Abstract
All examined isolates of the Lyme disease spirochete, Borrelia burgdorferi, naturally maintain numerous variants of a prophage family as circular cp32 episomes. Each cp32 carries a locus encoding one or two different Erp outer membrane, surface-exposed lipoproteins. Many of the Erp proteins bind a host complement regulator, factor H, which is hypothesized to protect the spirochete from complement-mediated killing. We now describe the isolation and characterization of a novel, chromosomally encoded protein, EbfC, that binds specific DNA sequences located immediately 5′ of all erp loci. This is one of the first site-specific DNA-binding proteins to be identified in any spirochete. The location of the ebfC gene on the B. burgdorferi chromosome suggests that the cp32 prophages have evolved to use this bacterial host protein for their own benefit and that EbfC probably plays additional roles in the bacterium. A wide range of other bacteria encode homologs of EbfC, none of which have been well characterized, so demonstration that B. burgdorferi EbfC is a site-specific DNA-binding protein has broad implications across the eubacterial kingdom.
Every Lyme disease spirochete isolated from nature has been found to contain multiple members of a DNA element family that replicate as circular plasmids (57). Those plasmids are designated “cp32s” in light of their circular plasmid nature and approximate size of 32 kb (11, 12, 56, 57). All cp32s are essentially identical to each other except in three loci: one that is involved in plasmid replication and segregation and two loci that encode surface-exposed lipoproteins (18, 48, 52, 57, 63). Intriguingly, cp32s appear to be the genomes of lysogenic bacteriophages, which evidently infect all Lyme spirochetes (12, 15, 19, 20, 57, 62).
Each cp32 element carries a mono- or bicistronic erp locus, which often varies in sequence between plasmids (51). The Erp lipoproteins are among the specific repertoire of proteins synthesized by Borrelia burgdorferi during mammalian infection (3, 16, 21, 26, 28, 43-46, 50, 51, 57). Synthesis of Erp proteins during mammalian infection is consistent with a known function of some Erp proteins: the binding of host serum protein factor H, an important fluid phase regulator of the alternative pathway of complement activation (4, 29, 32, 33, 42, 53). Factor H is normally bound by receptors on surfaces of host cells, where it protects those cells by inhibiting C3 cleavage and promoting C3b degradation. Binding of factor H via Erp and other B. burgdorferi outer-surface proteins is hypothesized to likewise protect the pathogen from complement-mediated destruction (34, 35, 37).
To date, the cp32 family members and erp loci of four different B. burgdorferi isolates have been characterized. The B. burgdorferi species type strain, B31, contains 10 different cp32 family members and 17 erp genes. Since one bicistronic locus is present in three identical copies and another locus, erpH, is naturally defective, individual bacteria of strain B31 can simultaneously produce up to 13 distinct Erp lipoproteins (11, 12). Strains 297 and Sh-2-82 proved to be nearly clones of each other, holding 9 cp32s and 13 erp genes in common, including 3 copies of a bicistronic locus (2, 54). As evidence of naturally occurring transduction of cp32s among B. burgdorferi strains, strain Sh-2-82 also contains an additional cp32 that appears to be identical to a cp32 of strain B31 (54). Strain N40 carries six cp32s and nine unique erp genes (54). Considerable diversity occurs between erp gene sequences, both within an individual bacterium and between strains, giving each Erp protein unique functional and antigenic characteristics (2, 51, 54, 57). However, a unifying feature of all erp genes is that each locus is preceded by a highly conserved DNA sequence. Within that region are the transcriptional promoter and two separate sites that specifically bind different bacterial cytoplasmic proteins (Fig. 1) (7, 22, 41, 51, 56, 57). Our laboratory previously demonstrated that the protein-binding region closest to the erp promoter, designated operator 2, is required for proper regulation of erp transcription (7). Continuing those studies, we used DNA affinity chromatography to purify a B. burgdorferi protein that binds specific DNA sequences within the boundaries of erp operator 2, which we identified as the novel protein EbfC. Characterization of the DNA sequences specifically recognized by that chromosomally encoded protein are also presented.
FIG. 1.
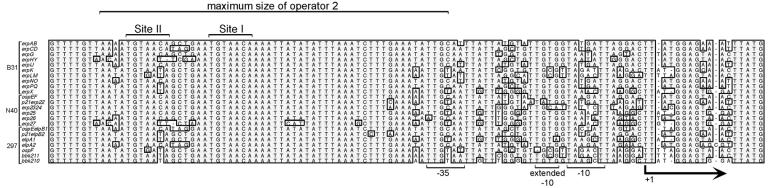
Alignment of noncoding DNA sequences located immediately 5′ of the determined erp loci of B. burgdorferi strains B31, N40, and 297 (the erp loci of strain Sh-2-82 are identical to those of strain 297 plus the erpNO locus of strain B31). Identical nucleotides found in the majority of loci are boxed and shaded. Initiation methionine codons of the first gene in each locus are to the far right. The maximum size of erp operator 2, as determined previously using competitive EMSA and transcriptional fusions to reporter genes (7), is indicated. The two EbfC-binding sites (TGT[A/T]ACA) determined by the present work are indicated as Site I and Site II above the alignment.
MATERIALS AND METHODS
Bacteria.
All studies used a virulent, clonal subculture of the B. burgdorferi species type strain, B31-MI-16 (46). Spirochetes were cultured at 34°C in Barbour-Stoenner-Kelly II (BSK-II) medium (8). Soluble protein extract was prepared from B. burgdorferi as previously described (6, 7).
DNA affinity purification.
A segment of 5′ noncoding DNA from the strain B31 erpG locus, containing the promoter and both identified operator sites, was amplified by PCR using oligonucleotide primers R8 and biotinylated E43 (Table 1) (6, 7, 56). Due to the modification of oligonucleotide E43, the resulting amplicon contained a biotin moiety at the end distal to the erp promoter. Reaction mixtures were separated by agarose gel electrophoresis followed by extraction and purification of the amplicon. The biotin-conjugated amplicon was then affixed to streptavidin magnetic beads (Dynal, Brown Deer, WI) as follows. Magnetic beads were washed twice in equal volumes of 2× binding and wash buffer (2× B&W; 10 mM Tris [pH 7.5], 1 mM EDTA, 2 M NaCl) using a magnetic stand (Dynal) to adhere the beads to the side of the tube during removal of the wash buffer. Beads were then resuspended in 550 μl 1× B&W containing the biotinylated PCR amplicon, incubated at room temperature for 30 min, and then washed three times with equal volumes of 2× B&W buffer. The beads were next washed twice in BS/THES buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 50 mM KCl, 20% glycerol, 20 mM Tris, 5 mM EDTA, 90 mM NaCl, and protease and phosphatase inhibitor cocktails [both from Sigma, St. Louis, MO] at concentrations of 6.7 μg and 1.7 μg per ml, respectively) and then once in BS/THES buffer containing 30 μg/ml poly(dI-dC). Following these treatments, beads were incubated for 15 min on ice with 7.5 mg B. burgdorferi soluble protein extract in BS/THES buffer plus 200 μg poly(dI-dC), the supernatant was removed, and the beads were then incubated again with a second, similar aliquot of protein extract plus poly(dI-dC). Next, the beads were washed twice with BS/THES buffer containing 30 μg/ml poly(dI-dC). Bound proteins were sequentially eluted by washing with 50 mM Tris [pH 7.5] and 10 mM EDTA plus either 500 mM, 750 mM, or 1 M NaCl. Aliquots of eluted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by staining with SYPRO-Ruby (Molecular Probes, Eugene, OR). Protein bands were extracted and analyzed by matrix-assisted laser desorption ionization-time of flight (mass spectrometry) (University of Louisville, Louisville, KY). Spectrometry results were compared with the known sequence of B. burgdorferi strain B31 using Mascot (Matrix Science, Boston, MA).
TABLE 1.
Oligonucleotides used in this work (all sequences are shown 5′ to 3′)
| Function or name | Sequence |
|---|---|
| DNA affinity purification and EMSA probe production | |
| Bio-E43 | Biotin-AAAATTTTAGTCAAATTTGGAGTG |
| R8 | GCAATATTTCAAAGATTTAAA |
| Bio-G14A | Biotin-TTGTAATGAGTAGAGCATTTG |
| VLSF-27B | Biotin-AATAGTTTGCCTAAGGAAAAAAGCG |
| VLSR-8 | CAACTTCTCCAATTGCAGCAGTAC |
| Cloning of ebfC | |
| BATA-1 | GCTTCAAAATTATAAAGG |
| BATA-2 | AATTATAAAGCACTTAAC |
| 462-M | CACCATGGCAGTAAATCCGTTAG |
| 462-L | CACCTTGGAGCAAGTGAAGTTCTG |
| 462-R | CTACATTCCAAAAGGAAGAACTCC |
| EMSA competitor production | |
| R8 | GCAATATTTCAAAGATTTAAA |
| G14A | TTGTAATGAGTAGAGCATTTG |
| 104F | TGTTAAAATGTAACAGCTGAATGTAACAAA |
| 104R | TTTGTTACATTCAGCTGTTACATTTTAACA |
| 100F | TAAAATGTAACAGCTGAATGTAACAAAATTATAT |
| 100R | ATATATAATTTTGTTACATTCAGCTGTTACATTTTA |
| 100NCF | AAATGTAACAGCTGAATGTAACAAAAT |
| 100NCR | ATTTTGTTACATTCAGCTGTTACATTT |
| 100NC-2F | GAATGTAACAAAAT |
| 100NC-2R | ATTTTGTTACATTC |
| 100-1F | ATGTAACAGCTGAATGTA |
| 100-1R | TACATTCAGCTGTTACAT |
| 100-2F | GAATGTAACAAAATTATAT |
| 100-2R | ATATAATTTTGTTACATTC |
| 100-3F | AAATGTAACAATCAGGTGTAACAAAAT |
| 100-3R | ATTTTGTTACACCTGATTGTTACATTT |
| 100-4F | AAACACAACAGCTGAATGTAACAAAAT |
| 100-4R | ATTTTGTTACATTCAGCTGTTGTGTTT |
| 100-5F | AAATGTAACAGCTGAACACAACAAAAT |
| 100-5R | ATTTTGTTGTGTTCAGCTGTTACATTT |
| 100-6F | AAATGTAACAAAATAATGTAACAAAAT |
| 100-6R | ATTTTGTTACATTATTTTGTTACATTT |
| 100-7F | AAATGTAACAGCTGAATGTAACAGCTG |
| 100-7R | CAGCTGTTACATTCAGCTGTTACATTT |
| 100-8F | GAATGTAACAGCTG |
| 100-8R | CAGCTGTTACATTC |
| 100-9F | GCCTGTAACAAAAT |
| 100-9R | ATTTTGTTACAGGC |
| 100-10F | GAATGTAACAGCTGTATA |
| 100-10R | TATACAGCTGTTACATTC |
| 64F | ACAAAATTATATATTTAAATCTTTGAAATA |
| 64R | TATTTCAAAGATTTAAATATATAATTTTGT |
| FLA-6 | TTCAGGGTCTCAAGCGTCTTG |
| FLA-7 | GCATTTTCAATTTTAGCAAGTG |
Recombinant EbfC.
As a first step, a region of the B. burgdorferi strain B31-MI-16 chromosome extending from approximately 100 bp 5′ of ebfC through approximately 60 bp 3′ of the gene was PCR amplified using oligonucleotide primers BATA-1 and BATA-2 (Table 1). The resulting amplicon was cloned into pCR2.1 (Invitrogen) and the insert completely sequenced. Using this plasmid clone as a template, the ebfC open reading frame (ORF) was amplified using either oligonucleotide primer 462-L (for the presumed lysine start site) or 462-M (for the presumed methionine start site) in conjunction with 462-R (reverse primer). Each of the two PCR products was cloned into the pET200 Champion TOPO expression vector (Invitrogen). The insert of one clone of each was completely sequenced to confirm that errors had not been introduced during the PCR or cloning processes. Recombinant proteins were produced in Escherichia coli BL-21 Star (pLysS) (Novagen, San Diego, CA) and overnight dual media (Zymo Research, Orange, CA). Bacteria were lysed by lysozyme treatment and sonication, debris was cleared by centrifugation, and recombinant EbfC was purified using Ni-nitrilotriacetic acid spin kits (QIAGEN). Recombinant protein quality and purity were assessed by SDS-PAGE followed by either staining with Coomassie brilliant blue or immunoblotting with horseradish peroxidase-conjugated anti-His-tag antibodies (QIAGEN) and chemiluminescence.
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays (EMSAs) were performed using a biotin-labeled probe and light shift chemiluminescence (Pierce), as previously described (7). The probe used for specific binding studies was a 124-bp fragment of the B. burgdorferi strain B31 erpG 5′ noncoding DNA, PCR amplified from a recombinant plasmid template (56) using oligonucleotide primers Bio-G14A and R8 (Table 1). With the exception of competitor 1 (a nonbiotinylated version of the 124-bp erp probe DNA) and the PflaB competitor, double-stranded competitor DNAs were produced from complementary single-stranded DNAs by heating the two DNAs together and then slowly cooling them to room temperature (7). Oligonucleotides used to produce competitor DNAs are listed in Table 1, with each competitor being produced from the two oligonucleotides bearing the same number-letter designation and either “F” or “R” (e.g., c100 from 100F plus 100R, c104 from 104F plus 104R, etc.). Competitor 1 was produced in the same manner as the biotinylated erp probe, except with the use of nonbiotinylated oligonucleotide G14A. The PflaB competitor was produced by PCR from a cloned flaB gene using oligonucleotides FLA-6 plus FLA-7 and purified as previously described (6). All competitor DNAs were used in 100-fold excess over the labeled probe.
An internal fragment of the B. burgdorferi strain B31 vlsE gene was amplified from a plasmid clone of that gene, using oligonucleotides VLSF27-B and VLSR8, and used as a labeled EMSA probe for examination of nonspecific DNA binding by EbfC.
Size fractionation chromatography.
The ability of the polyhistidine-tagged recombinant EbfC protein to form multimers was determined by gel filtration chromatography, using a Waters 600 pump and controller equipped with a Waters 996 photodiode array UV/Vis detector (Waters, Milford, MA). A Superdex 75 10/300 GL column (GE Healthcare) was prepared with a mobile phase consisting of 200 mM NaCl, 50 mM Tris-HCl (pH 7.5), and 1% (vol/vol) glycerol. The column was run with a flow rate of 0.20 ml per min. The elution of each standard was determined by monitoring the A280. A calibration curve was created using an MW-GF-70 low-molecular-weight calibration kit (Sigma-Aldrich), and the void volume, V0, was determined by injection of 200 μl of 1 mg/ml blue dextran in elution buffer with 5% glycerol. The remaining protein standards, bovine lung aprotinin (6.5 kDa), horse heart cytochrome c (12.4 kDa), bovine carbonic anhydrase (29 kDa), and bovine serum albumin (66 kDa), were individually prepared in elution buffer with 5% glycerol to a total concentration of 0.3 mg/ml each. The molecular-mass calibration curve was generated by plotting the log (molecular mass) versus V0/Ve (5). A 200-μl sample of recombinant EbfC (approximately 0.2 mg/ml) was then injected and its elution compared to the established curve.
Protein cross-linking.
Aliquots (10 μg) of purified recombinant EbfC in band shift buffer (10 mM HEPES [pH 8.0], 50 mM KCl, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride) (6) were incubated at room temperature for 20 min, and then formaldehyde was added to a final concentration of 2% (vol/vol) and the aliquots were incubated for an additional 20 min at room temperature. As a control, protein was also incubated without added formaldehyde. Proteins were then separated by SDS-PAGE, transferred to nitrocellulose membranes, and detected by immunoblotting using horseradish peroxidase-conjugated anti-His-tag antibodies (QIAGEN) and chemiluminescence.
RESULTS
Identification of EbfC.
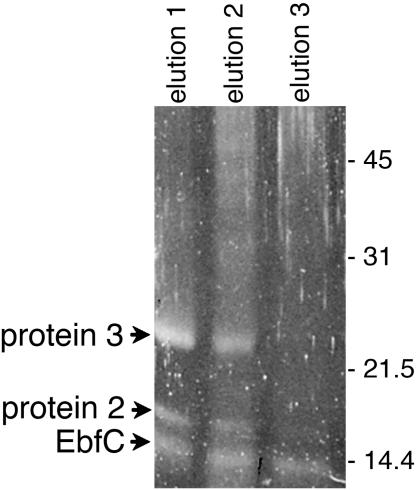
We previously used a fragment of the strain B31 erpG 5′ noncoding region in EMSAs to identify DNA sequences that specifically bound cytoplasmic proteins. A similar DNA fragment, containing all identified protein-binding regions, was therefore used as bait to fish for those proteins by DNA affinity chromatography. Biotinylated DNA was affixed to avidin-linked magnetic beads, incubated with B. burgdorferi cytoplasmic extract, and washed extensively, and then bound proteins were eluted. This protocol yielded three proteins which eluted under high-salt-concentration wash conditions, having molecular masses of approximately 11, 16, and 25 kDa (Fig. 2). Eluted proteins were separated by SDS-PAGE, extracted from gels, and analyzed by matrix-assisted laser desorption ionization-time of flight (mass spectrometry). Results were compared with the determined genome sequence of strain B31 to provide potential identities to the erp operator-binding proteins. The 11-kDa protein was further characterized and designated as “EbfC” (erp-binding factor, chromosomal), as described below. The other two purified proteins have yet to be fully characterized.
FIG. 2.
B. burgdorferi cytoplasmic proteins purified by affinity chromatography using erpG 5′ noncoding DNA as bait. Proteins in elutions 1, 2, and 3 were eluted with NaCl at concentrations of 500, 750, and 1,000 mM, respectively. Proteins were separated by SDS-PAGE and visualized with SYPRO-Ruby. Numbers on the right indicate positions of molecular mass standards.
Four peptide fragments of the 11-kDa protein matched the predicted amino acid sequence of a chromosomal ORF of strain B31, providing 30% coverage (data not shown). This ORF is annotated as ORF BB0462 in the strain B31 genome database and ORF BG0475 in the sequence database of the related Lyme disease spirochete Borrelia garinii strain PBi (25, 27). Intriguingly, many other bacteria encode orthologs of this protein, which have been grouped as “domain of unknown function” (DUF)-149, Pfam 2575, COG-0718, “YbaB”-like, and “YaaK”-like proteins (40). None of those related proteins appear to have been functionally defined prior to the current work. Serendipitously, the Haemophilus influenzae ortholog (Fig. 3) was crystallized and its three-dimensional structure solved by the University of Maryland Center for Advanced Research in Biotechnology as part of their “Structure to Function” project to solve structures of otherwise uncharacterized proteins (38) (http://s2f.umbi.umd.edu). The H. influenzae DUF-149 family member forms a homodimer containing a pair of protruding, parallel alpha helices that form a “pincerlike” shape. The gap between the two “pincer” arms is approximately the same width as the diameter of a DNA double helix, although the recombinant H. influenzae protein did not bind DNA nonspecifically (38). To the best of our knowledge, no further information on that protein or any other EbfC homolog has been published.
FIG. 3.
Alignment of the predicted amino acid sequences of B. burgdorferi EbfC and a homologous protein encoded by H. influenzae. Identical amino acids found in both proteins are boxed in black and similar residues in gray. The three-dimensional structure of the H. influenzae protein has been determined, but it has not been otherwise characterized (38).
We next assessed whether the B. burgdorferi EbfC protein could specifically bind erp operator DNA. Our examination of the B. burgdorferi strain B31 chromosome sequence suggested a 99-codon ebfC gene that could encode an 11.0-kDa protein, initiating with an AUG methionine codon and preceded by a near-consensus GGAGGGA Shine-Delgarno sequence. Use of that translational initiation site yields an amino-terminal protein sequence comparable to those of EbfC homologs of other bacterial species (Fig. 3 and data not shown). However, annotation of strain B31 by the Institute for Genomic Research proposed a sequence with a leucine initiation codon 11 residues further 5′, with a very weak AGA Shine-Delgarno sequence, encoding a 12.5-kDa protein (25). The Institute for Genomic Research annotation did not provide a rationale for suggesting use of that weak translation initiation site. Two recombinant, N-terminal polyhistidine-tagged proteins were produced from strain B31 DNA, one beginning at the methionine codon and a second, longer protein extending from the leucine codon. Both purified proteins were analyzed by EMSA, and both bound comparably well to erpG operator/promoter DNA (Fig. 4 and data not shown), suggesting that the residues between the postulated leucine and the first methionine do not contribute to DNA binding. The three-dimensional structure of the H. influenzae EbfC homolog places the protein's amino terminus at the end of an alpha helix, apart from the remainder of the protein and distal to the proposed DNA-binding site (38). These data, together with the DNA sequence comparisons described above, suggest that the methionine is the most likely initiation amino acid of native EbfC. The shorter recombinant protein was used for all further studies of EbfC function.
FIG. 4.
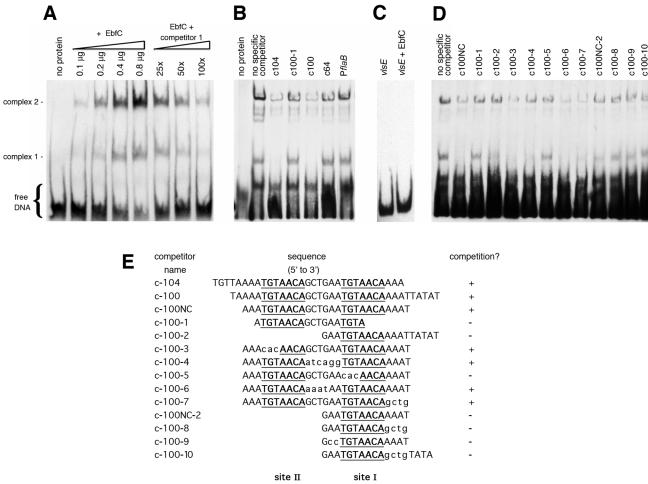
EMSA using a labeled 124-bp fragment of erpG 5′ noncoding DNA, recombinant EbfC protein, and various double-stranded DNA competitors. DNA alone is labeled “free.” Addition of EbfC resulted in two major protein-DNA complexes. Additional complexes are also visible, possibly due to EbfC-mediated aggregation of DNA (see the text). (A) Effects of addition of increasing concentrations of EbfC or unlabeled competitor on formation of EbfC-DNA complexes. Second through fifth lanes, addition of 0.1, 0.2, 0.4, or 0.8 μg EbfC to labeled erpG promoter/operator DNA; sixth through eighth lanes, labeled erpG DNA plus 0.8 μg EbfC plus 25-fold, 50-fold, or 100-fold excesses, respectively, of unlabeled DNA competitor 1 (the same 124 bp of erpG promoter/operator DNA). (B) Competition studies using large DNAs spanning EbfC-binding sites I and II (c104 and c100) or DNAs lacking those two sites (c100-1 and c64). The promoter region of the constitutively expressed B. burgdorferi flaB gene was included as a competitor for nonspecific protein binding. (C) Analysis of a 184-bp labeled fragment of B. burgdorferi vlsE, without and with added EbfC (0.8 μg), further demonstrating the DNA specificity of EbfC binding. (D) Competition studies using smaller DNAs containing either wild-type EbfC-binding sites or mutants thereof. (E) Sequences of DNA competitors used in the EMSAs shown in panels B and D. Consensus EbfC-binding sites I and II are illustrated in boldface type and are underlined. Nucleotides within competitors that differ from those of the erpG promoter/operator are indicated by lowercase letters. The ability of a competitor to prevent EbfC binding to the biotinylated probe was scored as plus (inhibition of complex 1 and >50% inhibition of complex 2) or minus (no detectable inhibition of either complex).
EMSA using purified EbfC yielded two specific DNA-protein complexes (Fig. 4). EMSA signals from both complexes increased in intensity with the addition of increasing concentrations of EbfC (Fig. 4A). Similarly, EMSA signals of both complexes diminished as increasing levels of unlabeled, specific competitor were added (Fig. 4A). This may have been due to the presence of two identical, adjacent EbfC-binding sites in the erp operator DNA (see below). Alternatively, DNA-EbfC complexes may form higher-ordered structures, as suggested by the ability of EbfC to form homotetramers in solution (see below). The more slowly migrating complex 2 appeared to represent a somewhat higher affinity DNA-protein complex than did complex 1.
Competition analyses using 100-fold excesses of unrelated DNAs, including the 5′ noncoding region of the constitutively expressed B. burgdorferi flaB gene, indicated that EbfC binding to the erp operator DNA was sequence specific (Fig. 4B). EMSA using a labeled 184-bp fragment of the B. burgdorferi vlsE gene as the probe did not detect any binding by EbfC, further indicating the specificity of that DNA-binding protein (Fig. 4C). As additional controls, EMSAs were performed using cellular extracts from E. coli, but an electrophoretic mobility shift was not observed in those studies (data not shown), demonstrating that it was the recombinant proteins and not potential contaminating E. coli proteins that were responsible for the mobility shifts.
EbfC forms dimers and higher-ordered multimers.
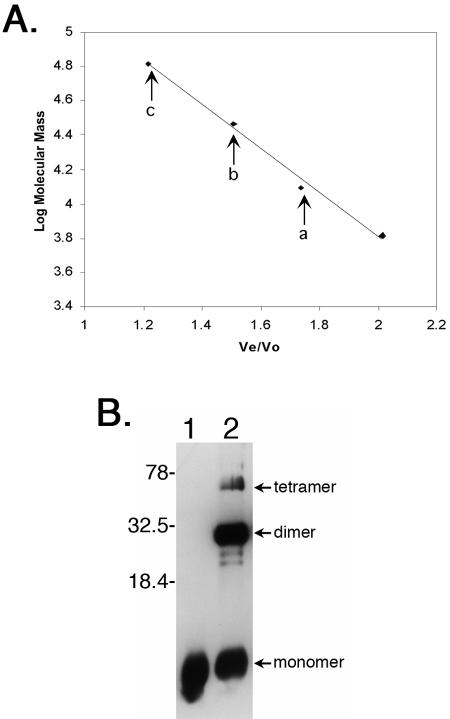
Since the EbfC homolog produced by H. influenzae formed a dimer when crystallized (38), we examined the B. burgdorferi EbfC for its ability to multimerize. To that end, purified recombinant EbfC protein was applied to a size fractionation column, and apparent molecular mass was determined. While a proportion of EbfC eluted with an apparent molecular mass of 13.8 kDa (corresponding with the calculated mass of EbfC plus the fusion partner polypeptide), additional peaks were obtained having apparent masses of 26.9 and 64.1 kDa, corresponding with the calculated masses of EbfC dimers and tetramers (Fig. 5A).
FIG. 5.
EbfC forms dimers and higher-ordered multimers in solution. (A) Size fractionation analysis of recombinant EbfC, with arrows denoting Ve/V0 values of three 280-nm-absorbing peaks corresponding to monomer (a), dimer (b), and tetramer (c) forms of the protein, having apparent molecular masses of 13,800, 26,900, and 64,100 Da, respectively. Diamonds indicate elution positions of molecular mass standards, left to right: bovine serum albumin (66,000 Da), bovine carbonic anhydrase (29,000 Da), horse heart cytochrome c (12,400 Da), and bovine lung aprotinin (6,500 Da). (B) Cross-linking of purified recombinant EbfC in solution. Lane 1, no formaldehyde cross-linking agent added; lane 2, protein incubated with formaldehyde. Protein bands with molecular masses corresponding to monomeric, dimeric, and tetrameric EbfC are indicated. Note that the recombinant protein is larger than wild-type EbfC due to the inclusion of the N-linked polyhistidine tag and linker residues (approximately 14 kDa versus 11 kDa for native EbfC). Numbers on the right indicate positions of molecular mass standards.
Multimerization of EbfC was also examined by incubation of purified recombinant protein with formaldehyde, which causes cross-linking between closely associated proteins (36), followed by separation by SDS-PAGE. In addition to the EbfC monomer, a band having the size of a dimer was detected (Fig. 5B). Both those protein bands were of comparable intensity, indicating a strong propensity of EbfC to dimerize in solution. Furthermore, a significant protein band of a size corresponding with 4 EbfC subunits was also observed.
Identification of EbfC-binding DNA sequences.
A variety of unlabeled, double-stranded DNAs were next used as EMSA competitors to identify DNA sequences capable of binding EbfC. For each competitor, two complementary oligonucleotides were synthesized and then annealed together, allowing us to efficiently test various fragments of erp 5′ DNA and mutants thereof. A 100-fold excess of each competitor DNA was used for each analysis.
Both the 31-bp c104 and the 34-bp c100 DNAs effectively competed away EbfC from the labeled erp operator DNA probe (Fig. 4B and D). A shorter, 27-bp DNA fragment, c100NC, consisting of a sequence contained within c100, also competed away EbfC binding (Fig. 4D). All three competitor DNAs completely inhibited formation of complex 1 and reduced complex 2 formation by greater than 50%. These data indicate that a specific EbfC-binding site(s) is located within the sequences shared by c100, c100NC, and c104. This DNA is within the maximum boundaries of erp operator 2 that we previously mapped through use of EMSA with larger DNA competitors and transcriptional fusions to a reporter gene (7). Deletion of erp operator 2 resulted in constitutive expression, indicating that a DNA sequence(s) within that region is necessary for regulation of transcription (7).
The ability of EbfC to form dimers and the known structure of the homologous H. influenzae protein provided important clues for the further characterization of the B. burgdorferi EbfC-binding site. Homodimers generally interact with palindromic DNA sequences, with each subunit binding one side of the palindrome. Two different palindromes are contained within the sequence of competitor c-100NC: CAGCTG and two copies of TGT(A/T)ACA (Fig. 4E). The CAGCTG sequence within c-100NC was mutated to either CAATCA (c100-4) or CAAAAT (c100-6), but both still effectively competed for EbfC binding, indicating that the CAGCTG palindrome is not directly involved in EbfC interactions (Fig. 4).
Mutation of the 3′ TGTaACA sequence to CACAACA in c100-5 prevented competition (Fig. 4). In contrast, that same mutation of the identical 5′ TGTAACA palindrome in c100-3 did not detectably influence EbfC competition. These results indicate that the TGT(A/T)ACA palindrome is involved in the binding of EbfC, although flanking DNA is also important. We designated the 3′ TGTAACA sequence of the erp operator as site I and the 5′ sequence as site II (Fig. 1 and 4). Alterations of the sequences immediately 5′ or 3′ of site I on competitors c100-4 and c100-7, respectively, did not perceptibly affect EbfC binding, indicating that specific flanking DNA sequences are not required for interactions with EbfC. DNAs that contained 4 or more base pairs on either side of the palindrome effectively competed for EbfC binding, while those palindromes with only 3 flanking base pairs did not compete (Fig. 4). Together, these data suggest that efficient EbfC binding requires a TGTaACA sequence bordered by at least 4 nonspecific bases. Those additional bases may be important for correct formation of the DNA double helix in the competitor or may nonspecifically interact with the EbfC protein.
All erp loci from every examined strain of B. burgdorferi contain an absolutely conserved EbfC site I (Fig. 1), suggesting that this sequence is critical to the bacterium and/or the cp32 prophage. On the other hand, considerable variation was observed among site II sequences, with several loci containing sequences in that location that our data predict to be unable to efficiently bind EbfC.
Searches of the B. burgdorferi strain B31 genome indicated that the palindrome TGT(A/T)ACA occurs only 4 to 7 times in each cp32 element and 92 times on the chromosome, a frequency of approximately 1 site every 5 to 10 kb. Other than the two palindromes described above, there are no further copies of the sequence within several kilobases of erp loci. erp operator 2 is the only locus in the B. burgdorferi strain B31 genome in which two EbfC-binding palindromes were found in close proximity.
Additional borrelial proteins bind erp operator 2.
In a previous study, we demonstrated that one or more proteins in B. burgdorferi cell extracts bind to erp operator 2 DNA (7). Moreover, EMSAs revealed a strong DNA-protein complex whose intensity was inversely proportional to the levels of erp transcription (7), suggesting that the observed complex represented a transcriptional repression mechanism. Comparing EMSAs of labeled erp operator DNA incubated with either B. burgdorferi cell lysates or purified recombinant EbfC revealed different band shift patterns for each (Fig. 6). Noting that the B. burgdorferi cell extract includes EbfC (Fig. 2), we conclude that at least one other borrelial protein binds erp operator 2 DNA to repress erp transcription levels.
FIG. 6.
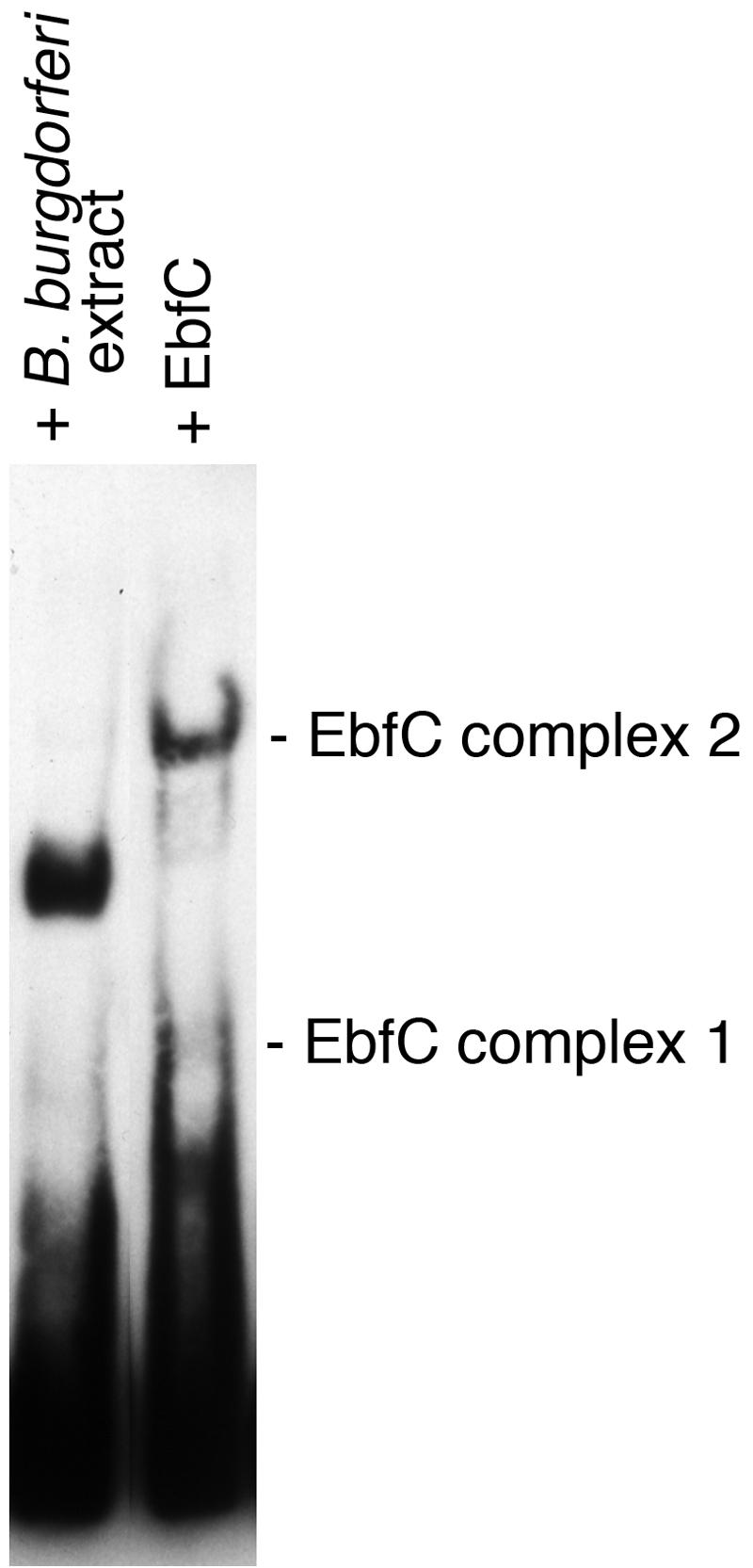
EMSAs of B. burgdorferi cell extract and purified recombinant EbfC binding to labeled erp operator DNA. Although the bacterial extract contains EbfC (Fig. 2), it appears that at least one additional protein contributes to the DNA-protein complex observed in the left lane.
DISCUSSION
All natural isolates of the Lyme disease spirochete contain multiple cp32 elements, each of which carries an erp locus. Two DNA regions immediately 5′ of erp promoters specifically bind B. burgdorferi cytoplasmic proteins, with operator 2 being involved in transcriptional regulation. We have now identified a novel DNA-binding protein, EbfC, as binding to operator 2, interacting with the interrupted palindromic sequence TGT(A/T)ACA. EbfC forms dimers in solution, consistent with its recognition of an inverted-repeat DNA sequence. Additional studies of EbfC, including creation of B. burgdorferi ebfC mutants and bacteria carrying erp loci with specifically mutated EbfC-binding sites, are currently under way to precisely elucidate the function(s) of this chromosomally encoded protein.
Our previous studies that mapped the maximum boundaries of erp operator 2 utilized large (>100-bp) EMSA competitor DNAs and deletion mutations spanning 100 or more bp (7). For this reason, the actual operator 2 region may be much smaller than illustrated in Fig. 1. The two EbfC-binding sites are located within the determined maximum boundaries of erp operator 2. In the present studies, binding of purified EbfC to erp operator 2 was effectively blocked with the 31-bp competitor c104, while our earlier experiments with this competitor found that it did not effectively compete for binding of proteins from total B. burgdorferi cytoplasmic extracts (7). This may mean that the wild-type protein has a greater affinity for long stretches of DNA than does the recombinant fusion protein and cannot be as effectively competed away with short linear DNA fragments. Alternatively, additional cytoplasmic factors may interact with EbfC to facilitate binding of a larger DNA region which cannot be competed with small DNAs such as c104. Comparisons of EMSA results using purified EbfC and B. burgdorferi cell lysates support the hypothesis that additional borrelial proteins also bind erp operator 2. Our DNA affinity purification techniques identified two other proteins that bind erp 5′ noncoding DNA, one or both of which are likely to be the hypothesized additional protein. We are continuing to characterize these as yet undefined proteins to determine the roles they and EbfC play in control of erp gene expression.
As mentioned above, there is a significant body of evidence demonstrating that the cp32 elements are in fact bacteriophage genomes (12, 15, 19, 20, 57, 62). While relapsing-fever spirochetes such as Borrelia hermsii also contain cp32 elements similar to those of B. burgdorferi, the B. hermsii cp32s lack erp loci, yet every single identified cp32 from a Lyme disease spirochete contains such a locus (51, 54, 55, 57). This suggests that B. burgdorferi cp32 bacteriophages have acquired and maintain erp genes for reasons specific to that host bacterium. The role of some Erp proteins in the binding of vertebrate factor H and subsequent protection from complement-mediated killing illustrates one reason for the phages to carry these genes, since the Erp proteins' contribution to survival of the bacterium also enhances the probability of phage survival. Bacteriophages of many other bacterial species have also acquired such “moron” DNAs, including the serum-resistance-encoding bor gene of E. coli phage lambda and the toxin-producing genes of Vibrio cholerae (9, 10, 30, 31, 59, 60). It is intriguing to note that the B. burgdorferi cp32 elements have evolved to use a DNA-binding protein encoded by the bacterial host chromosome, possibly tapping into a bacterial regulatory network for their own benefit. Some phages of other bacterial species use host regulatory machinery to control viral gene expression (13, 59). Alternatively, since bacterial DNAs are constrained into highly organized structures by DNA-binding proteins (58), it is possible that EbfC is used to mold both B. burgdorferi genetic material and the cp32 prophage genomes. The ability of EbfC to form homomultimers supports such a hypothetical role in DNA topological modulation. Further exploration of the interrelationships between B. burgdorferi, cp32s, erp genes, and EbfC will undoubtedly shed additional light not only on B. burgdorferi pathogenesis but on the interplay between bacteria and phages in general.
A broad spectrum of other bacterial species contain genes homologous to B. burgdorferi ebfC, but surprisingly little is known about either those genes or their encoded proteins. Most information has stemmed from characterization of neighboring genes. A physical linkage of dnaX immediately 5′ and recR immediately 3′ of the ebfC homolog is frequently found, leading to speculation that EbfC homologs may be involved in either DNA replication or repair, although those possibilities appear never to have been tested. Arguing against that circumstantial evidence, ebfC homologs are not always found adjacent to either dnaX or recR: B. burgdorferi ebfC is bordered to the 3′ by a probable ndk gene, and this bacterium does not even contain a homolog of recR (25), while the H. influenzae homolog is flanked on the 5′ side by genes involved with DNA uptake (17). Studied organisms both cotranscribe dnaX and the ebfC homolog and transcribe the second gene alone from promoters located within the dnaX open reading frame, thereby allowing independent expression of the EbfC homolog. There are at least two separate promoters within the E. coli dnaX gene that independently drive transcription of its ebfC homolog, one of which is controlled by the stress response sigma factor σE, while Corynebacterium glutamicum regulates a promoter within dnaX as part of the ClgR regulon (14, 23, 24, 39, 49, 61). Of relevance to those observations, B. burgdorferi lacks homologs of both σE and ClgR (25). Mutants of H. influenzae and Streptomyces coelicolor with deleted ebfC homologs are viable (1, 17, 47), raising our hopes that ΔebfC mutants of B. burgdorferi can also be obtained and the effects of that lesion can be studied. Our studies were the first to determine a function for an EbfC family member, that of a site-specific DNA-binding protein, placing the spirochete B. burgdorferi in the unusual position of being the model organism for studying this widely dispersed but barely characterized protein.
In conclusion, we purified and characterized a novel, chromosomally encoded DNA-binding protein of B. burgdorferi. The location of ebfC on the B. burgdorferi main chromosome suggests that the EbfC protein performs bacterial host functions in addition to those postulated for the cp32 prophages. Competition binding analyses demonstrated that EbfC binds the broken palindrome TGT(A/T)ACA. The EbfC-binding sites of erp loci are within the previously mapped maximum boundaries of operator 2, which is required for regulation of erp transcription (7). Two closely linked TGTAACA sequences are found within the operator, one of which is absolutely conserved in every erp locus yet examined. This localization suggests that EbfC may play a role in the regulation of erp transcription. Alternatively, EbfC may affect DNA conformation of the cp32 prophages and the bacterial host's genome. Homologs of EbfC are encoded by a wide range of other bacterial species, where they presumably also function as site-specific DNA-binding proteins. Continued characterization of EbfC and its effects on cp32 elements and the erp and other B. burgdorferi genes will continue to provide insight into the biology of the Lyme disease spirochete as well as many other medically and environmentally important bacteria.
Acknowledgments
These studies were funded by National Institutes of Health grant R01-AI44254. Sean Riley was supported in part by NIH Training Grant in Microbial Pathogenesis T32-AI49795 and a University of Kentucky Academic Excellence Scholarship.
We thank Wolfram Zückert and Sherwood Casjens for helpful advice and Sara Bair, Anne Cooley, Sarah Kearns, Natalie Mickelsen, Jennifer Miller, Ashutosh Verma, Kate von Lackum, and Michael Woodman for assistance in this work.
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins, D. R., M. J. Caimano, X. Yang, F. Cerna, M. V. Norgard, and J. D. Radolf. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 67:1526-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins, D. R., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18:507-520. [DOI] [PubMed] [Google Scholar]
- 4.Alitalo, A., T. Meri, H. Lankinen, I. Seppälä, P. Lahdenne, P. S. Hefty, D. Akins, and S. Meri. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847-3853. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, P. 1964. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem. J. 91:222-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babb, K., N. El-Hage, J. C. Miller, J. A. Carroll, and B. Stevenson. 2001. Distinct regulatory pathways control the synthesis of Borrelia burgdorferi infection-associated OspC and Erp surface proteins. Infect. Immun. 69:4146-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babb, K., J. D. McAlister, J. C. Miller, and B. Stevenson. 2004. Molecular characterization of Borrelia burgdorferi erp promoter/operator elements. J. Bacteriol. 186:2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 9.Barondess, J. J., and J. Beckwith. 1995. bor gene of phage λ, involved in serum resistance, encodes a widely conserved outer membrane lipoprotein. J. Bacteriol. 177:1247-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd, E. F., and H. Brüssow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 11.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 12.Casjens, S., R. van Vugt, K. Tilly, P. A. Rosa, and B. Stevenson. 1997. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 179:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casjens, S. R., E. B. Gilcrease, W. M. Huang, K. L. Bunny, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2004. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 186:1818-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, K., P. Saxena, and J. R. Walker. 1993. Expression of the Escherichia coli dnaX gene. J. Bacteriol. 175:6663-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damman, C. J., C. H. Eggers, D. S. Samuels, and D. B. Oliver. 2000. Characterization of Borrelia burgdorferi BlyA and BlyB proteins: a prophage-encoded holin-like system. J. Bacteriol. 182:6791-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, S., S. W. Barthold, S. S. Giles, R. R. Montgomery, S. R. Telford III, and E. Fikrig. 1997. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J. Clin. Investig. 99:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dougherty, B. A., and H. O. Smith. 1999. Identification of Haemophilus influenzae Rd transformation genes using cassette mutagenesis. Microbiology 145:401-409. [DOI] [PubMed] [Google Scholar]
- 18.Eggers, C. H., M. J. Caimano, M. L. Clawson, W. G. Miller, D. S. Samuels, and J. D. Radolf. 2002. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the Lyme disease spirochaete. Mol. Microbiol. 43:281-295. [DOI] [PubMed] [Google Scholar]
- 19.Eggers, C. H., S. Casjens, and D. S. Samuels. 2001. Bacteriophages of Borrelia burgdorferi and other spirochetes, p. 35-44. In M. H. Saier and J. García-Lara (ed.), The spirochetes: molecular and cellular biology. Horizon Press, Oxford, United Kingdom.
- 20.Eggers, C. H., B. J. Kimmel, J. L. Bono, A. F. Elias, P. Rosa, and D. S. Samuels. 2001. Transduction by φBB-1, a bacteriophage of Borrelia burgdorferi. J. Bacteriol. 183:4771-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Hage, N., K. Babb, J. A. Carroll, N. Lindstrom, E. R. Fischer, J. C. Miller, R. D. Gilmore, Jr., M. L. Mbow, and B. Stevenson. 2001. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147:821-830. [DOI] [PubMed] [Google Scholar]
- 22.El-Hage, N., and B. Stevenson. 2002. Simultaneous coexpression of Borrelia burgdorferi Erp proteins occurs through a specific, erp locus-directed regulatory mechanism. J. Bacteriol. 184:4536-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engels, S., C. Ludwig, J. Schweitzer, C. Mack, M. Bott, and S. Schaffer. 2005. The transcriptional activator ClgR controls transcription of genes involved in proteolysis and DNA repair in Corynebacterium glutamicum. Mol. Microbiol. 57:576-591. [DOI] [PubMed] [Google Scholar]
- 24.Flower, A. M., and C. S. McHenry. 1991. Transcriptional organization of the Escherichia coli dnaX gene. J. Mol. Biol. 220:649-658. [DOI] [PubMed] [Google Scholar]
- 25.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidmann, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 26.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3:799-808. [DOI] [PubMed] [Google Scholar]
- 27.Glöckner, G., R. Lehmann, A. Romualdi, S. Pradella, U. Schulte-Spechtel, M. Schilhabel, B. Wilske, J. Sühnel, and M. Platzer. 2004. Comparative analysis of the Borrelia garinii genome. Nucleic Acids Res. 32:6038-6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellwage, J., T. Meri, T. Heikkilä, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppälä, and S. Meri. 2001. The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 30.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 31.Karaolis, D. K. R., S. Somara, D. R. Maneval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 32.Kraiczy, P., K. Hartmann, J. Hellwage, C. Skerka, M. Kirschfink, V. Brade, P. F. Zipfel, R. Wallich, and B. Stevenson. 2004. Immunological characterization of the complement regulator factor H-binding CRASP and Erp proteins of Borrelia burgdorferi. Int. J. Med. Microbiol. 293(Suppl. 37):152-157. [DOI] [PubMed] [Google Scholar]
- 33.Kraiczy, P., C. Skerka, V. Brade, and P. F. Zipfel. 2001. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect. Immun. 69:7800-7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraiczy, P., C. Skerka, M. Kirschfink, P. F. Zipfel, and V. Brade. 2001. Mechanism of complement resistance of pathogenic Borrelia burgdorferi isolates. Int. Immunopharmacol. 1:393-401. [DOI] [PubMed] [Google Scholar]
- 35.Kraiczy, P., and R. Würzner. 2006. Complement escape of human pathogenic bacteria by acquisition of complement regulators. Mol. Immunol. 43:31-44. [DOI] [PubMed] [Google Scholar]
- 36.Kunkel, G. R., M. Mehrabian, and H. G. Martinson. 1981. Contact-site cross-linking agents. Mol. Cell. Biochem. 34:3-13. [DOI] [PubMed] [Google Scholar]
- 37.Kurtenbach, K., S. De Michelis, S. Etti, S. M. Schäfer, H.-S. Sewell, V. Brade, and P. Kraiczy. 2002. Host association of Borrelia burgdorferi sensu lato—the key role of host complement. Trends Microbiol. 10:74-79. [DOI] [PubMed] [Google Scholar]
- 38.Lim, K., A. Tempczyk, J. F. Parsons, N. Bonander, J. Toedt, Z. Kelman, A. Howard, E. Eisenstein, and O. Herzberg. 2003. Crystal structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function coexpressed with the recombinational DNA repair protein RecR. Proteins 50:375-379. [DOI] [PubMed] [Google Scholar]
- 39.Mahdi, A. A., and R. G. Lloyd. 1989. The recR locus of Escherichia coli K-12: molecular cloning, DNA sequencing and identification of the gene product. Nucleic Acids Res. 17:6781-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marconi, R. T., S. Y. Sung, C. A. N. Hughes, and J. A. Carlyon. 1996. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J. Bacteriol. 178:5615-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metts, M. S., J. V. McDowell, M. Theisen, P. R. Hansen, and R. T. Marconi. 2003. Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection. Infect. Immun. 71:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, J. C., N. El-Hage, K. Babb, and B. Stevenson. 2000. Borrelia burgdorferi B31 Erp proteins that are dominant immunoblot antigens of animals infected with isolate B31 are recognized by only a subset of human Lyme disease patient sera. J. Clin. Microbiol. 38:1569-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, J. C., K. Narayan, B. Stevenson, and A. R. Pachner. 2005. Expression of Borrelia burgdorferi erp genes during infection of non-human primates. Microb. Pathog. 39:27-33. [DOI] [PubMed] [Google Scholar]
- 45.Miller, J. C., and B. Stevenson. Borrelia burgdorferi erp genes are expressed at different levels within tissues of chronically infected mammalian hosts. Int. J. Med. Microbiol., in press. [DOI] [PubMed]
- 46.Miller, J. C., K. von Lackum, K. Babb, J. D. McAlister, and B. Stevenson. 2003. Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle. Infect. Immun. 71:6943-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelaez, A. I., R. M. Ribas-Aparicio, A. Gomez, and M. R. Rodicio. 2001. Structural and functional characterization of the recR gene of Streptomyces. Mol. Genet. Genomics 265:663-672. [DOI] [PubMed] [Google Scholar]
- 48.Porcella, S. F., T. G. Popova, D. R. Akins, M. Li, J. D. Radolf, and M. V. Norgard. 1996. Borrelia burgdorferi supercoiled plasmids encode multicopy tandem open reading frames and a lipoprotein gene family. J. Bacteriol. 178:3293-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rezuchova, B., H. Miticka, D. Homerova, M. Roberts, and J. Kormanec. 2003. New members of the Escherichia coli σE regulon identified by a two-plasmid system. FEMS Microbiol. Lett. 225:1-7. [DOI] [PubMed] [Google Scholar]
- 50.Stevenson, B., J. L. Bono, T. G. Schwan, and P. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stevenson, B., T. Bykowski, A. E. Cooley, K. Babb, J. C. Miller, M. E. Woodman, K. von Lackum, and S. P. Riley. The Lyme disease spirochete Erp lipoprotein family: structure, function and regulation of expression. In F. C. Cabello, H. P. Godfrey, and D. Hulinska (ed.), Molecular biology of spirochetes, in press. IOS Press, Amsterdam, The Netherlands.
- 52.Stevenson, B., S. Casjens, and P. Rosa. 1998. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology 144:1869-1879. [DOI] [PubMed] [Google Scholar]
- 53.Stevenson, B., N. El-Hage, M. A. Hines, J. C. Miller, and K. Babb. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stevenson, B., and J. C. Miller. 2003. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol. 57:309-324. [DOI] [PubMed] [Google Scholar]
- 55.Stevenson, B., S. F. Porcella, K. L. Oie, C. A. Fitzpatrick, S. J. Raffel, L. Lubke, M. E. Schrumpf, and T. G. Schwan. 2000. The relapsing fever spirochete Borrelia hermsii contains multiple, antigen-encoding circular plasmids that are homologous to the cp32 plasmids of Lyme disease spirochetes. Infect. Immun. 68:3900-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stevenson, B., W. R. Zückert, and D. R. Akins. 2001. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species, p. 87-100. In M. H. Saier and J. García-Lara (ed.), The spirochetes: molecular and cellular biology. Horizon Press, Oxford, United Kingdom. [PubMed]
- 58.Thanbichler, M., S. C. Wang, and L. Shapiro. 2005. The bacterial nucleoid: a highly organized and dynamic structure. J. Cell Biol. 96:506-521. [DOI] [PubMed] [Google Scholar]
- 59.Wagner, P. L., and M. K. Waldor. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 70:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 61.Yeung, T., D. A. Mullin, K. Chen, E. A. Craig, J. C. A. Bardwell, and J. R. Walker. 1990. Sequence and expression of the Escherichia coli recR locus. J. Bacteriol. 172:6042-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, H., and R. T. Marconi. 2005. Demonstration of cotranscription and 1-methyl-3-nitroso-nitroguanidine induction of a 30-gene operon of Borrelia burgdorferi: evidence that the 32-kilobase circular plasmids are prophages. J. Bacteriol. 187:7985-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zückert, W. R., and J. Meyer. 1996. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J. Bacteriol. 178:2287-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]