Abstract
The Spx protein of Bacillus subtilis exerts both positive and negative transcriptional control in response to oxidative stress by interacting with the C-terminal domain of the RNA polymerase (RNAP) alpha subunit (αCTD). Thus, transcription of the srf operon at the onset of competence development, which requires the ComA response regulator of the ComPA signal transduction system, is repressed by Spx-αCTD interaction. Previous genetic and structural analyses have determined that an Spx-binding surface resides in and around the α1 region of αCTD. Alanine-scanning mutagenesis of B. subtilis αCTD uncovered residue positions required for Spx function and ComA-dependent srf transcriptional activation. Analysis of srf-lacZ fusion expression, DNase I footprinting, and solid-phase promoter retention experiments indicate that Spx interferes with ComA-αCTD interaction and that residues Y263, C265, and K267 of the α1 region lie within overlapping ComA- and Spx-binding sites for αCTD interaction. Evidence is also presented that oxidized Spx, while enhancing interference of activator-RNAP interaction, is not essential for negative control.
The spx gene of Bacillus subtilis was identified as the site of mutations that overcome the requirement for the protease ClpXP in the expression of genes that are transcriptionally activated by response regulator proteins (16, 25). The srf operon of B. subtilis, which contains genes encoding products that function in the control of competence development and in nonribosomal peptide synthesis (4, 11, 17, 33), is activated by the ComPA two-component signal transduction system (5, 6). ComA is a response regulator that becomes phosphorylated by interaction with the histidine kinase ComP when the latter autophosphorylates in response to the peptide pheromone ComX (9). This quorum-sensing system converts ComA to active ComA phosphate (ComA∼P), which interacts as two dimers with the two ComA box elements residing upstream of the srf operon promoter (21, 30). ComA-dependent transcriptional activation is one of the regulatory events in B. subtilis that are negatively affected by Spx (25).
Competence development, as well as several other transition state processes of B. subtilis, is severely impaired in strains bearing mutations in clpX or clpP (12, 15, 16). Likewise, srf operon expression is diminished in clpX and clpP mutant cells (16, 19). Some of the suppressor mutations resulting in restored srf expression in a clpX background mapped to the rpoA gene, which encodes the RNAP α subunit. Codon substitutions in the region encoding the C-terminal domain of α (αCTD) were uncovered through this suppressor analysis (19). One codon change conferring the clpX suppressor phenotype was Y263C, in the α1 helix of the αCTD. The other clpX suppressor locus is the spx gene (16), the product of which interacts with the RNAP to affect transcription initiation (25, 27). Subsequent structural analysis confirmed that Spx interacts with the αCTD of RNAP and that the binding surface includes residue Y263 of the α subunit (27). This interaction is necessary for Spx-dependent repression of srf operon transcription (25). Unlike other negative transcriptional regulators, however, Spx does not exhibit sequence-specific DNA binding activity (23).
Spx, while exerting negative control on activator-stimulated transcription, positively controls transcription of the thioredoxin (trxA) and thioredoxin reductase (trxB) genes as well as several genes that function in the oxidative stress response and in cysteine synthesis (23, 24). Positive control is observed after thiol-specific oxidative stress and requires disulfide formation at the highly conserved N-terminal CXXC motif. The detailed mechanism of Spx-dependent transcriptional activation is not known at this time.
The repression of the srf operon by Spx during oxidative stress is at least partly the result of a higher Spx protein concentration and interaction with the αCTD of RNAP (24, 25). It is not known if oxidized Spx is required for repression. The mechanism of repression has been proposed to involve the interference of ComA interaction with RNAP by Spx (25, 35). As with other prokaryotic transcriptional activators, ComA-dependent activation involves interaction of activated ComA with RNAP αCTD (this report). Alanine-scanning mutagenesis of RNAP αCTD and the identification of rpoA mutant alleles that affect both Spx- and ComA-RNAP interaction are reported herein. The evidence presented supports the interference model of Spx-dependent repression.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacillus subtilis strains used in the study are listed in Table 1. B. subtilis strains constructed with alanine-scanning αCTD alleles are listed in Table S1 in the supplemental material. Oligonucleotides used in the study, including those used for alanine-scanning mutagenesis of rpoA, are listed in Table S2 in the supplemental material. To express mutant spx(C10A) from the isopropyl-β-thiogalactopyranoside (IPTG)-inducible Phyperspank (Pspank-hy) promoter (1), plasmid pZY14 was constructed. Plasmid pSN56 (24) was digested with BclI and SalI to obtain the 201-bp fragment containing the 3′ half of the spxLDD allele, which encodes the ClpXP-resistant form of Spx. The plasmid pSN95 (23) was digested with HindIII and BclI to obtain a 281-bp fragment containing the N-terminal portion of the spx(C10A) allele. Ligation of the two fragments with pUC18, which was digested with SalI and HindIII, was followed by transformation of Escherichia coli DH5α competent cells with the ligation mix. This resulted in construction of plasmid pZY11, which encodes the mutant SpxLDD(C10A). Plasmid pZY11 was digested with SalI and HindIII to obtain the 482-bp SpxLDD(C10A)-encoding fragment for ligation with pDR111, which was digested with SalI and HindIII, to yield plasmid pZY14 [Pspank-hy-spxLDD(C10A)]. Plasmid pZY14 was used to transform B. subtilis strain LAB545 (srfA-lacZ, contains pMMN92 [21] integrated into SPβc2del2::Tn917::pSK10Δ6 prophage [36]) to obtain ORB6307. SpxLDD-expressing strains having mutations in rpoA were obtained by transforming ORB5259 [rpoA(C265A)] and ORB5262 [rpoA(K267A)] with chromosomal DNA from ORB4342 [amyE::pSN56 (Pspank-hy-spxLDD)] to yield ORB6127 and ORB6128. The srf-lacZ fusion was introduced into the resulting strains and into ORB4342 (amyE::pSN56 rpoA+) by transduction with the SPβ phage lysate (36) carrying srf-lacZ (pMMN92) (21) with selection for chloramphenicol resistance (5 μg/ml) to yield ORB6127, ORB6128, and ORB6129. Construction of a srf-35 promoter mutant plasmid (−34,−35TG to CC) was carried out by site-directed mutagenesis (23). Upstream and downstream fragments were synthesized via PCR by using the mutagenic oligonucleotides oYZ02-6 and oYZ02-3a along with the upstream primer oYZ02-3(−347) and downstream primer oYZ02-4 (+65). The two resulting PCR fragments were used as templates for PCR with primers oYZ02-3 and oYZ02-4. The PCR fragment digested with BamHI and HindIII was inserted into pUC18 to obtain plasmid pZY6. The plasmid pMMN101, containing the mutant srf promoter fragment bearing a ComA box 2 mutation, was previously described (21).
TABLE 1.
Bacillus subtilis strains
| Strain | Genotype | Reference |
|---|---|---|
| LAB545 | trpC2 pheA SPβc2del2::Tn917::pMMN92(srfA-lacZ) | 21, this study |
| MH5636 | trpC2 pheA1 His10rpoC | 29 |
| OKB167 | trpC2 pheA1 comPA::Erm srfB::Tn917 | 20 |
| ORB3621 | trpC2 pheA rpoA(Y263C) | This study |
| ORB4123 | trpC2 pheA His10rpoC rpoAcxs-1 | 23 |
| ORB4342 | trpC2 pheA amyE::pSN56 | 24 |
| ORB4343 | trpC2 pheA rpoAY263C amyE::pSN56 | This study |
| ORB5501 | trpC2 pheA rpoA(C265A) His10rpoC | This study |
| ORB5259 | trpC2 pheA rpoA(C265A) | This study |
| ORB5262 | trpC2 pheA rpoA(K267A) | This study |
| ORB5327 | trpC2 pheA rpoA(K267A) SPβc2del2::Tn917::pMMN92(srfA-lacZ) | This study |
| ORB5422 | trpC2 pheA rpoA(C265A) SPβc2del2::Tn917::pMMN92(srfA-lacZ) | This study |
| ORB5553 | trpC2 pheA rpoA(Y263C) SPβc2del2::Tn917::pMMN92(srfA-lacZ) | This study |
| ORB5661 | trpC2 pheA1 comPA:Erm SPβc2del2::Tn917::pMMN92(srfA-lacZ) | This study |
| ORB6116 | trpC2 pheA rpoA(K267A) His102 rpoC | This study |
| ORB6127 | trpC2 pheA rpoA(C265A) amyE::pSN56 (pDR111-spxLDD) | This study |
| ORB6128 | trpC2 pheA rpoA(K267A) amyE::pSN56 (pDR111-spxLDD) | This study |
| ORB6129 | trpC2 pheA amyE::pSN56 SPβc2del2::Tn917::pMMN92(srfA-lacZ) | This study |
| ORB6130 | trpC2 pheA rpoA(Y263C) amyE::pSN56 SPβc2del2::Tn917::pMMN92(srfA-lacZ) | This study |
| ORB6131 | trpC2 pheA rpoA(C265A) amyE::pSN56 SPβc2del2::Tn917::pMMN92(srfA-lacZ) | This study |
| ORB6132 | trpC2 pheA rpoA(K267A) amyE::pSN56 SPβc2del2::Tn917::pMMN92(srfA-lacZ) | This study |
| ORB6137 | trpC2 pheA amyE::pSN56 (pDR111-spxLDD) SPβc2del2::Tn917::pTMH112(rpsD-lacZ) | This study |
| ORB6138 | trpC2 pheA rpoA(Y263C) amyE::pSN56 (pDR111-spxLDD) SPβc2del2::Tn917::pTMH112(rpsD-lacZ) | This study |
| ORB6139 | trpC2 pheA rpoA(C265A) amyE::pSN56 (pDR111-spxLDD) SPβc2del2::Tn917::pTMH112(rpsD-lacZ) | This study |
| ORB6140 | trpC2 pheA rpoA(K267A) amyE::pSN56 (pDR111-spxLDD) SPβc2del2::Tn917::pTMH112(rpsD-lacZ) | This study |
| ORB6303 | trpC2 pheA1 amyE::pDR111 | This study |
| ORB6304 | trpC2 pheA1 amyE::pZY14(spxLDD(C10A)) | This study |
| ORB6305 | trpC2 pheA1 amyE::pDR111 SPβc2del2::Tn917::pMMN92(srfA-lacZ) | This study |
| ORB6307 | trpC2 pheA1 amyE::pZY14(spxLDD(C10A)) SPβc2del2::Tn917::pMMN92(srfA-lacZ) | This study |
Alanine-scanning mutagenesis of the rpoA CTD region.
The rpoA-rplQ region of the B. subtilis chromosome was amplified by PCR using the primers oMN99-91 and oMN001-106 (19). The fragment was inserted into HindIII- and XbaI-cleaved pAG58-ble-1 (34) to yield pSN108. Alanine-scanning mutagenesis was conducted using mutagenic primers and the PCR amplification method of site-directed mutagenesis. Two primers (forward and reverse) specifying a single mutation were used to perform inverse PCR on whole pSN108 plasmid DNA. The mutation also introduced an additional restriction site in the mutated DNA insert. The PCR product was extracted with phenol-chloroform-isoamyl alcohol and precipitated with ethanol using yeast RNA as the carrier. The DNA was cleaved with DpnI to eliminate template DNA, and the restriction reaction was used to directly transform competent cells of E. coli strain DH5α. Introduction of the mutation into the rpoA-rplQ fragment was confirmed by nucleotide sequencing (Oregon National Primate Research Center, Core Facility, Beaverton). The primers used for alanine-scanning mutagenesis are listed in Table S2 in the supplemental material. The pSN108 derivative bearing the alanine codon substitution was used to transform competent cells of strain JH642. The plasmid integrated into the rpoA locus of the chromosome by a Campbell recombination mechanism. The selection for elimination of the plasmid vector DNA by loop-out recombination, thus leaving the alanine codon substitution in the rpoA gene, was accomplished according to a previous published procedure (19). The presence of the mutation was confirmed by PCR of the rpoA CTD region followed by cleavage with the restriction enzyme that recognizes the mutated sequence.
Diamide sensitivity.
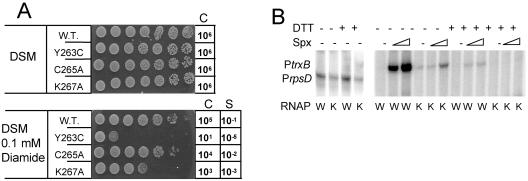
Wild-type B. subtilis strain JH642, ORB3621 (rpoAcxs-1), ORB5259 [rpoA(C265A)], and ORB5262 [rpoA(K267A)] were grown in Difco sporulation medium (DSM) at 37°C with shaking until mid-log phase (optical density at 600 nm = 0.5). Viable-cell numbers were measured by plating 5 μl of cells from a dilution series onto DSM agar medium with or without 0.1 mM diamide. Cells were also spotted onto DSM plates without drug in the same way as above.
Protein purification.
RNAP containing a His10-tagged RpoC (β′) subunit was purified from B. subtilis MH5636 (wild type [WT]), ORB4123 (rpoAcxs-1), ORB5501 [rpoA(C265A)], or ORB6116 [rpoA(K267A)] by using a procedure described previously (29). Intein-tagged ComA was purified using a procedure described previously (25). The self-cleavable affinity tag system IMPACT (New England Biolabs) was used to purify ComA from E. coli strain BL21(DE3)(pLysS). The ComA proteins obtained have a Pro-Gly extension at the C termini and were further purified by elution with a 100-to-600 mM KCl gradient from a High Q column (Bio-Rad). Intein-tagged Spx was purified by using a procedure described previously (26). His6-tagged wild-type, Cxs-16, C10A, and C13A Spx proteins were purified using a previously published procedure (23).
In vitro transcription reactions.
Linear DNA fragments for templates of promoters PrpsD (from −115 to about +71), PsrfA (−347 to about +104) and PtrxB (−220 to about +88) for in vitro transcription were generated by PCR. The oligonucleotides used in PCR to generate promoter fragments are listed in Table S4 in the supplemental material. The transcription reaction mixtures (20 μl) contained 40 mM Tris HCl (pH 7.9), 10 mM NaCl, 6 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol (DTT) (unless otherwise indicated), 10 units RNasin (Promega), 50 nM Psrf or 20 nM PtrxB or 50 nM PrpsD template, 0.05 μM RNAP, and 1.6 μM ComA phosphorylated by treatment with acetyl phosphate as previously described (25). The mixtures were incubated at 37°C for 10 min with or without Spx before the addition of 40 μM ATP, CTP, and GTP, 10 μM UTP, and 5 μCi [α-32P]UTP. After incubation (times are indicated in figure legends and text), the reactions were stopped by addition of 10 μl stop buffer (1 M ammonium acetate, 0.1 mg/ml yeast RNA, 0.03 M EDTA) and then precipitated with 75 μl ethanol at −80°C. Electrophoresis was performed on 6% urea gel as described previously (13).
Assay of β-galactosidase activity.
β-Galactosidase activity was determined as previously described (18) and is presented as Miller units (14).
DNase I footprinting experiment.
A radioactively end-labeled fragment of the srf promoter (from −138 to +65) was made by PCR amplification using primers o-MN02-195 and o-YZ02-4 and JH642 chromosomal DNA as a template. To end-label the template or coding strand, one member of each primer set was treated with T4 polynucleotide kinase and [γ-32P]ATP. The PCR products were separated on a nondenaturing polyacrylamide gel and purified with Elutip-d columns (Schleicher and Schuell). Dideoxy sequencing ladders were obtained using a Thermo Sequenase cycle sequencing kit (USB) with the primers used for the footprinting reactions. DNase I footprinting experiments were performed in 20 μl reaction buffer containing 10 mM Tris HCl (pH 7.9), 30 mM KCl, 10 mM MgCl2, and 0.5 mM β-mercaptoethanol. Proteins were incubated with labeled probe (50,000 cpm) at 37°C for 20 min. The reaction mixtures were treated with 3 μl of 0.02-mg/ml DNase I (diluted in 5 mM MgCl2, 5 mM CaCl2) at room temperature for 15 s (without proteins) or 30 s (with proteins). The reactions were then stopped with 10 μl stop buffer (6.25 mM EDTA [pH 8.0], 0.125% sodium dodecyl sulfate (SDS), 0.375 M sodium acetate, 62.5 μg/ml yeast RNA). After phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation, pellets were dissolved in loading dye and subjected to 6% polyacrylamide-8 M urea gel electrophoresis as previously described (23).
SPPR.
Solid-phase promoter retention (SPPR) experiments used streptavidin-attached agarose beads that bind to biotinylated DNA fragments along with any interacting proteins. Biotinylated DNA fragments were synthesized by PCR with biotinylated 5′ upstream oligonucleotides and underivatized downstream oligonucleotides (see Table S3 in the supplemental material). Streptavidin agarose beads were equilibrated with binding buffer (10 mM Tris-HCl [pH 8.0], 100 mM KCl, 10 mM MgCl2 and 0.5 mM β-mercaptoethanol). The beads were preincubated with the biotinylated DNA fragment for 30 min in binding buffer containing 1% Casamino Acids and 0.1 mg/ml bovine serum albumin (BSA) to shield the nonspecific binding sites on the agarose beads. After the unbound DNA fragment was washed out, protein mixtures were added to the beads in binding buffer containing 0.05 mg/ml yeast RNA, 0.08 mg/ml pUC18 plasmid, and 0.1 mg/ml BSA and then incubated for 1 h at room temperature with gentle shaking. After the unbound protein was washed out by suspension in and centrifugation from binding buffer, the beads were heated at 95°C in SDS loading dye to release the proteins from the agarose beads. SDS-polyacrylamide gel electrophoresis was performed to examine the proteins that were immobilized on the biotinylated DNA-streptavidin agarose complex. The 12% SDS-polyacrylamide gel was stained by colloidal Coomassie G (3), and the images were taken with a UV Transilluminator with a visible-spectrum conversion filter.
RESULTS
Spx-RNAP interaction reduces ComA-assisted binding of RNAP to the srf promoter.
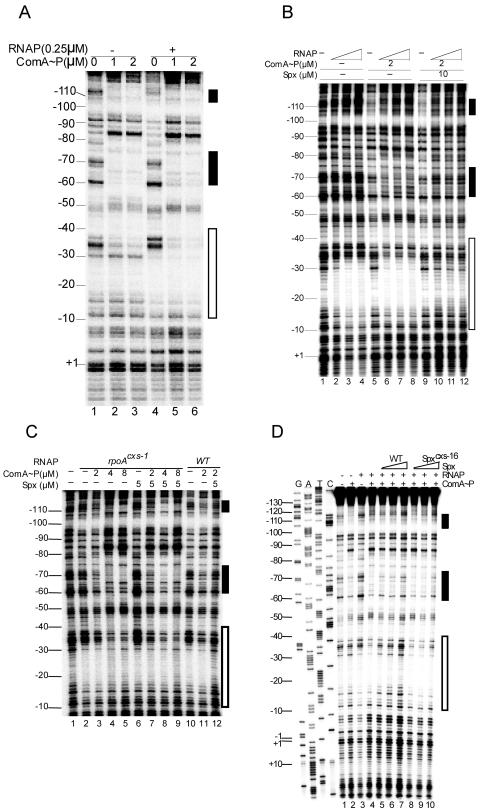
Previous studies provided evidence for a model of Spx-dependent transcriptional repression that involves the direct interference of interaction between the promoter-bound transcriptional activator and RNAP (reviewed in reference 35). This was based on in vitro transcription studies using purified srf promoter DNA and ComA protein phosphorylated by treatment with acetyl phosphate. ComA∼P had been shown to bind upstream of the srf promoter in two ComA boxes (30). However, electrophoretic mobility shift analysis (EMSA) suggested that ComA binding to srf promoter DNA requires its interaction with RNAP (25). The binding of ComA∼P to srf promoter DNA was reexamined, and the effect of Spx-RNAP interaction was investigated. DNase I footprinting showed that ComA∼P is able to bind to ComA boxes 1 and 2 upstream of the srf −35 sequence (Fig. 1A) as previously shown (30). Higher concentrations resulted in protection near the −35 region (Fig. 1A, lane 3). RNAP alone protected sequences between −70 and −90 and between −10 and −30, but protection was extended upstream to the −35 region when ComA∼P was included in the reaction (Fig. 1A, lanes 4 to 6). The ComA-assisted binding of RNAP to the −35 region is observed again in Fig. 1B. While higher concentrations of RNAP seemed to interact with DNA in the region between −10 to −30 in the absence of ComA (Fig. 1B, lanes 2 to 4), protection in the −30 to −40 region was observed when ComA∼P was present (Fig. 1B, lanes 6 to 8). Figure 1B also shows that the addition of Spx protein significantly reduced RNAP binding and weakened ComA∼P binding (lanes 10 to 12).
FIG. 1.
Effect of Spx on binding of ComA∼P and RNAP to the srf promoter region. (A) RNAP and ComA∼P (25) were added to DNase I footprinting reaction mixtures containing the srf promoter fragment synthesized by PCR and end labeled on the noncoding strand. Concentrations of RNAP and ComA are indicated. Reaction conditions are described in Materials and Methods. The two black rectangles indicate the locations of the ComA binding elements, box 1 (upper) and box 2 (lower). The white rectangle marks the site of RNAP-promoter interaction. (B) RNAP and ComA∼P were combined with end-labeled srf promoter DNA as described for panel A, but a gradient of RNAP concentration was tested in the footprinting reaction. RNAP concentrations (from left to right and marked by the white ramped triangle) are 0.25, 0.5, and 1 μM. Spx is included in the rightmost reactions at the indicated concentration. ComA box 2 is indicated by the black rectangle (box 1 is obscured at the top of the gel image). (C) Footprinting reactions containing either WT or rpoAcxs-1 RNAP with and without ComA∼P and Spx. White and black rectangles indicate RNAP and ComA binding sites as in panel A. Protein reaction components were applied in the concentrations indicated. (D) WT RNAP and ComA∼P were added to footprinting reaction mixtures in the absence or presence of Spx or the mutant Spxcxs-16 protein. RNAP, 0.1 μM; ComA, 2 μM; Spx, 5, 10, and 20 μM; Spxcxs-16, 5, 10, and 20 μM.
The rpoAcxs-1 and spxcxs-16 mutations block Spx-dependent inhibition of ComA-assisted RNAP binding to the srf promoter. A concentration of Spx (5 μM) that prevents binding to WT RNAP to the srf promoter (Fig. 1C, lanes 11 and 12) did not have a significant negative effect on binding of RNAP bearing the mutant RpoAcxs-1 subunit (Fig. 1C, lanes 7 to 9). The binding of WT RNAP to the srf promoter in the presence of ComA∼P, while inhibited by WT Spx (Fig. 1D, lanes 5 to 7), was not affected by the inclusion of the mutant inactive Spxcxs-16, which was previously shown to confer reduced interaction between Spx and αCTD (Fig. 1D, lanes 8 to 10). Recently reported structural analysis has shown that the amino acid positions altered by the rpoAcxs-1 and spxcxs-16 mutations define part of the αCTD-Spx interaction interface (27).
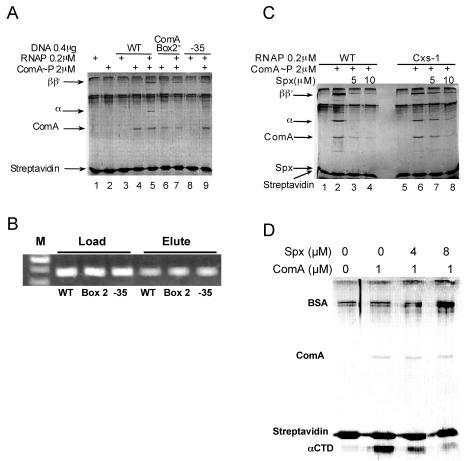
The fact that ComA∼P is able to interact with the srf promoter region and, in doing so, assists RNAP interaction contradicts previously reported EMSA results, which showed poor ComA interaction in the absence of RNAP. Hence, we employed a third method to examine the function of ComA and Spx in RNAP-promoter interaction. The method, the SPPR method, involves immobilization of biotinylated promoter DNA to a streptavidin-agarose bead support. Proteins are added to the bead-bound DNA, and the proteins retained after washing are examined by SDS-polyacrylamide electrophoresis and a colloidal Coomassie blue staining protocol (3). A blocking solution containing a mixture of BSA and Casamino Acids (amino acid solution) was used to prevent nonspecific binding of protein to the streptavidin-agarose beads (Fig. 2A). RNAP β, β′ and α subunits are visible on the gels, but σ is obscured by the BSA band. RNAP binds poorly to the bead-bound srf promoter DNA (Fig. 2A, lane 3), and the addition of ComA∼P to the mixture enhances RNAP retention (lanes 4 and 5). A ComA box 2 mutant version of srf promoter reduces the amount of ComA binding and reduces RNAP retention (Fig. 2A, lanes 6 and 7). The same outcome was observed with a −35 mutant form of the srf promoter (Fig. 2A, lanes 8 and 9), which has been shown to eliminate ComA-stimulated srf transcription (data not shown). This is consistent with the result that the protection observed in the −35 region of srf promoter DNA in footprinting reactions containing RNAP and ComA∼P (Fig. 1) is due to RNAP binding. Figure 2B shows that equal amounts of DNA were applied to the streptavidin-agarose beads and could be recovered by phenol-chloroform extraction from the beads. Thus, the SPPR method provides an authentic picture of ComA/RNAP interaction at the srf promoter.
FIG. 2.
Binding of ComA∼P and RNAP to the srf promoter as observed using SPPR analysis. (A) Biotinylated srf or mutant srf promoter DNA bound to streptavidin beads was combined with RNAP and/or ComA. Bound protein was analyzed by SDS-polyacrylamide gel electrophoresis as outlined in Materials and Methods. WT, box 2 mutant, and −35 mutant srf promoter DNA was used in the indicated reactions. Protein concentrations: RNAP, 0.2 μM; ComA, 2 μM; Spx, 10 μM. (B) Ethidium bromide-stained DNA on a 1% agarose gel. Biotinylated DNA fragments of WT, box 2 mutant, and −35 mutant DNA were applied to the streptavidin beads and extracted with phenol-chloroform from streptavidin agarose beads. (C) Effect of Spx on RNAP and ComA binding to srf promoter DNA as determined by SPPR analysis. RNAP of WT and rpoAcxs-1 (Cxs-1) strains was used in the reactions. ComA∼P phosphorylated by acetyl phosphate and Spx were added in the concentrations indicated. (D) SPPR reactions with purified αCTD and ComA, untreated or treated with Spx. Amounts of proteins in reactions are indicated. The SPPR method is described in Materials and Methods.
The addition of Spx to the SPPR reaction mixture containing ComA∼P and RNAP reduces binding of RNAP to the bead-bound srf promoter DNA (Fig. 2C). When mutant rpoAcxs-1 RNAP was used in the reaction in place of WT RNAP, a reduction in Spx-dependent RNAP release was observed (Fig. 2C, lanes 6 to 8), a result consistent with the DNase footprinting data of Fig. 1 and previously published data (24). In both footprinting and SPPR experiments, Spx substantially reduced RNAP binding to the srf promoter but also reduced ComA-DNA interaction. We conclude from these experiments that ComA is capable of interacting with srf promoter DNA, as was previously shown, but that this interaction is strengthened when RNAP is present, as was observed in the footprinting experiments of Fig. 1.
The hypothesis that ComA interacts with RNAP by binding to αCTD was supported by the observation that ComA can recruit purified αCTD protein to the bead-bound srf promoter DNA. The SPPR experiment of Fig. 2D shows that αCTD cannot interact with srf promoter DNA unless ComA∼P is present. Spx addition causes the release of αCTD. This indicates that promoter-bound ComA∼P can interact with αCTD and this interaction is sensitive to Spx.
Alanine-scanning mutagenesis of RNAP αCTD uncovers residues required for ComA-dependent activation of srf transcription.
We sought to employ lacZ fusion expression, in vitro transcription, footprinting, and SPPR analyses to study the effects of αCTD mutations on ComA and Spx function and to gain a better understanding of how Spx represses transcription. Our objective was to determine if ComA and Spx have overlapping binding surfaces on αCTD, indicating that Spx sterically hinders ComA-RNAP interaction.
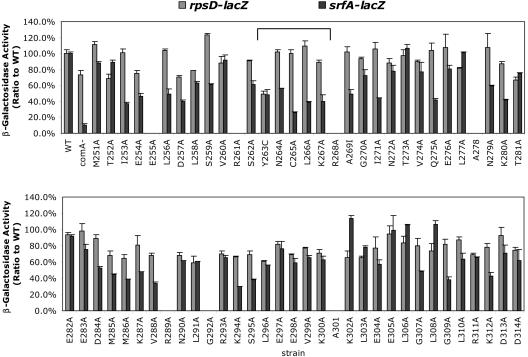
The αCTD-coding region of the B. subtilis rpoA gene was subjected to alanine-scanning mutagenesis, and the resulting mutant alleles were introduced into the rpoA locus by a previously reported procedure (19). The expression of a lacZ fusion controlled by the srf promoter was examined in the αCTD mutants (Fig. 3). The activity of srf-lacZ was tested at the onset of stationary phase, while the mid-log expression of rpsD-lacZ was also monitored as a ComA- and Spx-independent control fusion. The rpsD gene encodes ribosomal protein S4 (10), and its expression is maximal during the middle of exponential phase.
FIG. 3.
Measurement of lacZ fusion activity in B. subtilis strains bearing alanine codon substitutions in the αCTD-coding region of the rpoA gene. β-Galactosidase activity was measured in culture samples collected at the beginning of stationary phase for srf-lacZ-bearing cells and mid-log phase for rpsD-lacZ cells. Activity is expressed as a percentage of the activity measured in rpoA+ cells. The bracket shows the region around α1 of αCTD.
The C265, L266, K267A, K287A, and G307A residues are required for optimal ComA-dependent srf-lacZ expression (Fig. 3). The Y263C mutation, previously shown to confer reduced interaction between Spx and αCTD, has a negative effect on both srf-lacZ and rpsD-lacZ fusions.
Since the region around α1 of αCTD (Fig. 3, bracket) contains the binding interface between RNAP αCTD and Spx, as shown by crystal structure analysis (27), the effects of mutations altering residues C265 and K267 on ComA- and Spx-dependent control of the srf promoter were further examined.
The rpoA(C265A) mutation affects ComA-activated srf transcription and RNAP binding to the srf promoter.
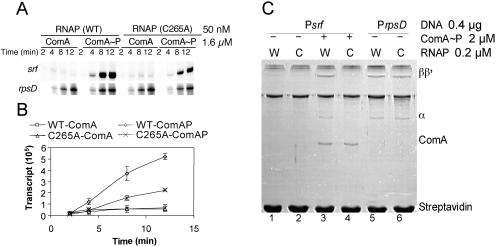
The effect of the C265A mutation in rpoA on srf promoter utilization was examined using in vitro transcription and SPPR analysis. RNAP was purified from the rpoA(C265A) mutant cells and combined with ComA∼P for time course transcription experiments. The reaction mixtures containing mutant RNAP and ComA∼P showed a reduced rate of transcript accumulation (Fig. 4A and B), which was in keeping with the reduced expression of srf-lacZ in rpoA(C265A) mutant cells (Fig. 3). The mutant polymerase showed a level of rpsD transcript accumulation similar to that of wild-type RNAP. SPPR analysis shows that ComA-assisted binding of rpoA(C265A) RNAP to the srf promoter is defective (Fig. 4C, lanes 3 and 4), while no defect in binding of mutant RNAP to the rpsD promoter is observed (Fig. 4C, lanes 5 and 6). The mutation has no detectable effect on Spx repression (see below). The C265A substitution has no effect on ResD-dependent transcriptional activation in vivo and in vitro (H. Geng and M. M. Nakano, unpublished data), indicating that the mutation has a specific effect on ComA-RNAP interaction and does not confer a general defect on RNAP activity.
FIG. 4.
Effect of rpoA(C265A) mutation on srf transcription and on ComA and RNAP binding to srf promoter DNA. (A) Time course in vitro runoff transcription experiment using srf promoter DNA as the template and untreated ComA (ComA) or ComA treated with acetyl phosphate (ComA∼P), plus RNAP or rpoA(C265A) RNAP. (B) Plot of band intensities derived from three repeats of the experiment shown in panel A against time of incubation. (C) Binding of WT and mutant rpoA(C265A) RNAP with ComA∼P to the srf promoter as determined by SPPR analysis. Reactions containing wild-type RNAP (W) and mutant rpoA(C265A) RNAP (C) are indicated.
The rpoA(K267A) mutation affects ComA- and Spx-activated transcription and the Spx-dependent negative control.
Based on the recently published crystal structure of the αCTD-Spx complex, the K267 residue of αCTD contacts the conserved R47 of Spx (27) and might be important for stable RNAP-Spx interaction. A B. subtilis strain bearing the rpoA allele with a K267A codon substitution is hypersensitive to diamide-induced thiol-specific oxidative stress (Fig. 5A), which is the phenotype associated with defective RNAP-Spx interaction. The C265A mutation has no significant effect on diamide resistance, and the Y263C mutation, as shown previously (24), confers hypersensitivity to diamide due to reduced Spx-RNAP interaction and consequent defective oxidative stress response. In vitro transcription analysis of the mutant rpoA(K267A) RNAP in the presence of Spx showed that Spx-stimulated transcription from the trxB promoter is reduced compared to the reaction containing WT RNAP (Fig. 5B). The mutation also appears to have a modest effect on rpsD-lacZ expression (Fig. 3) and transcription from the rpsD promoter in vitro (Fig. 5B).
FIG. 5.
In vivo and in vitro phenotypes of rpoA mutants. (A) Sensitivity of rpoAcxs-1, rpoA(C265A), and rpoA(K267A) mutants to the thiol-specific oxidant diamide. Cultures of WT cells and those of each mutant grown in DSM to mid-log phase were serially diluted to 10−6, and 5 μl of each dilution was spotted onto DSM agar and DSM agar containing diamide. C, control indicating the final dilution spotted that showed growth; S, sensitivity, shown as the approximate fraction of total cells surviving exposure to diamide [(C + diamide)/C − diamide)]. (B) Transcription from the trxB promoter catalyzed by WT RNAP (W) and mutant rpoA(K267A) RNAP (K) in the absence (−) and presence (+) of Spx (0.4 μM and 0.8 μM). Where used, DTT was added to a final concentration of 5 mM. The control transcription reaction mixture contained rpsD gene promoter DNA.
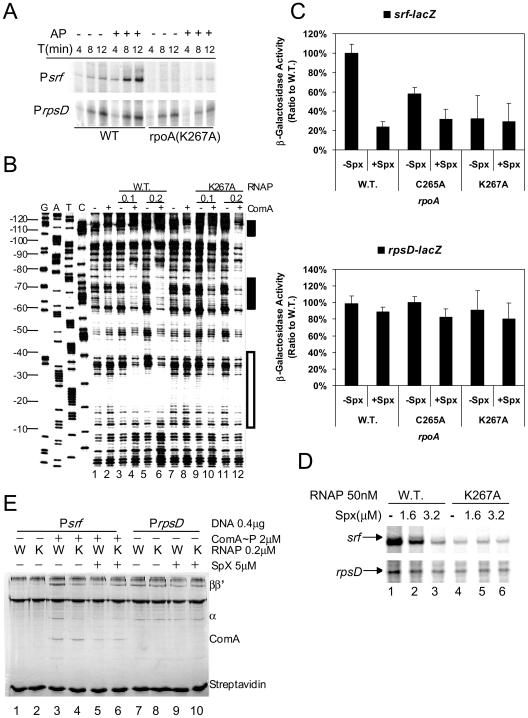
The effect of the rpoA(K267A) mutation on ComA-activated srf transcription and its repression by Spx was examined. In vitro transcription analysis of time course reaction mixtures containing mutant RpoA(K267A) RNAP showed that ComA∼P stimulates transcription catalyzed by the mutant RNAP, but the rate of transcript accumulation was significantly lower than that of reactions containing WT RNAP (Fig. 6A). Defective binding of mutant rpoA(K267A) holoenzyme in the presence of ComA∼P was observed in gels of footprinting reactions. While protection in the −35 region was observed in footprint reactions containing Psrf, wild-type RNAP, and ComA∼P (Fig. 6B, lanes 3 to 6), reduced protection in the −35 region is apparent in the reactions in which the mutant RpoA(K267A) RNAP was added in place of wild-type RNAP (lanes 9 to 12).
FIG. 6.
Effect of rpoA(K267A) mutation on ComA-dependent srf transcription and Spx-dependent repression. (A) Time course in vitro transcription experiment showing the accumulation of srf transcript in reaction mixtures containing untreated (−) or acetyl phosphate (1.6 mM)-treated (+) ComA, and either wild-type RNAP (WT) or rpoA(K267A) RNAP (50 nM). (B) Denaturing gel analysis of DNase I footprint reactions containing Psrf DNA with (+) and without (−) ComA∼P in the presence of wild-type RNAP (WT) or RpoA(K267A) RNAP. RNAP was used at 0.1 μM and 0.2 μM as indicated. (C) Effect of SpxLDD production on levels of β-galactosidase activity in srf-lacZ (ORB6129, ORB6131, and ORB6132) and rpsD-lacZ (ORB6137, ORB6139 and ORB6140) cells bearing the wild-type rpoA allele or the rpoA(C265A) or rpoA(K267A) mutant allele. srf-lacZ fusion-bearing cells were collected from cultures at the end of exponential growth, while rpsD-lacZ cells were collected from mid-log-phase cultures. (D) Effect of Spx (1.6 μM) on transcription of srf in reaction mixtures containing ComA (1.6 μM), acetyl phosphate (1.6 mM), and either wild-type (WT) or rpoA(K267A) RNAP (50 nM). Control transcripts from reactions containing rpsD promoter DNA are shown at the bottom. (E) Effect of rpoA mutations on ComA-dependent RNAP binding to the srf and rpsD promoter and on Spx-dependent RNAP release from promoter DNA. SPPR reaction mixtures contained ComA (+) and either wild-type (W) or rpoA(K267A) (K) RNAP in the presence (+) or absence of Spx.
The reduced in vitro transcriptional activity and ComA-assisted promoter binding of the mutant RpoA(K267A) RNAP is in keeping with the reduced in vivo activity observed in srf-lacZ strains bearing the rpoA(K267A) mutation (Fig. 6C). This low activity was not affected by Spx, as expression of the spxLDD allele (which encodes the protease-resistant form of Spx), while causing repression in rpoA+ srf-lacZ cells, did not result in repression of srf-lacZ in rpoA(K267A) cells. No significant effect of SpxLDD expression or rpoA(K267A) mutation on the expression of rpsD-lacZ (Fig. 6C and D) was observed. The rpoA(C265A) mutation did not prevent Spx-dependent repression, in that reduced expression of srf-lacZ was observed when SpxLDD is produced (Fig. 6C and D), and ComA-dependent srf transcription in vitro was repressed when Spx protein was added to the transcription reaction mixture containing the mutant RpoA(C265A) form of RNAP (data not shown).
SPPR analysis shows that ComA-assisted RNAP binding to Psrf is impaired by the mutant RpoA(K267A) subunit (Fig. 6E, compare lanes 3 and 4). However, interaction of RNAP with the rpsD promoter was not affected by the rpoA(K267A) mutation (Fig. 6E, lanes 7 to 10). Spx disrupted the ComA-RNAP complex at the srf promoter, as shown in SPPR reactions (Fig. 6E, lanes 3 and 5). However, Spx had no significant effect on ComA-assisted promoter binding of RpoA(K267A) RNAP (Fig. 6E, lanes 4 and 6). The K267 amino acid position in the αCTD is important for ComA-dependent transcriptional activation, for Spx-dependent repression, and for Spx-dependent transcriptional activation.
The CXXC motif of Spx is not essential for repression of srf transcription.
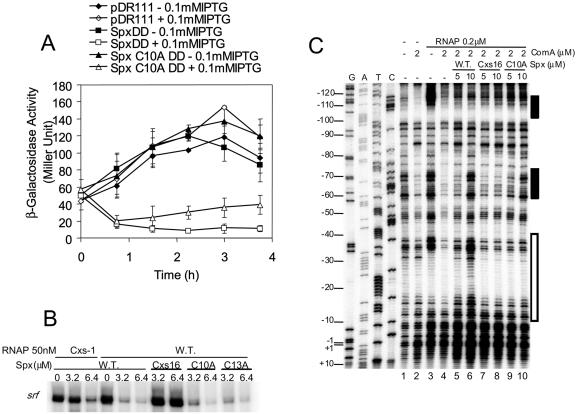
Transcriptional activation by B. subtilis Spx at the trxA and trxB promoters requires the oxidized form of Spx having an intrachain disulfide at the N-terminal CXXC motif (23). It was not known if the CXXC motif was also required for Spx-dependent repression. A protease-resistant form of Spx (24) bearing a C10A substitution in the CXXC motif was produced from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression system in a B. subtilis strain bearing a srf-lacZ fusion. Separate cultures expressing wild-type and C10A mutant forms of SpxLDD were analyzed by Western blotting, and equal amounts of SpxLDD protein were observed in each strain (data not shown). When the SpxLDD protein with the C10A substitution was expressed in IPTG-treated cells, srf-lacZ was repressed to nearly the same level as observed in cells expressing the parental spxLDD construct (Fig. 7A). An attempt was made to express a C13A mutant form of SpxLDD in srf-lacZ cells, but the product was unstable in B. subtilis, and only low levels of protein were detected (data not shown).
FIG. 7.
Effect of amino acid substitutions in the CXXC motif of Spx on Spx-dependent repression of srf transcription. (A) Measurement of β-galactosidase activity in a time course experiment of cultures of srf-lacZ bearing cells expressing either SpxLDD or SpxLDD(C10A). spxLDD and mutant spxLDD(C10A) alleles were expressed from an IPTG-inducible construct derived from pDR111. Vector control cultures are indicated (pDR111), as are spxLDD and mutant spxLDD(C10A) cultures. Data are from two experiments. (B) In vitro transcription data from reactions with WT or rpoAcxs-1 RNAP, ComA∼P (1.6 μM), and increasing concentrations of either Spx, Spxcxs-16, Spx(C10A), or Spx(C13A). (C) Image of denaturing polyacrylamide gel of DNase I footprinting reactions containing end-labeled srf promoter DNA, RNAP, ComA∼P, and either Spx, Spxcxs-16, or Spx(C10A), at the concentrations indicated.
The activity of Spx(C10A) and Spx(C13A) in vitro was examined in transcription reactions and by DNase I footprinting (Fig. 7B and C). In vitro transcription reaction mixtures containing srf promoter DNA, ComA∼P, RNAP, and Spx were assembled to examine Spx-dependent repression. The WT Spx repressed transcription from the srf promoter, while the negative control, Spxcxs-16, showed reduced repressing activity. Both Spx(C10A) and Spx(C13A) repressed transcription nearly to the level of WT Spx (Fig. 7B). Analysis of RNAP-ComA binding to the srf promoter in footprinting reactions showed that the mutant Spx(C10A) had a reduced ability to displace RNAP and ComA compared to WT Spx (Fig. 7C, compare lanes 5 and 6 with lanes 9 and 10). It was concluded that the CXXC motif, while enhancing repression, is not essential for the repressor activity of Spx.
DISCUSSION
Among the genes repressed by Spx, the ComA regulon genes, particularly those of the srf operon, were found to undergo the greatest reduction in transcript levels when Spx interacted with RNAP in cells overexpressing Spx (24). ComA activates transcription of the srf operon by interacting with two regions of dyad symmetry residing upstream from the srf promoter −35 sequence. Transcription requires these interactions as well as RNAP contact with the −35 region, which is assisted by ComA∼P. Our data suggest that ComA interacts with the RNAP αCTD in a region previously shown to contact Spx (27). Thus, Spx blocks productive interaction between ComA and RNAP at the srf promoter by occupying an overlapping site on αCTD.
The Y263C mutation of αCTD reduced rpsD (ribosomal S4) and ComA-dependent srf transcription to nearly the same extent (Fig. 3). This residue is also necessary for functional Spx-RNAP interaction and in response regulator ResD-stimulated transcription (Geng and Nakano, unpublished), which induces anaerobic-specific gene transcription in response to oxygen limitation (22). As detailed previously, this residue is highly conserved in low-GC gram-positive bacteria that also carry spx. These observations reinforce the view that the Y263 residue is an important feature of RNAP in gram-positive organisms.
The footprinting and SPPR data indicate that ComA-RNAP interaction is necessary for ComA-assisted recruitment of RNAP to the srf promoter to form a stable promoter complex. ComA is capable of binding to the srf promoter without RNAP, as observed in previous studies, but RNAP-promoter binding appears to solidify ComA-srf promoter interaction by interaction with ComA. Spx interferes with ComA-RNAP interaction, since addition of Spx to the EMSA (25), footprinting, and SPPR reaction mixtures weakens both RNAP-promoter and ComA-promoter complexes. Footprinting shows protection in the −35 region, particularly nucleotide −30, that is attributable to ComA-assisted RNAP binding. Despite little change in DNase I protection in the −10 region, the interactions observed in the footprinting result represent a productive complex, as shown by in vitro transcription data.
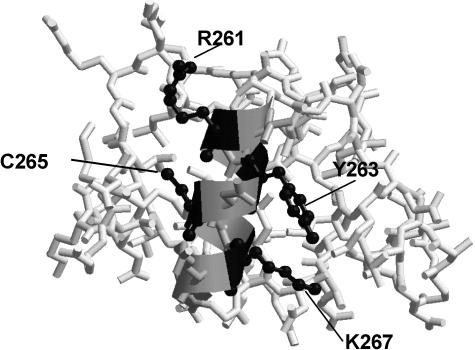
C265 and K267, along with Y263, reside in the α1 helix of αCTD (Fig. 8). The C and K residues correspond to residues C269 and K271 in the E. coli RNAP αCTD. The two residues are located C-terminal to the α1 residues that constitute part of the DNA-binding “265 determinant” of αCTD (2, 7, 31), which includes R265 (R261 in B. subtilis), V264 (V260), and N268 (N264). The conservation of the three residues in B. subtilis and the overall structural similarity between E. coli and B. subtilis αCTD (27) suggests that the B. subtilis αCTD also contains the analogous 265 determinant that binds to extended promoter DNA. The residues required for ComA and Spx interaction with RNAP lie adjacent to the 265 determinant sequences of B. subtilis αCTD. The residues N264, K294, and S295 in B. subtilis αCTD, which correspond to the 265 determinant residues of E. coli αCTD, are required for optimal srf and have a modest effect on rpsD expression. These confer a 40 to 70% reduction in srf-lacZ expression. These data suggest that upstream promoter binding by αCTD is necessary for ComA-activated srf transcription. Alanine substitutions of residues G292 and R261 in B. subtilis αCTD were not recovered in our screen after 20 attempts to obtain mutant rpoA recombinants, raising the possibility that these substitutions were lethal.
FIG. 8.
Structure of B. subtilis RNA polymerase αCTD (27). Stick structures denote peptide backbone and amino acid side chains. The ribbon indicates the α1 helix. The side chains of residues R261, Y263, C265, and K267 are presented as black ball-and-stick structures.
Crystal structure analysis of Spx reveals the two-domain structure of the ArsC homolog (27), a central domain that interacts with RNAP α and the redox domain formed by the N- and C-terminal sequences of Spx and containing the CXXC motif. Two peptide coils connect the central domain with the redox domain. The central domain contacts αCTD at the α1 region and involves the participation of helices α2 and α5 of the Spx central domain. The fact that the CXXC motif is necessary for positive transcriptional control, yet is some distance from the αCTD binding surface of Spx, suggests that Spx may contact other components of RNAP holoenzyme. The transcription factor AsiA of phage T4 contacts σ70 of E. coli RNAP holoenzyme and the flap domain of the β subunit. In doing so, the distance between regions 4 and 2 of σ70 is altered, and σ70 region 4 is now in a position to contact MotA, which is bound to phage T4-specific promoters (8, 28, 32). It is possible that the oxidized form of Spx also contacts σA and, perhaps, β as part of the mechanism of positive transcription control. Preliminary studies indicate that residues in σA region 4.2 are important for Spx-dependent transcriptional activation and repression (Y. Zhang and P. Zuber, unpublished data).
Our results show that the redox disulfide center of Spx, while enhancing the repressor activity, is not essential for negative control. This finding highlights the importance of control mechanisms affecting Spx concentration, which would seem to determine in large part when and under what conditions Spx negative control is exerted. An increase in Spx concentration is observed upon oxidative stress (24), which is the result of increased spx gene transcription (M. Leelakriangsak and P. Zuber, unpublished data) and enhanced Spx stability (24).
. . . . . . .
Supplementary Material
Acknowledgments
We thank H. Geng for assisting in construction of αCTD alanine-scanning mutant library and M. M. Nakano for valuable discussion and critical reading of the manuscript.
Research was supported by grant GM45898 (to P.Z.) from the National Institutes of Health and a grant from the Medical Research Foundation of Oregon (to P.Z.). This work was also supported by a Korea Research Foundation grant (KRF-2004-013-F00001) to S.Y.C.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 3.Candiano, G., M. Bruschi, L. Musante, L. Santucci, G. M. Ghiggeri, B. Carnemolla, P. Orecchia, L. Zardi, and P. G. Righetti. 2004. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25:1327-1333. [DOI] [PubMed] [Google Scholar]
- 4.D'Souza, C., M. M. Nakano, and P. Zuber. 1994. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 91:9397-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubnau, D., J. Hahn, M. Roggiani, R. Piazza, and Y. Weinrauch. 1994. Two-component regulators and genetic competence in Bacillus subtilis. Res. Microbiol. 145:403-411. [DOI] [PubMed] [Google Scholar]
- 6.Dubnau, D., and C. M. Lovett, Jr. 2002. Transformation and recombination, p. 453-471. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 7.Gaal, T., W. Ross, E. E. Blatter, H. Tang, X. Jia, V. V. Krishnan, N. Assa-Munt, R. H. Ebright, and R. L. Gourse. 1996. DNA-binding determinants of the alpha subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 10:16-26. [DOI] [PubMed] [Google Scholar]
- 8.Gregory, B. D., B. E. Nickels, S. J. Garrity, E. Severinova, L. Minakhin, R. J. Urbauer, J. L. Urbauer, T. Heyduk, K. Severinov, and A. Hochschild. 2004. A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc. Natl. Acad. Sci. USA 101:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477-508. [DOI] [PubMed] [Google Scholar]
- 10.Grundy, F. J., and T. M. Henkin. 1992. Characterization of the Bacillus subtilis rpsD regulatory target site. J. Bacteriol. 174:6763-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamoen, L. W., H. Eshuis, J. Jongbloed, G. Venema, and D. van Sinderen. 1995. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol. Microbiol. 15:55-63. [DOI] [PubMed] [Google Scholar]
- 12.Liu, J., W. M. Cosby, and P. Zuber. 1999. Role of Lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation of Bacillus subtilis. Mol. Microbiol. 33:415-428. [DOI] [PubMed] [Google Scholar]
- 13.Liu, J., and P. Zuber. 2000. The ClpX protein of Bacillus subtilis indirectly influences RNA polymerase holoenzyme composition and directly stimulates sigmaH-dependent transcription. Mol. Microbiol. 37:885-897. [DOI] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 15.Msadek, T., V. Dartois, F. Kunst, M. L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27:899-914. [DOI] [PubMed] [Google Scholar]
- 16.Nakano, M. M., F. Hajarizadeh, Y. Zhu, and P. Zuber. 2001. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 42:383-394. [DOI] [PubMed] [Google Scholar]
- 17.Nakano, M. M., R. Magnuson, A. Myers, J. Curry, A. D. Grossman, and P. Zuber. 1991. srfA is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis. J. Bacteriol. 173:1770-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano, M. M., M. A. Marahiel, and P. Zuber. 1988. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 170:5662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano, M. M., Y. Zhu, J. Liu, D. Y. Reyes, H. Yoshikawa, and P. Zuber. 2000. Mutations conferring amino acid residue substitutions in the carboxy-terminal domain of RNA polymerase α can suppress clpX and clpP with respect to developmentally regulated transcription in Bacillus subtilis. Mol. Microbiol. 37:869-884. [DOI] [PubMed] [Google Scholar]
- 20.Nakano, M. M., and P. Zuber. 1989. Cloning and characterization of srfB: a regulatory gene involved in surfactin production and competence in Bacillus subtilis. J. Bacteriol. 171:5347-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano, M. M., and P. Zuber. 1993. Mutational analysis of the regulatory region of the srfA operon in Bacillus subtilis. J. Bacteriol. 175:3188-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano, M. M., P. Zuber, P. Glaser, A. Danchin, and F. M. Hulett. 1996. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J. Bacteriol. 178:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakano, S., K. N. Erwin, M. Ralle, and P. Zuber. 2005. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 55:498-510. [DOI] [PubMed] [Google Scholar]
- 24.Nakano, S., E. Küster-Schöck, A. D. Grossman, and P. Zuber. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 100:13603-13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano, S., M. M. Nakano, Y. Zhang, M. Leelakriangsak, and P. Zuber. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. USA 100:4233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakano, S., G. Zheng, M. M. Nakano, and P. Zuber. 2002. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J. Bacteriol. 184:3664-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newberry, K. J., S. Nakano, P. Zuber, and R. G. Brennan. 2005. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. USA 102:15839-15844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pande, S., A. Makela, S. L. Dove, B. E. Nickels, A. Hochschild, and D. M. Hinton. 2002. The bacteriophage T4 transcription activator MotA interacts with the far-C-terminal region of the sigma70 subunit of Escherichia coli RNA polymerase. J. Bacteriol. 184:3957-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi, Y., and F. M. Hulett. 1998. PhoP-P and RNA polymerase sigmaA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Mol. Microbiol. 28:1187-1197. [DOI] [PubMed] [Google Scholar]
- 30.Roggiani, M., and D. Dubnau. 1993. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J. Bacteriol. 175:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savery, N. J., G. S. Lloyd, S. J. Busby, M. S. Thomas, R. H. Ebright, and R. L. Gourse. 2002. Determinants of the C-terminal domain of the Escherichia coli RNA polymerase alpha subunit important for transcription at class I cyclic AMP receptor protein-dependent promoters. J. Bacteriol. 184:2273-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simeonov, M. F., R. J. Bieber Urbauer, J. M. Gilmore, K. Adelman, E. N. Brody, A. Niedziela-Majka, L. Minakhin, T. Heyduk, and J. L. Urbauer. 2003. Characterization of the interactions between the bacteriophage T4 AsiA protein and RNA polymerase. Biochemistry 42:7717-7726. [DOI] [PubMed] [Google Scholar]
- 33.van Sinderen, D., G. Galli, P. Cosmina, F. de Ferra, S. Withoff, G. Venema, and G. Grandi. 1993. Characterization of the srfA locus of Bacillus subtilis: only the valine-activating domain of srfA is involved in the establishment of genetic competence. Mol. Microbiol. 8:833-841. [DOI] [PubMed] [Google Scholar]
- 34.Youngman, P., H. Poth, B. Green, K. York, G. Olmedo, and K. Smith. 1989. Methods for genetic manipulation, cloning, and functional analysis of sporulation genes in Bacillus subtilis, p. 65-87. In I. Smith, R. A. Slepecky, and P. Setlow (ed.), Regulation of procaryotic development. American Society for Microbiology, Washington, D.C.
- 35.Zuber, P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186:1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuber, P., and R. Losick. 1987. Role of AbrB in the Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J. Bacteriol. 169:2223-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.