Abstract
Tryptophan is an essential amino acid that is required for normal development in Chlamydia species, and tryptophan metabolism has been implicated in chlamydial persistence and tissue tropism. The ability to synthesize tryptophan is not universal among the Chlamydiaceae, but species that have a predicted tryptophan biosynthetic pathway also encode an ortholog of TrpR, a regulator of tryptophan metabolism in many gram-negative bacteria. We show that in Chlamydia trachomatis serovar D, TrpR regulates its own gene and trpB and trpA, the genes for the two subunits of tryptophan synthase. These three genes form an operon that is transcribed by the major form of chlamydial RNA polymerase. TrpR acts as a tryptophan-dependent aporepressor that binds specifically to operator sequences upstream of the trpRBA operon. We also found that TrpR repressed in vitro transcription of trpRBA in a promoter-specific manner, and the level of repression was dependent upon the concentrations of TrpR and tryptophan. Our findings provide a mechanism for chlamydiae to sense changes in tryptophan levels and to respond by modulating expression of the tryptophan biosynthesis genes, and we present a unified model that shows how C. trachomatis can combine transcriptional repression and attenuation to regulate intrachlamydial tryptophan levels. In the face of host defense mechanisms that limit tryptophan availability from the infected cell, the ability to maintain homeostatic control of intrachlamydial tryptophan levels is likely to play an important role in chlamydial pathogenesis.
There is accumulating evidence that tryptophan plays a pivotal role in intracellular chlamydial growth and pathogenesis, which has led to interest in the mechanisms that regulate intrachlamydial tryptophan levels. It is known that infected host cells can limit the supply of tryptophan through a gamma interferon (IFN-γ)-mediated mechanism (23). IFN-γ transcriptionally activates a cellular enzyme, indoleamine-2-3-dioxygenase, which degrades the intracellular tryptophan pool, leading to inhibition of chlamydial growth and replication (31). In tissue culture, low tryptophan levels cause chlamydiae to enter a persistent state that can last for months, with altered growth characterized by large aberrant organisms and a failure to produce infectious progeny (2). Chlamydial gene regulation has been shown to be dysregulated during this persistent cell culture state (1, 3, 8, 9). In particular, there is transcriptional upregulation of the tryptophan biosynthesis genes (35, 36), indicating a compensatory response by chlamydiae to increase tryptophan biosynthesis in the face of decreased availability of this essential amino acid from the host cell.
In Escherichia coli, the tryptophan biosynthesis genes are regulated by an aporepressor, TrpR, and tryptophan is required as a corepressor (22). The role of tryptophan is to induce a conformational change in the helix-turn-helix DNA binding domain of TrpR, which enables TrpR dimers to bind the cognate operator upstream of the trp operon and repress transcription (42). In contrast, when tryptophan levels are low, TrpR is unable to bind the operator, and expression of trp genes is increased through derepression. This control mechanism allows the bacterium to sense and respond to intracellular tryptophan levels.
Homologs of TrpR have been predicted in some species of Chlamydia, suggesting that this tryptophan-responsive mechanism of gene regulation may be utilized to homeostatically regulate intrachlamydial tryptophan levels. Intriguingly, only the chlamydial species predicted to encode TrpR also encode components of a tryptophan biosynthetic pathway (21, 27). For example, Chlamydia caviae (formerly known as Chlamydia psittaci strain GPIC) has an almost complete tryptophan biosynthetic pathway and a TrpR homolog (21). In Chlamydia trachomatis, the pathway is incomplete, but this species includes genes encoding both tryptophan synthase, the last enzyme in the pathway, and TrpR (7, 27). In contrast, Chlamydia pneumoniae has neither genes for tryptophan biosynthesis nor an identifiable TrpR homolog (20). These observations have led to the hypothesis that TrpR is the regulator of the tryptophan biosynthesis genes in Chlamydia (27, 35, 39).
In this study, we provide functional evidence that TrpR is an aporepressor that regulates tryptophan biosynthesis genes in C. trachomatis. We have identified a TrpR operator upstream of an operon containing trpR and trpBA. Recombinant TrpR was able to bind this operator, but binding was observed only when tryptophan was present. In addition, we show that TrpR is able to repress transcription of trpRBA in a promoter-specific and tryptophan-dependent manner.
MATERIALS AND METHODS
Reverse transcriptase PCR (RT-PCR).
HeLa cell monolayers were pretreated with 5 ng/ml human IFN-γ and then infected with C. trachomatis serovar D at a multiplicity of infection of 15. Infected cells were incubated for 48 h at 37°C in RPMI 1640 supplemented with 5 ng/ml IFN-γ, and total RNA was prepared using RNA STAT-60 (Tel-Test, Friendswood, Tex.). Ten micrograms of RNA treated with DNase I (Ambion, Austin, Tex.) was used for cDNA synthesis with avian myeloblastosis virus reverse transcriptase (Fisher Scientific, Pittsburgh, Pa.) and a specific 3′ primer that anneals to sequences within the open reading frame regions of trpA, T796 (5′-CACCATTTATTCCCGCGTCT). Negative control reactions were performed in the absence of reverse transcriptase. A trpR cDNA product was amplified by PCR with primers within trpR, T264 (5′-AAAAATCAAGAGGAGTCTGGCT) and T263 (5′-GCGGTACCTCAGATCTCTTTTTGTAAAAACTCTTTAAA). A trpB cDNA product was amplified with primers within trpB, T328 (5′-TGGGCAACAACACACTCATT) and T329 (5′-GCTCGTAACGCCTCTTCATC). A trpA cDNA product was amplified with primers within trpA, T795 (5′-GCGGCAAAAGCTCTGATTCAA) and T796. A trpR-B cDNA product was amplified by PCR with a 5′ primer within trpR, T264, and a 3′-primer within trpB, T330 (5′-CCCCCAAAGGATGTTTTATG). A trpBA cDNA product was amplified with a 5′ primer within trpB, T328, and a 3′ primer within trpA, T796.
Primer extension.
Primer extension was performed as described previously (6) using primer T450 (5′-GTAAAAACTTTTCTTTTTGCATTTTAG), specific for the 5′ end of the trpR open reading frame, and end labeled with 3,000 Ci/mmol [α-32P]ATP (MP Biomedical, Irvine, Calif.). The radiolabeled primer was annealed to 10 μg of chlamydial RNA, and cDNA was synthesized with avian myeloblastosis virus reverse transcriptase. The reaction mixture was incubated at 42°C for 50 min, and the cDNA products were electrophoresed on a 6% polyacrylamide-urea gel. The size of the primer extension product was determined by comparison to an M13 DNA sequencing ladder primed with the M13 forward primer.
Cloning of chlamydial trpR.
C. trachomatis trpR was cloned into the expression vector pRSET-C (Invitrogen, Carlsbad, Calif.) to produce plasmid pMT1182, which expressed full-length TrpR (except for the ATG start codon) with an N-terminal six-histidine tag. trpR was amplified by PCR with Tgo DNA polymerase (Roche Diagnostics, Indianapolis, Ind.) using C. trachomatis serovar D genomic DNA and primers T264 and T263. The PCR product was digested with KpnI and cloned into pRSET-C between KpnI and blunted BamHI sites. pMT1182 was sequenced to ensure that the coding region of TrpR matched the published nucleotide sequence (27).
Overexpression and purification of TrpR protein.
His6-TrpR was overexpressed in E. coli BL21(DE3) (Stratagene, La Jolla, Calif.) freshly transformed with pMT1182. Two hundred fifty milliliters of cells was grown at 37°C to an optical density at 600 nm of 0.5 and induced with 1 mM isopropyl-β-d-thiogalactosidase (IPTG). After 2.5 h, cells were collected by centrifugation; resuspended in 10 ml buffer N (10 mM Tris-HCl [pH 8.0], 300 mM NaCl, 10 mM 2-mercaptoethanol) containing 50 mM imidazole, 1 mM phenylmethylsulfonyl fluoride, and 10 μg/ml of pepstatin A; and disrupted with a Branson Sonifier 450 (30 s for three times) in the presence of 0.2% Sarkosyl. Soluble protein was separated from cell debris by centrifugation at 10,000 × g for 10 min at 4°C (Beckman JA-17 rotor), and 2% Triton X-100 was added to sequester the Sarkosyl. The lysate was then loaded onto a 1-ml Ni2+-charged HiTrap chelating column (Amersham Biosciences, Piscataway, N.J.). The column was washed with 40 ml of buffer N containing 100 mM imidazole. His-tagged protein was eluted with 5 ml buffer N containing 150 mM imidazole in 1-ml fractions. Fractions 3 to 5 were pooled, and the volume was brought up to 10 ml with buffer N to prevent precipitation during dialysis against storage buffer (10 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 100 μM EDTA, 10 mM 2-mercaptoethanol, 100 mM NaCl, 30% glycerol) overnight and again for 4 h. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by silver staining and Western blot analysis using anti-RGS-His6 antibody (QIAGEN, Valencia, Calif.). The protein concentration was determined using the Bio-Rad protein assay (Hercules, Calif.).
Production of polyclonal anti-TrpR antibodies.
Recombinant TrpR protein was gel purified by SDS-PAGE and used to generate rabbit polyclonal antibodies (Harlan Bioproducts for Science, Madison, Wis.). Antibodies were purified with a protein A agarose column according to the manufacturer's instructions (Bio-Rad).
DNA templates for the EMSA.
A 110-bp restriction fragment containing the trpRBA promoter region and a putative trp operator was used as a probe for electrophoretic mobility shift assay (EMSA). This fragment was amplified by PCR from C. trachomatis serovar D genomic DNA with primers T566 (5′-GAGGGGAGAATTCTAAGAAAAGA) and T567 (5′-GCCAGCCAGACTCCTCTT) and cloned into the SmaI site of pGEM-7ZF(+) (Promega Biotech, Madison, Wis.) to produce pMT1283. The EMSA restriction fragment template was digested from pMT1283 with EcoRI and BamHI, followed by gel purification from a 2% agarose gel. A 156-bp C. pneumoniae glnPQ fragment was used as a nonspecific competitor fragment (24). Double-stranded oligonucleotide competitors were prepared as previously described by annealing two complementary primers (32). Primers T699 (5′-AATTCTTATGAAATGTTGTAATATTATAGCATTACAAAAAGGTGCGA) and T700 (5′-GATCCTCGCACCTTTTTGTAATGCTATAATATTACAACATTTCATAA) were annealed to produce a specific oligonucleotide competitor containing the C. trachomatis trp operator. T690 (5′-GATCCTAATTGCATAAATATGATTTCATTATAAATAAATATGCATAAG) and T691 (5′-CTAGACTTATGCATATTTATTTATAATGAAATCATATTTATGCAATTA) were annealed to produce a nonspecific oligonucleotide containing tandem C. pneumoniae ArgR operators (24).
EMSA.
The DNA restriction fragment containing the putative trp operator was labeled with [α-32P]dATP using the Klenow fragment of E. coli DNA polymerase. Free nucleotides were removed using a Mini Quick Spin DNA column (Roche Diagnostics), and the activity of the probe was quantified using a scintillation counter. The EMSA analysis was based on the method of Carey (4). An 0.5 nM concentration of labeled probe was mixed with His6-TrpR in buffer containing 12.5 mM sodium phosphate buffer (pH 6.8), 25 mM NaCl, 1 mM EDTA, 1 mg/ml bovine serum albumin, 10% (vol/vol) glycerol, and 4 mM l-tryptophan and incubated at room temperature for 20 min. A range of concentrations of His6-TrpR from 0 to 10 nM (monomer) was tested, with details provided in the legend to Fig. 3. Some reaction mixtures also included unlabeled competitor DNA fragments or oligonucleotide or polyclonal anti-TrpR antibodies. Samples were loaded under tension onto a 10% polyacrylamide gel, containing 0.1 mM sodium phosphate buffer (pH 6.8) and 1 mM l-tryptophan that had been prerun at 220 V for 1 h at 4°C. The gel was electrophoresed at 300 V for 5 h at 4°C in TrpR EMSA buffer (0.1 mM sodium phosphate [pH 6.8] and 1 mM l-tryptophan) with recirculation. After electrophoresis, the gel was dried down and exposed to a phosphorimager screen. The screen was scanned with a Bio-Rad Personal FX scanner, and the data were analyzed with Quantity One software (Bio-Rad). The dissociation constant (Kd) was calculated using KaleidaGraph software (Synergy Software, Reading, Pa.).
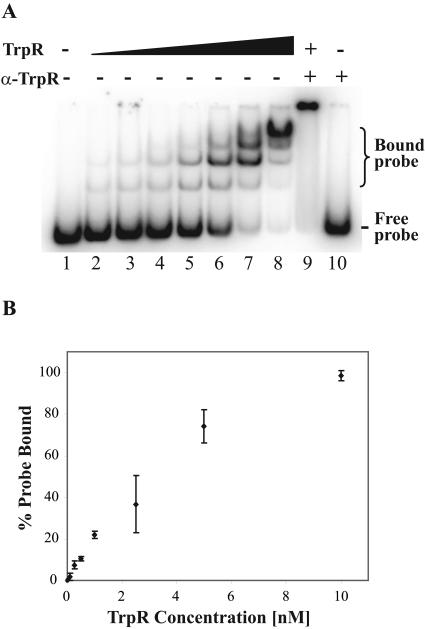
FIG. 3.
EMSA with C. trachomatis trpR probe titrated with recombinant TrpR. EMSA reactions and electrophoresis were performed in the presence of l-tryptophan. (A) The positions of bound and free probe are indicated. Lane 1, labeled probe alone; lanes 2 to 8, addition of increasing concentrations of TrpR (0.1, 0.25, 0.5, 1, 2.5, 5, and 10 nM, respectively); lane 9, addition of 10 nM TrpR and polyclonal anti-TrpR antibody; lane 10, addition of polyclonal anti-TrpR antibody alone. (B) Quantification of EMSA. EMSA reactions with various concentrations of recombinant TrpR were performed in triplicate and quantified by phosphorimager analysis. Standard deviations are marked by error bars.
Construction of transcription plasmids.
Plasmid pMT1250 contains the promoter region of trpRBA (+152 to −10), which was amplified from C. trachomatis serovar D genomic DNA by PCR with primers T521 (5′-GCGAGAACGAATTTATGGGTTTTAGAT) and T522 (5′-TTTTTGTAATGCTATAATATTACAACATTTCA). The trpRBA promoter insert was cloned upstream of a promoterless G-less cassette transcription template in pMT1125 (32). Construction of control plasmid pMT1198 containing the C. trachomatis omcB promoter has been previously described (24).
In vitro transcription.
Transcription reactions were performed with heparin-agarose-purified C. trachomatis RNA polymerase as previously described (28) and 25 nM (each) transcription plasmid. In some reactions, His6-TrpR (over a range of concentrations from 0 to 500 nM) and/or l-tryptophan (from 0 to 50 μM) was added, with more details given in the legend to Fig. 5.
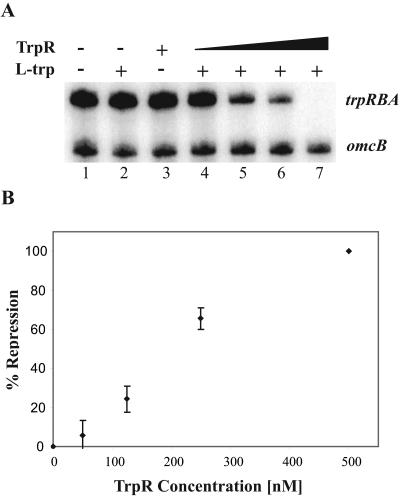
FIG. 5.
In vitro transcription with C. trachomatis RNA polymerase and recombinant TrpR. The upper band is the transcript from the trpRBA promoter, and the lower band is the transcript from the omcB promoter. (A) Lane 1, no TrpR, no l-tryptophan; lane 2, no TrpR, 50 μM l-tryptophan; lane 3, 500 nM TrpR, no l-tryptophan; lanes 4 to 7, increasing concentrations of TrpR (50, 125, 250, and 500 nM, respectively), 50 μM l-tryptophan. (Β) Quantification of TrpR-mediated transcriptional repression. Reactions were performed in triplicate and quantified by phosphorimager analysis. Standard deviations are marked by error bars. The degree of transcriptional repression was calculated by normalizing it to the amount of transcription in the absence of TrpR and the presence of l-tryptophan.
RESULTS
Characterization of the trp operon in C. trachomatis serovar D.
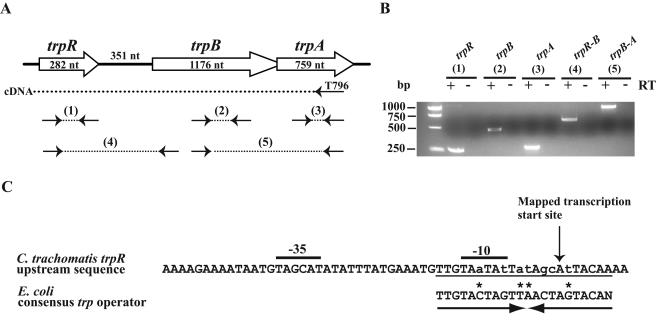
As trpR has an unusual location in the C. trachomatis serovar D genome adjacent to the tryptophan biosynthesis genes, trpB and trpA (38), we examined the transcriptional organization to determine if these genes are part of an operon. Using RT-PCR, we synthesized a cDNA fragment using a primer in trpA and then individually amplified cDNA from trpR, trpB, and trpA and the overlapping regions between trpR-trpB and trpB-trpA (Fig. 1A and B). These results indicate that trpR, trpB, and trpA are cotranscribed as a polycistronic message and establish these genes as part of an operon.
FIG. 1.
Transcriptional organization of tryptophan biosynthesis genes in C. trachomatis. (A) Schematic diagram of genes involved in tryptophan biosynthesis showing predicted RT-PCR products. nt, nucleotides. (B) RT-PCR analysis. cDNA synthesized using a primer that annealed in trpA (T796) was amplified using primers for trpR (product 1, shown in part A), trpB (product 2), trpA (product 3), trpR-trpB (product 4), and trpB-trpA (product 5). A no-RT control is included for each reaction. (C) Sequence upstream of trpR. The transcription start site determined by primer extension is marked by a vertical arrow. Predicted σ66 −35 and −10 promoter elements are marked. A putative trp operator overlapping the −10 promoter element and the transcription start site is underlined and aligned with the E. coli consensus trp operator. The operator forms an inverted repeat as shown by a pair of arrows. Lowercase letters indicate mismatches with the consensus, and an asterisk marks a position in the chlamydial sequence where the mismatch is accompanied by a complementary change in the inverted repeat.
To study the regulation of this tryptophan biosynthesis operon, we mapped the transcription start site of trpRBA by primer extension. The single primer extension product mapped to an adenine located 18 nucleotides upstream of the predicted translation start site for trpR (data not shown). Immediately upstream of this start site, we located a candidate promoter containing −35 and −10 elements (TAGCAT and TAATAT spaced 17 nucleotides apart; Fig. 1C) with a 2/6 and a 5/6 match to the respective optimal promoter elements recognized by C. trachomatis σ66 RNA polymerase (25, 29). We tested this predicted trpRBA promoter with our C. trachomatis in vitro transcription assay and found that it was transcribed by σ66 RNA polymerase (see Fig. 5A, lane 1).
By sequence inspection, we also identified a 22-bp inverted repeat that overlapped the −10 promoter element and the transcription start site of trpRBA (Fig. 1C). This predicted trp operator matched the E. coli consensus trp operator at 15 out of 22 positions (18). Two of the mismatches in the first repeat were accompanied by compensatory base substitutions in the inverted repeat sequence.
Purification of chlamydial TrpR.
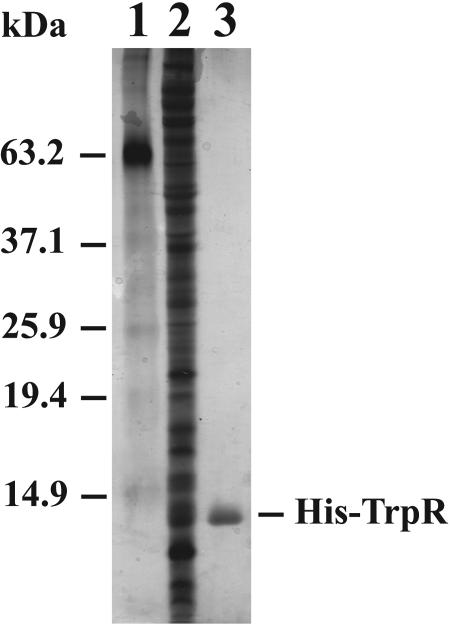
We purified C. trachomatis TrpR as a recombinant protein with an N-terminal six-histidine tag. The trpR coding sequence was cloned into the E. coli protein expression vector pRSET and overexpressed in E. coli strain BL21(DE3). Soluble His6-TrpR was purified by nickel affinity chromatography in a one-step purification to near-homogeneity (Fig. 2) at a concentration of 9.8 μM.
FIG. 2.
Silver stain of SDS-PAGE gel showing purification of recombinant His-TrpR. Lane 1, molecular mass markers; lane 2, lysate of E. coli overexpressing C. trachomatis TrpR; lane 3, TrpR after purification with a Ni column.
TrpR binds to the upstream region of C. trachomatis trpR in vitro.
We used an EMSA to determine if TrpR can bind to the predicted trp operator located upstream of trpR. EMSA conditions were adapted from previous studies with E. coli TrpR (4), but the pH of the electrophoresis buffer had to be increased from pH 6.0 to pH 6.8 to allow the TrpR-DNA complex to enter the gel. As l-tryptophan has been shown to be a necessary cofactor for TrpR in other bacteria (22), we performed the EMSA reactions in the presence and absence of l-tryptophan. When l-tryptophan was present, TrpR bound the trpR probe in a concentration-dependent manner, with complete binding of probe by 10 nM TrpR (Fig. 3A, lanes 1 to 8). The gel shift complex consisted of at least four lower-mobility bands, which is consistent with the observation in E. coli that there is oligomerization of TrpR dimers at the trp operator (4, 15, 40). Addition of polyclonal anti-TrpR antibodies produced a supershift, indicating that the binding was due to TrpR (Fig. 3A, lane 9). There was no gel shift with antibodies alone (Fig. 3A, lane 10) or in the absence of l-tryptophan (data not shown). To measure the kinetics of binding, EMSA reactions were performed over a range of TrpR concentrations (0.1, 0.25, 0.5, 1, 2.5, 5, and 10 nM) and an apparent dissociation constant (Kd) of 2 nM was calculated (Fig. 3B).
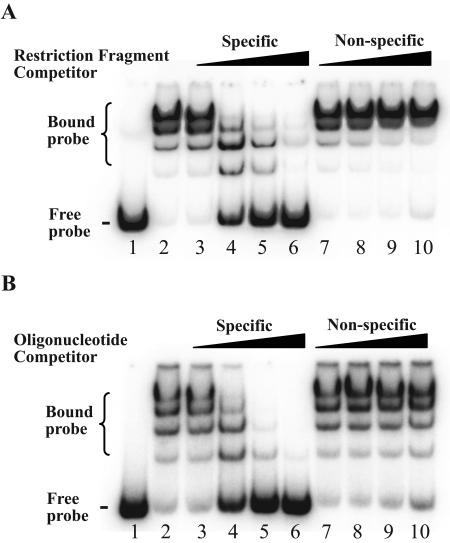
Competitive EMSA experiments were performed to confirm that TrpR binding was sequence specific. A molar excess of unlabeled trpR DNA fragment was able to compete for TrpR binding in a concentration-dependent manner, with almost complete loss of the gel shift at a 64-fold excess of competitor (Fig. 4A, lanes 3 to 6). In contrast, a nonspecific DNA restriction fragment did not compete for TrpR binding (Fig. 4A, lanes 7 to 10).
FIG. 4.
Competitive EMSA with trpR probe and recombinant TrpR. All reactions, except those with probe alone, were performed with 10 nM TrpR. (A) Competition with DNA fragment containing trp operator. Lane 1, probe alone; lane 2, no competitor; lanes 3 to 6, increasing amounts of molar excess of specific competitor relative to labeled probe (1-, 4-, 16-, and 64-fold, respectively); lanes 7 to 10, molar excess of nonspecific competitor (1-, 4-, 16-, and 64-fold, respectively). (B) Competition with double-stranded DNA oligonucleotide containing trp operator. Lane 1, probe alone; lane 2, no competitor; lanes 3 to 6, increasing amount of molar excess of specific competitor (10-, 50-, 250-, and 1,250-fold, respectively); lanes 7 to 10, molar excess of nonspecific competitor (10-, 50-, 250-, and 1,250-fold, respectively).
We used double-stranded oligonucleotide competitors to narrow the site of binding to the predicted trp operator. A short, unlabeled oligonucleotide, containing the trp operator and 10 nucleotides of flanking DNA on each end, was able to compete with the labeled trp DNA fragment for TrpR binding (Fig. 4B, lanes 3 to 7). In contrast, a nonspecific double-stranded oligonucleotide competitor did not compete for TrpR binding (Fig. 4B, lanes 7 to 10). These results demonstrate that in C. trachomatis, the predicted trp operator of the trpRBA operon is a likely binding site for TrpR.
TrpR represses transcription of the trpRBA promoter in vitro.
To determine if TrpR functions as a transcriptional repressor, we measured the effect of TrpR on transcription of the trpRBA promoter. In an in vitro assay, TrpR repressed transcription from the trpRBA promoter in the presence of l-tryptophan (Fig. 5A, lanes 4 to 7). Neither TrpR nor l-tryptophan alone had a significant effect on transcription (Fig. 5A, lanes 2 and 3). TrpR-mediated repression was concentration dependent with complete repression by 500 nM TrpR (Fig. 5B). This concentration of TrpR is 50 times higher than that required in the EMSA experiments, which correlates with the 50-fold-higher DNA concentration used in the transcription reactions. Transcription of the internal control omcB promoter was not affected (Fig. 5A), demonstrating that the activity of TrpR was promoter specific.
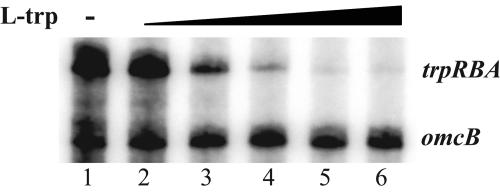
Next, we tested the role of l-tryptophan as a cofactor for TrpR-mediated repression. Keeping the concentration of TrpR constant, we titrated the amount of l-tryptophan used for transcription of the trpRBA promoter. We found that repression was dependent on the concentration of l-tryptophan and maximum repression was not reached until at least a 100:1 ratio of l-tryptophan to TrpR (Fig. 6). This finding is consistent with the observations in other bacteria that a large molar excess of the corepressor, l-tryptophan, is necessary for negative regulation by TrpR (10, 38). We also tested other representative amino acids but found that neither l-tyrosine, l-phenylalanine, l-alanine, l-arginine, l-serine, l-methionine, nor l-proline could function as a corepressor for TrpR (data not shown). Our results indicate that TrpR functions as an aporepressor that modulates trpRBA promoter activity in an l-tryptophan-dependent manner.
FIG. 6.
In vitro transcription assaying the effect of l-tryptophan titration on TrpR-mediated repression. All reactions were performed with 250 nM recombinant TrpR. Lane 1, no l-tryptophan; lanes 2 to 6, increasing concentrations of l-tryptophan (1, 5, 10, 25, and 50 μM, respectively).
DISCUSSION
In this report, we demonstrate that TrpR is an aporepressor that together with the corepressor l-tryptophan regulates transcription of tryptophan biosynthesis genes in C. trachomatis. Using in vitro assays, we have shown that C. trachomatis TrpR binds in the presence of tryptophan to its cognate operator upstream of trpRBA and represses transcription by the major form of chlamydial RNA polymerase. Our studies provide a mechanism for the homeostatic control of intrachlamydial tryptophan levels through transcriptional regulation of the tryptophan biosynthetic pathway.
We found that trpR and the adjacent tryptophan biosynthesis genes, trpBA, are cotranscribed as an operon in C. trachomatis serovar D. This is an unusual transcriptional organization in bacteria, and the more typical arrangement is seen in E. coli where trpR is physically separate from the operon containing the tryptophan biosynthesis genes, although both are regulated by TrpR (11, 22). The location of trpR adjacent to trpBA is conserved in all other serovars of C. trachomatis (7). One point of difference, though, is that trpR and trpBA were not found to be part of an operon in serovar L2 (35). However, in this serovar, trpBA is still induced at the transcriptional level by tryptophan limitation, and we infer that it is still under the control of a TrpR-dependent promoter. So far, there is no functional evidence of an independent trpBA promoter in serovar L2, and we have not been able to locate an operator for trpBA using TrpR binding studies (J. C. Akers and M. Tan, unpublished observation).
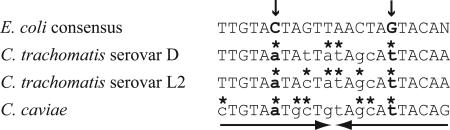
We predict that other chlamydial species can also sense tryptophan levels and regulate the expression of tryptophan biosynthesis genes through TrpR but that this mechanism is not conserved among all members of the family Chlamydiaceae. C. caviae includes a gene encoding a TrpR ortholog together with an almost complete tryptophan biosynthetic pathway in a single operon (36). Expression of this operon is induced in tissue culture by tryptophan limitation (36), providing support for tryptophan-dependent regulation by TrpR in this species. We have identified a candidate operator upstream of the C. caviae trp operon with sequence conservation to the C. trachomatis and consensus E. coli trp operators (Fig. 7). In contrast, trpR and trpBA have not been identified in the genomes of C. pneumoniae, Chlamydia muridarum (formerly known as C. trachomatis strain MoPn) (20), and Parachlamydia strain UWE25 (14), a Chlamydia-like endosymbiont of free-living amoebae. These findings indicate that TrpR-mediated transcriptional repression is not uniformly conserved among chlamydial species.
FIG. 7.
Alignment of the E. coli consensus trp operator with the C. trachomatis serovar D trp operator and predicted trp operators from C. trachomatis serovar L2 and C. caviae. An inverted repeat within the operator is marked by a pair of arrows. Lowercase letters indicate mismatches with the consensus, and an asterisk marks a position in the chlamydial sequence where the mismatch is accompanied by a complementary change in the inverted repeat. Vertical arrows and boldface mark invariant positions in the E. coli consensus operator that are not conserved in the chlamydial trp operators.
While the chlamydial trp operator shows conservation with the consensus E. coli trp operator, there are nucleotide differences especially towards the center of the operator (Fig. 7). The internal symmetry of the chlamydial trp operator is preserved, however, as several nucleotide substitutions in the first repeat are accompanied by compensatory changes in the inverted repeat sequence. Of these differences between the chlamydial and E. coli trp operators, the most noteworthy are at positions 6 and 11, which are invariably a C and G, respectively, in E. coli, but replaced by an A and T, respectively, in the three chlamydial trp operators.
An interesting paradox is that TrpR appears to be present in all C. trachomatis serovars even when tryptophan synthase, the product of TrpR regulation, is enzymatically inactive (serovar B is an anomaly because it has a large deletion involving the trpRBA locus) (7). The serovars of C. trachomatis that cause ocular disease all have mutations in trpA or trpB which result in a nonfunctional tryptophan synthase. Surprisingly, the expression of trpBA in these ocular serovars is still regulated in response to tryptophan levels (35), and we can identify a well-conserved operator upstream of trpRBA for each serovar (Akers and Tan, unpublished). The obvious question is why these serovars would maintain a mechanism to regulate the expression of a nonfunctional enzyme. Perhaps tryptophan synthase in C. trachomatis has other functions besides tryptophan biosynthesis. It is also possible, however, that it is the homeostatic regulation of trpR expression in response to tryptophan levels that is being conserved, which would in turn suggest that TrpR regulates other target genes in C. trachomatis.
A number of candidate TrpR target genes in Chlamydia can be considered because they are homologs of genes that are regulated by TrpR in E. coli. The only other tryptophan biosynthesis gene in C. trachomatis is trpC, which encodes an enzyme upstream of tryptophan synthase in the tryptophan biosynthetic pathway (27). trpC is located at a separate site from trpRBA in the C. trachomatis genome. In E. coli, TrpR also regulates aroH and aroL, the genes for aromatic amino acid biosynthesis (12, 43), and mtr, which encodes a tryptophan transporter (13). Homologs of all three genes can be identified in C. trachomatis. The mtr homolog is called tyrP, and it is predicted to encode a tyrosine/tryptophan transporter. There has been particular interest in tyrP because the number of gene copies in C. pneumoniae has been shown to vary according to tissue tropism and disease manifestation (10). So far, however, there is little evidence to indicate that any of these genes are regulated by TrpR in Chlamydia. The expression of neither C. trachomatis trpC, aroH, aroL, tyrP.1, nor tyrP.2 is induced by tryptophan limitation (35). In addition, we have not been able to identify sequences resembling a trp operator upstream of these genes in C. trachomatis (Akers and Tan, unpublished). One approach for recognizing additional TrpR-regulated genes would be to perform transcriptional profiling with a DNA microarray under specific conditions of tryptophan limitation to identify genes that are coregulated with the trpRBA operon.
It is also possible that the tryptophan biosynthesis operon is the only target of TrpR in Chlamydia. There is only one other example in eubacteria where trpR is adjacent to the tryptophan biosynthesis genes (38). Furthermore, TrpR is found only in the chlamydial species that have a tryptophan biosynthesis operon (20, 21, 27). An additional clue is provided by the location of trpR and the tryptophan biosynthesis genes in the plasticity zone, which is a region of genetic diversity among chlamydial species (20). It would appear that trpR and the tryptophan biosynthesis genes form a functional unit in Chlamydia that has been selectively acquired or lost during chlamydial evolution and divergence.
While TrpR provides a mechanism for regulation at the level of transcription initiation, there is evidence that the tryptophan biosynthesis genes in Chlamydia are also regulated by transcription attenuation, as is the case in E. coli. Using a bioinformatics approach, Merino and Yanofsky (17) have predicted an attenuator upstream of C. trachomatis trpBA. The presence of an attenuator could explain the relatively large intergenic region between trpR and trpBA even though they are part of an operon; intergenic regions are characteristically short in Chlamydia, especially for genes within an operon (16, 26, 30). We propose a unified model in which transcriptional repression and attenuation allow C. trachomatis to regulate intrachlamydial tryptophan levels. During chlamydial growth in the presence of sufficient tryptophan, the aporepressor, TrpR, forms a complex with the corepressor tryptophan and binds the cognate operator to repress transcription from the trp operon. In addition, formation of the transcription terminator structure in the trpR-trpBA intergenic region further reduces readthrough transcription of trpBA. However, in conditions of tryptophan limitation, such as during chlamydial persistence, TrpR is unable to bind its operator, leading to derepression of trpRBA expression. Low tryptophan levels would also decrease the availability of charged tRNATrp, favoring ribosome pausing and formation of the antiterminator structure, thus allowing RNA polymerase to read through and transcribe trpBA. The net effect would be that expression of trpR and that of trpBA would both be upregulated when tryptophan levels are low, allowing synthesis of tryptophan if the necessary substrate is available (7). This combination of control through transcriptional repression and attenuation may allow chlamydiae to respond quickly to changes in tryptophan availability by regulating tryptophan biosynthesis.
Even though chlamydiae are intracellular parasites that reside in what might be thought of as a relatively stable environment, it is clear that they maintain the capacity to respond to environmental stimuli by coordinately regulating gene expression. Among the recently described transcriptional regulators in Chlamydia that can sense and respond to environmental and metabolic conditions are a stress response repressor, HrcA (32-34); a metal-dependent repressor, DcrA (19, 37); and an arginine-responsive aporepressor, ArgR, that utilizes arginine as a corepressor to regulate expression of a putative arginine transporter (24). An alternative sigma factor, σ28, has been shown to be a stage-specific transcriptional regulator (41), although the signals that it responds to are not known. Understanding the response to tryptophan levels is crucial because tryptophan depletion has been shown to be a consequence of the host IFN-γ response in infected cells and to result in chlamydial persistence. TrpR provides a mechanism that allows chlamydiae to sense tryptophan limitation and to respond by upregulating tryptophan biosynthesis. It remains to be seen whether derepression of TrpR gene regulation is purely a marker of persistence or a clue to the underlying molecular mechanism of chlamydial persistence.
ADDENDUM
While the manuscript was in review, a complementary study by McClarty and colleagues was published that provides elegant genetic evidence that TrpR is a regulator of trp genes in C. trachomatis (5). In addition, the study identified an operator upstream of trpR and predicts a second operator upstream of trpBA.
Acknowledgments
We thank Eike Niehus, Hilda Hiu Yin Yu, Chris Schaumburg, Elizabeth Di Russo, and Allan Chen for critical reading of the manuscript and helpful advice.
This work is supported by a grant from the NIH (AI 44198). M.T. is supported by an NIH Independent Scientist Award (AI 057563). J.C.A. is supported by a predoctoral training grant from the NIH (National Research Service Award LM007443 from the National Library of Medicine).
REFERENCES
- 1.Beatty, W. L., G. I. Byrne, and R. P. Morrison. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. USA 90:3998-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beatty, W. L., R. P. Morrison, and G. I. Byrne. 1994. Persistent chlamydiae: from cell culture to a paradigm of chlamydial pathogenesis. Microbiol. Rev. 58:686-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belland, R. J., D. E. Nelson, D. Virok, D. D. Crane, D. Hogan, D. Sturdevant, W. L. Beatty, and H. D. Caldwell. 2003. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. USA 100:15971-15976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey, J. 1988. Gel retardation at low pH resolves trp repressor-DNA complexes for quantitative study. Proc. Natl. Acad. Sci. USA 85:975-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson, J. H., H. Wood, C. Roshick, H. D. Caldwell, and G. McClarty. 2006. In vivo and in vitro studies of Chlamydia trachomatis TrpR:DNA interactions. Mol. Microbiol. 59:1678-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engel, J. N., and D. Ganem. 1987. Chlamydial rRNA operons: gene organization and identification of putative tandem promoters. J. Bacteriol. 169:5678-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fehlner-Gardiner, C., C. Roshick, J. H. Carlson, S. Hughes, R. J. Belland, H. D. Caldwell, and G. McClarty. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893-26903. [DOI] [PubMed] [Google Scholar]
- 8.Gerard, H. C., L. Kohler, P. J. Branigan, H. Zeidler, H. R. Schumacher, and A. P. Hudson. 1998. Viability and gene expression in Chlamydia trachomatis during persistent infection of cultured human monocytes. Med. Microbiol. Immunol. 187:115-120. [DOI] [PubMed] [Google Scholar]
- 9.Gerard, H. C., B. Krausse-Opatz, Z. Wang, D. Rudy, J. P. Rao, H. Zeidler, H. R. Schumacher, J. A. Whittum-Hudson, L. Kohler, and A. P. Hudson. 2001. Expression of Chlamydia trachomatis genes encoding products required for DNA synthesis and cell division during active versus persistent infection. Mol. Microbiol. 41:731-741. [DOI] [PubMed] [Google Scholar]
- 10.Gieffers, J., L. Durling, S. P. Ouellette, J. Rupp, M. Maass, G. I. Byrne, H. D. Caldwell, and R. J. Belland. 2003. Genotypic differences in the Chlamydia pneumoniae tyrP locus related to vascular tropism and pathogenicity. J. Infect. Dis. 188:1085-1093. [DOI] [PubMed] [Google Scholar]
- 11.Gunsalus, R. P., and C. Yanofsky. 1980. Nucleotide sequence and expression of Escherichia coli trpR, the structural gene for the trp aporepressor. Proc. Natl. Acad. Sci. USA 77:7117-7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heatwole, V. M., and R. L. Somerville. 1992. Synergism between the Trp repressor and Tyr repressor in repression of the aroL promoter of Escherichia coli K-12. J. Bacteriol. 174:331-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heatwole, V. M., and R. L. Somerville. 1991. The tryptophan-specific permease gene, mtr, is differentially regulated by the tryptophan and tyrosine repressors in Escherichia coli K-12. J. Bacteriol. 173:3601-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horn, M., A. Collingro, S. Schmitz-Esser, C. L. Beier, U. Purkhold, B. Fartmann, P. Brandt, G. J. Nyakatura, M. Droege, D. Frishman, T. Rattei, H.-W. Mewes, and M. Wagner. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728-730. [DOI] [PubMed] [Google Scholar]
- 15.Jeeves, M., P. D. Evans, R. A. Parslow, M. Jaseja, and E. I. Hyde. 1999. Studies of the Escherichia coli Trp repressor binding to its five operators and to variant operator sequences. Eur. J. Biochem. 265:919-928. [DOI] [PubMed] [Google Scholar]
- 16.Mathews, S., and P. Timms. In silico identification of chlamydial promoters and their role in regulation of development. In P. M. Bavoil and P. B. Wyrick (ed.), Chlamydia: genomics, pathogenesis and implications for control, in press. Horizon Scientific Press, Wymondham, United Kingdom.
- 17.Merino, E., and C. Yanofsky. 2005. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 21:260-264. [DOI] [PubMed] [Google Scholar]
- 18.Mironov, A., E. Koonin, M. Roytberg, and M. Gelfand. 1999. Computer analysis of transcription regulatory patterns in completely sequenced bacterial genomes. Nucleic Acids Res. 27:2981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rau, A., S. Wyllie, J. Whittimore, and J. E. Raulston. 2005. Identification of Chlamydia trachomatis genomic sequences recognized by chlamydial divalent cation-dependent regulator A (DcrA). J. Bacteriol. 187:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Read, T. D., G. S. A. Myers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, J. Peterson, M. J. Beanan, O. White, S. L. Salzberg, R.-C. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose, J., C. Squires, C. Yanofsky, H. Yang, and G. Zubay. 1973. Regulation of in vitro transcription of the tryptophan operon by purified RNA polymerase in the presence of partially purified repressor and tryptophan. Nat. New Biol. 245:133-137. [DOI] [PubMed] [Google Scholar]
- 23.Rottenberg, M. E., A. Gigliotti-Rothfuchs, and H. Wigzell. 2002. The role of IFN-gamma in the outcome of chlamydial infection. Curr. Opin. Immunol. 14:444-451. [DOI] [PubMed] [Google Scholar]
- 24.Schaumburg, C. S., and M. Tan. 2006. Arginine-dependent gene regulation via the ArgR repressor is species specific in Chlamydia. J. Bacteriol. 188:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaumburg, C. S., and M. Tan. 2003. Mutational analysis of the Chlamydia trachomatis dnaK promoter defines the optimal −35 promoter element. Nucleic Acids Res. 31:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen, L., Y. Shi, A. L. Douglas, T. P. Hatch, C. M. O'Connell, J. M. Chen, and Y. X. Zhang. 2000. Identification and characterization of promoters regulating tuf expression in Chlamydia trachomatis serovar F. Arch. Biochem. Biophys. 379:46-56. [DOI] [PubMed] [Google Scholar]
- 27.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 28.Tan, M., and J. N. Engel. 1996. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter. J. Bacteriol. 178:6975-6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan, M., T. Gaal, R. L. Gourse, and J. N. Engel. 1998. Mutational analysis of the Chlamydia trachomatis rRNA P1 promoter defines four regions important for transcription in vitro. J. Bacteriol. 180:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan, M., B. Wong, and J. N. Engel. 1996. Transcriptional organization and regulation of the dnaK and groE operons of Chlamydia trachomatis. J. Bacteriol. 178:6983-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor, M., and G. Feng. 1991. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 5:2516-2522. [PubMed] [Google Scholar]
- 32.Wilson, A. C., and M. Tan. 2002. Functional analysis of the heat shock regulator HrcA of Chlamydia trachomatis. J. Bacteriol. 184:6566-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, A. C., and M. Tan. 2004. Stress response gene regulation in Chlamydia is dependent on HrcA-CIRCE interactions. J. Bacteriol. 186:3384-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson, A. C., C. C. Wu, J. R. Yates III, and M. Tan. 2005. Chlamydial GroEL autoregulates its own expression through direct interactions with the HrcA repressor protein. J. Bacteriol. 187:7535-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood, H., C. Fehlner-Gardner, J. Berry, E. Fischer, B. Graham, T. Hackstadt, C. Roshick, and G. McClarty. 2003. Regulation of tryptophan synthase gene expression in Chlamydia trachomatis. Mol. Microbiol. 49:1347-1359. [DOI] [PubMed] [Google Scholar]
- 36.Wood, H., C. Roshick, and G. McClarty. 2004. Tryptophan recycling is responsible for the interferon-gamma resistance of Chlamydia psittaci GPIC in indoleamine dioxygenase-expressing host cells. Mol. Microbiol. 52:903-916. [DOI] [PubMed] [Google Scholar]
- 37.Wyllie, S., and J. E. Raulston. 2001. Identifying regulators of transcription in an obligate intracellular pathogen: a metal-dependent repressor in Chlamydia trachomatis. Mol. Microbiol. 40:1027-1036. [DOI] [PubMed] [Google Scholar]
- 38.Xie, G., C. Bonner, T. Brettin, R. Gottardo, N. Keyhani, and R. Jensen. 2003. Lateral gene transfer and ancient paralogy of operons containing redundant copies of tryptophan-pathway genes in Xylella species and in heterocystous cyanobacteria. Genome Biol. 4:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie, G., C. A. Bonner, and R. A. Jensen. 2002. Dynamic diversity of the tryptophan pathway in chlamydiae: reductive evolution and a novel operon for tryptophan recapture. Genome Biol. 3:R0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, J., A. Gunasekera, T. A. Lavoie, L. Jin, D. E. Lewis, and J. Carey. 1996. In vivo and in vitro studies of TrpR-DNA interactions. J. Mol. Biol. 258:37-52. [DOI] [PubMed] [Google Scholar]
- 41.Yu, H. H. Y., and M. Tan. 2003. σ28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol. Microbiol. 50:577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, R. G., A. Joachimiak, C. L. Lawson, R. W. Schevitz, Z. Otwinowski, and P. B. Sigler. 1987. The crystal structure of trp aporepressor at 1.8 Å shows how binding tryptophan enhances DNA affinity. Nature 327:591-597. [DOI] [PubMed] [Google Scholar]
- 43.Zurawski, G., R. P. Gunsalus, K. D. Brown, and C. Yanofsky. 1981. Structure and regulation of aroH, the structural gene for the tryptophan-repressible 3-deoxy-d-arabino-heptulosonic acid-7-phosphate synthetase of Escherichia coli. J. Mol. Biol. 145:47-73. [DOI] [PubMed] [Google Scholar]