB promoter should also be subject to control by prpE. This was tested by using quantitative RT-PCR, and the results demonstrated clearly that the expression of this promoter also depends on the presence of normal levels of PrpE (Fig. 6).
FIG. 6.
Expression of the sspB gene, coding for the small acid-soluble protein, in the wild-type strain (wt) and a strain in which prpE was deleted. Bacteria were grown in Sterlini-Mandelstam medium at 37°C with shaking. At time zero, sporulation was initiated by replacing Sterlini-Mandelstam growth medium with sporulation medium. Total RNA was isolated after 3 h after the change of medium. The quantitative RT-PCRs were standardized relative to sigA mRNA.
Spore properties.
In order to obtain more information about a possible role for PrpE in the formation of spore protective layers, the resistance properties of spores produced from the deletion mutant were analyzed in several tests. We found that spores produced by a strain lacking prpE were slightly more resistant to wet heat, ethanol, and trichloroacetic acid but that their resistance to UV irradiation, chloroform, and lysozyme was comparable to that of wild-type spores. However, we observed a significant difference in the resistance of spores to drying between the wild type and the prpE deletion strain. The survival of ΔprpE spores was reduced by about 50% in the drying test (Table 4). The results suggest that the PrpE phosphatase is involved in the correct maturation of spores, since it decreases susceptibility to extreme desiccation.
TABLE 4.
Spore resistance to desiccationa
| Strain | % Survival of spores |
|---|---|
| 168 (wild type) | 100 |
| MM1325 (ΔprpE) | 55 |
| MM1502 (ΔprpE amyE::pSpac-prpE) | 98.5 |
A spore suspension in water was dried under a vacuum for 60 min. The pellet was resuspended in the original volume of water, and serial dilutions were made. Control spore suspensions were kept in water for 60 min at room temperature. IPTG was present at a 1 mM concentration during spore formation by strain MM1502.
DISCUSSION
The results obtained in this study suggest that under certain conditions, protein phosphorylation plays an important role in a mechanism that normally leads to a break in spore dormancy and a return to vegetative growth. The protein phosphatase PrpE was present at low levels in cells throughout the growth phase in sporulation medium, but the concentration, as detected by fluorescence of the PrpE-GFP fusion or even GFP alone (expressed from the prpE promoter), appeared to increase in the forespore compartment during sporulation, and PrpE was still detectable in dormant spores. After germination and return to vegetative growth, the amount of PrpE protein in the cells decreased, as revealed by fluorescence of the GFP-PrpE fusion. This may indicate that synthesis is switched off or reduced at this point, as was also apparent in early-log-phase cultures probed by Western blotting (Fig. 2C and D). In vegetative cells, prpE is likely expressed from a σA promoter; however, the sigma factor responsible for prpE expression in the forespore remains unclear.
Environmental signals that can break dormancy are presumably normally received through the Ger receptors, triggering spore germination. Thus, GerA receptors are responsible for sensing the presence of l-alanine, while GerB, together with GerK, is responsible for sensing the presence of sugars and amino acids (AGFK solution). We initially found that the prpE deletion strain appeared to show a low sporulation efficiency, as determined by counting of colonies after plating of spores on LB or NB. However, detailed studies revealed that the numbers of spores produced by the wild type and the mutant strain were in fact identical, while germination of the mutant spores was impaired. This defect was reversed by expressing prpE from an IPTG-inducible promoter (pSpac), confirming the specific requirement for PrpE. The spores produced by the strain lacking prpE were unable to germinate in the presence of l-alanine, a process that requires GerA receptors. Moreover, germination triggered by an AGFK solution, requiring GerK, was also partially reduced. Results obtained over a 5-day germination period on plates with spores produced by a strain lacking PrpE clearly demonstrate that such spores are defective in germination (see Fig. 3B). In contrast, germination of spores produced by the ΔprpE strain was triggered normally by the Ca2+-dipicolinic acid chelate or dodecylamine pathway; both these pathways are independent of known Ger receptors (25, 30).
Previous studies showed that overproduction of either the GerA or the GerK receptor from an sspB promoter blocked the sporulation process in some unknown way (4). We show here that when cells with PrpE overproduction (or deletion of prpE in some cases) are induced to overproduce germination receptors, the inhibition of sporulation is suppressed. These observations led us to suspect that PrpE may normally be involved in the expression of the function of Ger receptors. Using ger-lacZ transcriptional fusions, we indeed showed that deletion of prpE reduces transcription of gerA but not that of gerB and gerK. The sspB promoter that was used to overproduce GerA and GerK in these experiments is, like the native ger promoters, also dependent on σG and SpoVT, the sporulation-specific factors (2, 18), and it was interesting to confirm that expression from the sspB promoter during sporulation was also reduced in the prpE deletion strain (Fig. 6). It will be interesting to determine whether expression of other σG-dependent promoters depends on PrpE in some way.
The SspB protein is one of two major proteins essential for UV resistance of spores (18). Nevertheless, the spores produced by the prpE deletion strain showed normal resistance to UV radiation, despite reduced synthesis of SspB in the mutant cells. This observation may be explained in two ways: either the remaining level of SspB is still sufficient for normal protection of DNA or other Ssp proteins can compensate for the depleted level of SspB, or both. The second possibility is supported by earlier findings that the functions of the SspA and SspB proteins are interchangeable and their expression interdependent. Thus, depletion of SspB in a ΔprpE strain can be compensated for by SspA (19, 20). Moreover, our study has also shown that spores produced by an sspB deletion strain display normal UV resistance (18).
Finally, we conclude that a strain lacking PrpE shows greatly reduced expression of gerA. However, since overproduction of PrpE appeared to have an effect similar to that of prpE deletion, at least in the case of gerA expression, the results suggest that an optimal level of phosphorylation of a specific target(s) is required for function. Thus, the results obtained indicate that the expression of genes coding for GerA and GerK receptors, in addition to being controlled by transcription by σG, is regulated through the action, direct or indirect, of PrpE in response to unknown intra- or extracellular stimuli. Finally, we also noted that spores produced from the ΔprpE strain were more sensitive to dry heat than wild-type spores. This effect might be directly connected with the reduced level of Ger receptors, or it may be an indication that additional spore proteins are controlled in some way by PrpE.
Acknowledgments
We thank I. Barry Holland for critical reading of the manuscript. Special thanks to Monika Wiktosińska for assistance with microscope observation.
This work was supported by the Polish Ministry of Education and Science (2 P04A 008 27 and 2 P04A 039 30). K.H. was a recipient of a UNESCO-PAN fellowship.
REFERENCES
- 1.Anagnostopoulos, C., and I. P. Crawford. 1961. Transformation studies on the linkage of markers in the tryptophan pathway in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 47:378-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagyan, I., J. Hobot, and S. Cutting. 1996. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J. Bacteriol. 178:4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagyan, I., and P. Setlow. 2002. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J. Bacteriol. 184:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabrera-Martinez, R. M., F. Tovar-Rojo, V. R. Vepachedu, and P. Setlow. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 185:2457-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corfe, B. M., R. L. Sammons, D. A. Smith, and C. Mauel. 1994. The gerB region of the Bacillus subtilis 168 chromosome encodes a homologue of the gerA spore germination operon. Microbiology 140:471-478. [DOI] [PubMed] [Google Scholar]
- 6.Corfe, B. M., A. Moir, D. Popham, and P. Setlow. 1994. Analysis of the expression and regulation of the gerB spore germination operon of Bacillus subtilis 168. Microbiology 140:3079-3083. [DOI] [PubMed] [Google Scholar]
- 7.Dong, T. C., S. M. Cutting, and R. J. Lewis. 2004. DNA-binding studies on the Bacillus subtilis transcriptional regulator and AbrB homologue, SpoVT. FEMS Microbiol. Lett. 233:247-256. [DOI] [PubMed] [Google Scholar]
- 8.Feavers, I. M., J. Foulkes, B. Setlow, D. Sun, W. Nicholson, P. Setlow, and A. Moir. 1990. The regulation of transcription of the gerA spore germination operon of Bacillus subtilis. Mol. Microbiol. 4:275-282. [DOI] [PubMed] [Google Scholar]
- 9.Garsin, D. A., L. Duncan, D. M. Paskowitz, and R. Losick. 1998. The kinase activity of the antisigma factor SpoIIAB is required for activation as well as inhibition of transcription factor σF during sporulation in Bacillus subtilis. J. Mol. Biol. 284:569-578. [DOI] [PubMed] [Google Scholar]
- 10.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley and Sons, New York, N.Y.
- 11.Hilbert, D. W., and P. J. Piggot. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igarashi, T., B. Setlow, M. Paidhungat, and P. Setlow. 2004. Effects of a gerF (lgt) mutation on the germination of spores of Bacillus subtilis. J. Bacteriol. 186:2984-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwanicki, A., A. Herman-Antosiewicz, M. Pierechod, S. J. Seror, and M. Obuchowski. 2002. PrpE, a PPP protein phosphatase from Bacillus subtilis with unusual substrate specificity. Biochem. J. 366:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, M., W. Shao, M. Perego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535-542. [DOI] [PubMed] [Google Scholar]
- 15.Kaltwasser, M., T. Wiegert, and W. Schumann. 2002. Construction and application of epitope- and green fluorescent protein-tagging integration vectors for Bacillus subtilis. Appl. Environ Microbiol. 68:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kroos, L., B. Zhang, H. Ichikawa, and Y. T. Yu. 1999. Control of sigma factor activity during Bacillus subtilis sporulation. Mol. Microbiol. 31:1285-1294. [DOI] [PubMed] [Google Scholar]
- 17.Lord, M., D. Barilla, and M. D. Yudkin. 1999. Replacement of vegetative σA by sporulation-specific σF as a component of the RNA polymerase holoenzyme in sporulating Bacillus subtilis. J. Bacteriol. 181:2346-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mason, J. M., and P. Setlow. 1986. Essential role of small, acid-soluble spore proteins in resistance of Bacillus subtilis spores to UV light. J. Bacteriol. 167:174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason, J. M., and P. Setlow. 1987. Different small, acid-soluble proteins of the α/β type have interchangeable roles in the heat and UV radiation resistance of Bacillus subtilis spores. J. Bacteriol. 169:3633-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mason, J. M., R. H. Hackett, and P. Setlow. 1988. Regulation of expression of genes coding for small, acid-soluble proteins of Bacillus subtilis spores: studies using lacZ gene fusions. J. Bacteriol. 170:239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moir, A., B. M. Corfe, and J. Behravan. 2002. Spore germination. Cell. Mol. Life Sci. 59:403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moir, A., E. H. Kemp, C. Robinson, and B. M. Corfe. 1994. The genetic analysis of bacterial spore germination. Soc. Appl. Bacteriol. Symp. Ser. 23:9S-16S. [PubMed] [Google Scholar]
- 23.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paidhungat, M., and P. Setlow. 1999. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant d-alanine. J. Bacteriol. 181:3341-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paidhungat, M., and P. Setlow. 2000. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piggot, P. J., and D. W. Hilbert. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579-586. [DOI] [PubMed] [Google Scholar]
- 27.Riemann, H., and Z. J. Ordal. 1961. Germination of bacterial endospores with calcium and dipicolinic acid. Science 133:1703-1704. [DOI] [PubMed] [Google Scholar]
- 28.Rode, L. J., and J. W. Foster. 1960. Germination of bacterial spores by long-chain alkyl amines. Nature 188:1132-1134. [DOI] [PubMed] [Google Scholar]
- 29.Rode, L. J., and J. W. Foster. 1961. Germination of bacterial spores with alkyl primary amines. J. Bacteriol. 81:768-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Setlow, B., A. E. Cowan, and P. Setlow. 2003. Germination of spores of Bacillus subtilis with dodecylamine. J. Appl. Microbiol. 95:637-648. [DOI] [PubMed] [Google Scholar]
- 31.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550-556. [DOI] [PubMed] [Google Scholar]
- 32.Sonenshein, A. L. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561-566. [DOI] [PubMed] [Google Scholar]
- 33.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takamatsu, H., and K. Watabe. 2002. Assembly and genetics of spore protective structures. Cell. Mol. Life Sci. 59:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 36.Yuan, G., and S. L. Wong. 1995. Regulation of groE expression in Bacillus subtilis: the involvement of the σA-like promoter and the roles of the inverted repeat sequence (CIRCE). J. Bacteriol. 177:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yudkin, M. D., and J. Clarkson. 2005. Differential gene expression in genetically identical sister cells: the initiation of sporulation in Bacillus subtilis. Mol. Microbiol. 56:578-589. [DOI] [PubMed] [Google Scholar]
- 38.Zuberi, A. R., A. Moir, and I. M. Feavers. 1987. The nucleotide sequence and gene organization of the gerA spore germination operon of Bacillus subtilis 168. Gene 51:1-11. [DOI] [PubMed] [Google Scholar]