Abstract
The P ring of the bacterial flagellar motor consists of multiple copies of FlgI, a periplasmic protein. The intramolecular disulfide bond in FlgI has previously been reported to be essential for P-ring assembly in Escherichia coli, because the P ring was not assembled in a dsbB strain that was defective for disulfide bond formation in periplasmic proteins. We, however, found that the two Cys residues of FlgI are not conserved in other bacterial species. We then assessed the role of this intramolecular disulfide bond in FlgI. A Cys-eliminated FlgI derivative formed a P ring that complemented the flagellation defect of our ΔflgI strain when it was overproduced, suggesting that disulfide bond formation in FlgI is not absolutely required for P-ring assembly. The levels of the mature forms of the FlgI derivatives were significantly lower than that of wild-type FlgI, although the precursor protein levels were unchanged. Moreover, the FlgI derivatives were more susceptible to degradation than wild-type FlgI. Overproduction of FlgI suppressed the motility defect of ΔdsbB cells. Additionally, the low level of FlgI observed in the ΔdsbB strain increased in the presence of l-cystine, an oxidative agent. We propose that intramolecular disulfide bond formation facilitates the rapid folding of the FlgI monomer to protect against degradation in the periplasmic space, thereby allowing its efficient self-assembly into the P ring.
Many motile bacteria swim by rotating one or more helical flagella like a screw. Each flagellum consists of three substructures: a helical filament extending from the cell body, a motor that is embedded in the cell envelope, and a flexible hook that connects these two structures. In gram-negative bacteria such as Escherichia coli and Salmonella spp., the flagellar motor consists of several ring structures surrounding a rod that penetrates the cell envelope (for reviews, see references 20 and 21). The L, P, and MS rings are located in the outer membrane, the peptidoglycan layer, and the cytoplasmic membrane, respectively, whereas the C ring lies on the cytoplasmic side of the MS ring. The P and L rings together form a stiff cylindrical structure that acts as a bushing to hold the rotating rod (2). The MS ring/FliG complex is thought to rotate due to torque-generating units (the MotA/MotB complexes), which surround the rotor in a circular array. The rod acts as a driving shaft, transmitting the rotary power of the motor to the flagellar filament (for a review, see reference 6).
The P ring consists of approximately 26 copies of a single protein, FlgI. The precursor form of FlgI, which has a cleavable N-terminal 19-amino-acid signal sequence, is translocated via the Sec apparatus to the periplasmic space (13, 14). The Dsb system facilitates the formation of an intramolecular disulfide bond in the mature form of FlgI (8; see below). FlgA, a periplasmic chaperone, assists the copies of FlgI as they assemble into the P ring surrounding the growing rod (15, 19, 22, 31). Because flagellar assembly is a highly ordered process, flagellar morphogenesis fails to continue beyond the assembly of the P ring in the flgI mutant (19).
Many proteins require the formation of disulfide bonds for proper protein folding, stability, and function. In E. coli, disulfide bond formation in periplasmic proteins is catalyzed by the Dsb system (for a review, see reference 7); after the periplasmic protein DsbA oxidizes a newly exported target protein, the reduced form of DsbA is reoxidized by DsbB, a cytoplasmic membrane protein. Dailey and Berg (8) reported that when it was grown on M63 minimal medium, an E. coli dsbB strain which was not able to form disulfide bonds in periplasmic proteins showed a defect in flagellation that was similar to the one observed in the flgI strain. The LP-ring structure was not detected in the flagellar assemblies isolated from dsbB cells grown on M63 minimal medium, whereas it was seen when the cells were grown in the presence of l-cystine, an oxidative agent. Furthermore, during sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, the mobility of the FlgI protein from dsbB+ cells was changed by the addition of β-mercaptoethanol, a reducing agent. From these lines of evidence, the authors argued that the intramolecular disulfide bond in FlgI is essential for P-ring assembly. The mechanism by which the loss of this disulfide bond in FlgI results in defective P-ring assembly, however, has remained unclear.
In this study, we assessed the role of the disulfide bond in FlgI. Surprisingly, a Cys-eliminated FlgI variant formed a P ring that complemented the flagellation defect of our ΔflgI strain, suggesting that the disulfide bond in FlgI is not essential for P-ring assembly. In the periplasmic space, the Cys-eliminated FlgI variants were more susceptible to degradation than wild-type FlgI. The level of the FlgI protein in ΔdsbB cells was relatively low but was elevated in the presence of l-cystine. The motility defect of the ΔdsbB strain was suppressed by overproduction of the wild-type FlgI protein. Taking these findings together, we propose that intramolecular disulfide bond formation in FlgI is not absolutely required for P-ring assembly but is important to prevent the degradation of the protein.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and media.
The strains used in this work are listed in Table 1. The HK295 (dsb+) and HK320 (ΔdsbB) strains were kindly supplied by Hiroshi Kadokura and Jon Beckwith (Harvard Medical School). The ΔflgI::cat strain YZ1 was constructed by deleting the chromosomal flgI gene using the method described by Datsenko and Wanner (9) and the following primers: ΔflgI(+) (5′-CTGGTTGCAGCGTTTCTTCCTTAACCTGTCGCCAATGTAACATATGAATATCCTCCTTAG-3′) and ΔflgI(−) (5′-ATGATTTCCAGTTTTGCCCGCAGACATCCCGCACTTTGCAGTGTAGGCTGGAGCTGCTTC-3′). The constructed ΔflgI::cat region contained the complete flgH open reading frame and an approximately 40-bp region upstream of the initiation codon of flgJ, which presumably contained the ribosome-binding sequence of this gene. The ΔflgI::cat region of the chromosome was transduced into the RP437 strain by P1 phage (27). E. coli cells were cultured at 37°C in LB medium (1% Bacto tryptone, 0.5% yeast extract, and 0.5% NaCl), at 30°C in TG medium (1% Bacto tryptone, 0.5% NaCl, and 0.5% [wt/vol] glycerol), or in M63 minimal medium supplemented with 0.4% (wt/vol) glycerol as the sole source of carbon (27). When necessary, ampicillin and chloramphenicol were added to final concentrations of 50 μg/ml and 25 μg/ml, respectively.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| W3110 | Wild type | |
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 λ− Δ(lac-proAB) (F′ traD36 proAB lacIqlacZΔM15) | 33 |
| BL21(DE3) | T7 expression host | 32 |
| HK295 | MC1000 Δara-714 leu+ | 16 |
| HK320 | HK295 ΔdsbB | 16 |
| RP3098 | Δ(flhD-flhA)4 | 29 |
| RP437 | F−thi thr leu his met eda rpsL; wild type for chemotaxis | 25 |
| YZ1 | RP437 ΔflgI::cat | This work |
| Plasmids | ||
| pBAD24 | PBADaraC Ampr | 11 |
| pET-3a | T7 expression vector, Ampr | Novagen |
| pLysS | T7 lysozyme, Cmr | 32 |
| pYZ201 | pBAD24 flgI | This work |
| pYZ202 | pYZ201 C254A | This work |
| pYZ203 | pYZ201 C338A | This work |
| pYZ204 | pYZ201 C254A/C338A | This work |
| pYZ220 | pET-3a flgI (mature FlgI) | This work |
Ampr, ampicillin resistant; Cmr, chloramphenicol resistant; PBAD, araBAD promoter.
Construction of the plasmids.
Routine DNA manipulations were carried out according to standard procedures (26). The plasmids used in this work are listed in Table 1. The flgI gene was PCR amplified from purified chromosomal DNA of the W3110 strain using appropriate primers. The PCR product was digested with KpnI and SphI, and a 1.4-kb flgI fragment was inserted into the corresponding sites of the pBAD24 vector plasmid to obtain pYZ201. To construct pYZ220, the plasmid that encoded a mature form of FlgI under the control of a T7 promoter, we used the PCR and the NdeI and BamHI sites of the pET-3a vector plasmid (Novagen). The 5′ sequence of the insert was 5′-CAT(ATG)GAG(CGT)-3′, which encodes the initiator Met-Glu1-Arg2 (the NdeI site is underlined and the superscript numbers represent the amino acid residue positions in the mature form of FlgI), and the 3′ sequence was 5′-ATC(ATC)TAA(GGA)TCC-3′, which encodes Ile345-Ile346-ochre (the BamHI site is underlined). Site-directed mutagenesis was performed as described previously (18).
Motility assays.
The fraction of swimming cells was measured as follows. M63 minimal medium with 0.4% (wt/vol) glycerol was inoculated at a 100-fold dilution with an overnight culture (grown on LB medium at 37°C) and cultured at 30°C. When necessary, 25 μg/ml l-cystine was added at the time of inoculation, and 0.04% l-arabinose was added 2 h after the inoculation. At the exponential growth phase, the cultures were observed under a dark-field microscope. The swimming fraction was determined by counting the swimming cells among the total cells. The data in this report are the average values from at least three frames for each set of conditions.
For the swarm assay, a single colony from an LB plate was used to inoculate a soft-agar T broth plate (1% Bacto tryptone, 0.5% NaCl, and 0.27% Bacto agar) containing 50 μg/ml ampicillin and 0.04% l-arabinose. The plates were incubated at 30°C for 11 h.
Detection of FlgI and flagellin.
TG medium or M63 glycerol medium was inoculated at a 100-fold dilution with an overnight culture (grown on LB medium) and cultured at 30°C. When necessary, 50 μg/ml ampicillin was added at the time of inoculation and 0.04% l-arabinose was added 2 h after the inoculation. At the exponential growth phase, the cells from 50 μl of the cultures (optical density at 660 nm [OD660], 1.0) were harvested by centrifugation (9,500 × g, 5 min). Otherwise, trichloroacetic acid (TCA) was added to the cultures to a final concentration of 10% (wt/vol), and the insoluble material was recovered by centrifugation, washed with acetone, and dried. The centrifuged cells or the TCA-insoluble material was suspended in sodium dodecyl sulfate (SDS) loading buffer containing 5% (vol/vol) β-mercaptoethanol and then sonicated and heated at 100°C for 5 min. The proteins were separated by SDS-PAGE. Immunoblotting was performed using anti-FlgI (see below) or antiflagellin (laboratory stock) antibodies as described previously (34).
Antibodies.
The anti-E. coli FlgI antibodies (FlgI346) were raised against the mature form of FlgI. Mature FlgI was purified from the BL21(DE3)/pLysS strain that was harboring plasmid pYZ220. Cells were harvested by centrifugation, washed with buffer A (10 mM Tris-HCl, 150 mM NaCl, and 0.5 mM phenylmethylsulfonyl fluoride at pH 7.5), and resuspended in buffer A containing 5 mM MgCl2, 10 μg/ml DNase I, and 1% (wt/vol) Triton X-100. The cell suspension was twice passed through a French press at 500 kg/cm2. After the soluble fraction was removed by centrifugation (7,000 × g for 10 min), the insoluble fraction was suspended in buffer A containing 5 mM MgCl2, 10 μg/ml DNase I, and 1% (wt/vol) Triton X-100; sonicated for 1 min on ice; and centrifuged again. The insoluble fraction was then homogenized in buffer A containing 8 M urea and gently shaken at room temperature overnight. Urea-insoluble material was removed by ultracentrifugation (100,000 × g for 1 h at 25°C), and the remaining soluble material was passed through an anion-exchange column (Amersham). Partially purified mature FlgI was separated by SDS-PAGE and stained with Coomassie blue R250. The band corresponding to FlgI was excised and used to inoculate rabbits. Anti-FlgI rabbit antibodies were produced and affinity purified by Biogate Co., Ltd. (Gifu, Japan).
NaN3 treatment.
TG medium with 50 μg/ml ampicillin was inoculated at a 100-fold dilution with an overnight culture (grown on LB medium) and cultured at 30°C. Four hours later, 0.04% l-arabinose was added to the culture. The cells were incubated for 10 min at 30°C, and then 2 mM NaN3 was added to one of two aliquots. After a subsequent 10-min incubation, whole-cell extracts from 50 μl of the culture (OD660 = 1.0) were collected by TCA precipitation and analyzed by immunoblotting using anti-FlgI antibodies as described above.
Pulse-induction experiment.
TG medium with 50 μg/ml ampicillin was inoculated at a 100-fold dilution with an overnight culture (grown on LB medium) and cultured at 30°C. l-Arabinose (0.04%) was added 2 and 3 h after the inoculation for the RP3098 and YZ1 strains, respectively. The cells were incubated for 30 min and divided into two aliquots. Each aliquot was centrifuged (2,500 × g, 5 min, 25°C) to harvest the cells and then suspended in TG medium with or without 0.04% l-arabinose. Both samples were then incubated for 20 min at 30°C, and whole-cell extracts from 50 μl of the cultures (OD660 = 1.0) were collected by TCA precipitation as described above. The YZ1 strain was subjected to an additional 40-min cultivation. All samples were analyzed by immunoblotting using anti-FlgI antibodies as described above.
Isolation of flagellar structures.
The isolation of the flagellar structures was carried out as described previously (1, 10) with several modifications. TG medium was inoculated at a 100-fold dilution with an overnight culture (grown on LB medium) and cultured at 30°C. Arabinose was added to a final concentration of 0.04% 2 h after the inoculation. After a subsequent 3-h cultivation, the cells were harvested and converted into spheroplasts by adding lysozyme and EDTA to the final concentrations of 0.1 mg/ml and 2 mM, respectively. The spheroplasts were lysed by adding Triton X-100 to a final concentration of 1% (wt/vol). To reduce the viscosity of the samples after lysis was complete, MgSO4 and DNase I were added to the suspension to the final concentrations of 5 mM and 0.1 mg/ml, respectively. After the viscosity of the solution decreased, EDTA was added to a final concentration of 5 mM. Unlysed cells and cellular debris were removed by centrifugation at 8,000 × g for 20 min. Polyethylene glycol 6000 and NaCl were added to the lysate to the final concentrations of 3% and 0.15 M, respectively. The samples were then incubated at 4°C overnight to form bundled flagella (17). The suspension was centrifuged at 27,000 × g for 30 min at 4°C, and the pellet was resuspended in 3 ml of TET buffer (10 mM Tris-HCl, 5 mM EDTA, and 0.1% [wt/vol] Triton X-100 at pH 8.0). To remove cellular debris, the solution was centrifuged at 1,000 × g for 15 min at 4°C. The supernatant was centrifuged at 100,000 × g for 30 min, and the pellet was resuspended in 300 μl of TET buffer. To dissociate the flagellar filaments into monomeric flagellin, the preparation was diluted 10-fold in 50 mM glycine-HCl (pH 2.5) and shaken for 30 min at room temperature. The suspension was centrifuged at 150,000 × g for 40 min at 25°C, and the pellet was resuspended in 20 μl of TET buffer.
Electron microscopy.
The isolated flagellar structures were negatively stained with 2% phosphotungstic acid (pH 7.2) and observed with a JEM-1200EX electron microscope (JEOL, Japan).
RESULTS
The Cys residues in FlgI are not conserved in other bacterial species.
If the disulfide bond is essential for the structure and function of FlgI, the two Cys residues are likely to be conserved among various species. To address this, we analyzed the primary sequences of the FlgI proteins from various bacterial species that rely on flagellar motility (Table 2). Some FlgI proteins of the other species had either one or zero Cys residues in the predicted mature form of FlgI. Thus, the two Cys residues in E. coli FlgI are not conserved in most of the other motile bacteria.
TABLE 2.
Numbers of Cys residues in FlgI proteins
| Organism(s)d | No. of Cys residues
|
Identity to E. coli FlgI (%)c | |
|---|---|---|---|
| Full-length FlgI | Predicted mature FlgI | ||
| Escherichia coli | 2 | 2 | |
| Salmonella spp. | 2 | 2 | 92.3 |
| Yersinia pestis | 2 | 2 | 78.4 |
| Pseudomonas putida | 1 | 0 | 48.8 |
| Vibrio cholerae | 0 | 0 | 47.8 |
| Vibrio parahaemolyticusa | 0 | 0 | 46.7 |
| Aeromonas hydrophilaa,b | 4 | 2 | 44.4 |
| Aquifex aeolicus | 1 | 0 | 44.1 |
| Rhodobacter sphaeroides | 1 | 0 | 43.9 |
| Agrobacterium tumefaciens | 0 | 0 | 43.9 |
| Caulobacter crescentus | 2 | 1 | 43.7 |
| Sinorhizobium meliloti | 1 | 0 | 43.4 |
| Helicobacter pylori | 0 | 0 | 36.5 |
| Thermotoga maritima | 1 | 1 | 33.9 |
FlgI of polar flagella.
The cleavage site of the signal sequence was predicted by multiple-alignment analysis.
The identity between the full-length FlgI protein and E. coli FlgI was calculated with GENETYX version 10 (GENETYX).
UniProt accession numbers for the FlgI proteins are as follows: E. coli, P75941; Salmonella spp., P15930; Y. pestis, Q8ZFB1; P. putida, Q52082; V. cholerae, Q9KQ14; V. parahaemolyticus, Q9X9J4; A. hydrophila, Q8GLP1; A. aeolicus, O67608; R. sphaeroides, P58204; A. tumefaciens, Q44340; C. crescentus, P33979; S. meliloti, Q52948; H. pylori, O25028; and T. maritima, Q9X1M5.
Cys-eliminated FlgI variants can restore motility in the ΔflgI strain.
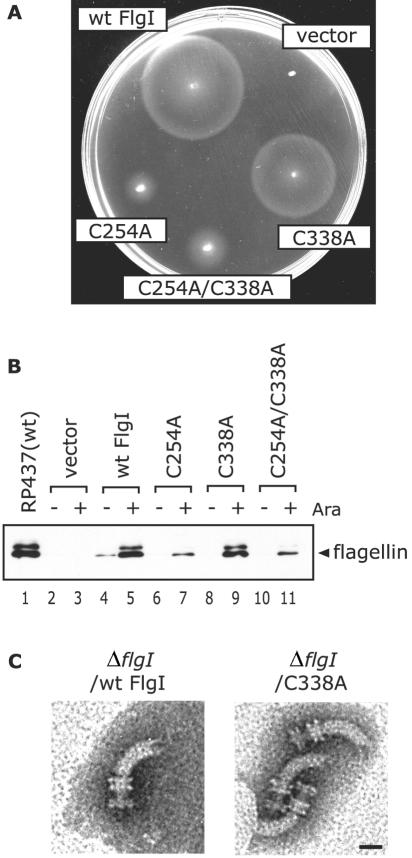
In order to confirm the importance of the two Cys residues in E. coli FlgI, we replaced them and characterized the resulting mutant proteins. First, we constructed the YZ1 flgI deletion strain of E. coli (ΔflgI::cat) using the gene disruption procedure described by Datsenko and Wanner (9). The ΔflgI strain lost the ability to swarm in semisolid agar (Fig. 1A), was nonmotile under a dark-field microscope (data not shown), and had no flagella, as was confirmed by electron microscopy (data not shown). The swarming (Fig. 1A) and swimming (data not shown) abilities of the bacteria were restored upon expression of wild-type FlgI from the pYZ201 plasmid. To address the importance of the disulfide bond, we then replaced the two Cys residues with Ala residues using site-directed mutagenesis. Hereinafter, these substitutions are referred to as C254A, C338A, and C254A/C338A (the numbers correspond to the positions of the amino acid residues in the mature form of FlgI). The ΔflgI strain was transformed with each of the plasmids encoding the Cys-eliminated FlgI variants, and the transformants were examined for their ability to spread in soft-agar medium. The ΔflgI strain with the C338A mutation was almost as motile as the strain that produced wild-type FlgI, whereas the C254A and C254A/C338A mutation-containing strains displayed a limited ability to spread in the agar (Fig. 1A). When the ΔflgI strain expressed either the C338A variant or wild-type FlgI, the fractions of cells that were observed to swim in liquid medium were roughly the same (data not shown).
FIG. 1.
Motility and the production of the flagellar structures by the ΔflgI strains producing Cys-eliminated FlgI. (A) Swarms of YZ1 (ΔflgI) cells harboring pYZ201 (wild-type [wt] FlgI), pBAD24 (vector), or the pYZ201 derivatives (C254A, C338A, and C254A/C338A substitutions). Soft-agar T broth containing l-arabinose was inoculated with single colonies and incubated at 30°C for 11 h. (B) Flagellin produced in RP437 or YZ1 cells harboring pBAD24, pYZ201, or the derivatives of pYZ201 was detected by immunoblotting using antiflagellin antibodies. The cells were cultured in the absence or presence of l-arabinose, harvested, and suspended in SDS loading buffer with β-mercaptoethanol. The position of flagellin is indicated on the right side of the panel. Ara, l-arabinose. (C) Electron micrographs of the flagellar structures isolated from YZ1 cells producing wild-type FlgI (left) and from YZ1 cells producing FlgI C338A (right). A P ring stacked with an L ring, an MS ring, and a hook can be seen. Bar, 20 nm. The defects in the flagellation and P-ring formation of the ΔflgI strain were complemented by the Cys-eliminated FlgI derivatives.
The formation of flagella in the strains was examined by immunoblotting analysis using antiflagellin antibodies. An intense band was detected in the whole-cell extract from cells expressing the C338A variant or wild-type FlgI. On the other hand, in the C254A and C254A/C338A preparations, significant but much weaker bands were detected (Fig. 1B). As judged by electron microscopy, the average number of flagella per ΔflgI cell was slightly smaller in cells producing the C338A variant than in cells producing wild-type FlgI (data not shown). The C254A and C254A/C338A substitutions significantly decreased the number of flagella to approximately 10% of the number observed in wild-type bacteria and resulted in the frequent occurrence of shorter filaments (data not shown). These results show that the C338A substitution had a very limited effect on flagellar formation but that C254A and C254A/C338A produced more-profound effects. Since flagellation without the LP ring in an flgI strain of E. coli has been observed through an overproduction of the FlgE hook protein (23), we examined P-ring formation in the C338A variant-producing ΔflgI strain. Isolated flagellar structures from ΔflgI cells producing wild-type FlgI or the C338A variant contained P-ring structures (Fig. 1C). No rivet-like structures, i.e., flagellar structures missing the LP ring (19), were observed in these preparations. These results strongly argue that the disulfide bond in the FlgI protein is not essential for the formation of the P ring by this protein.
The Cys-eliminated FlgI variants are degraded in the periplasmic space.
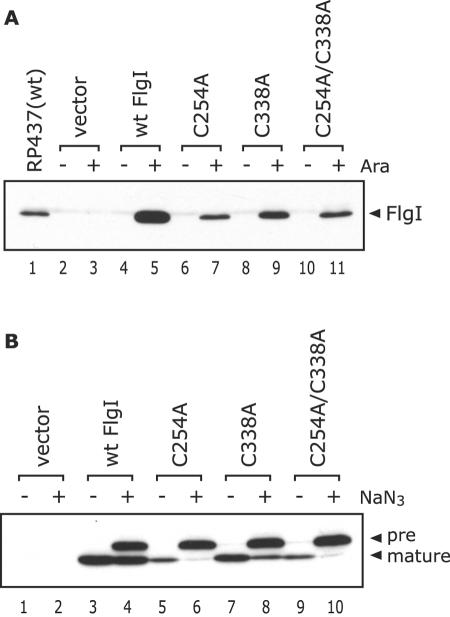
The mutant FlgI proteins expressed in the ΔflgI strain were examined by immunoblotting using anti-FlgI antibodies (Fig. 2A). Although the C338A protein and wild-type FlgI each restored the motility of the ΔflgI strain, the amount of the C338A protein was significantly less than the amount of wild-type FlgI (Fig. 2A, lanes 5 and 9). Additionally, the levels of the C254A and C254A/C338A proteins were lower than that of the C338A protein (Fig. 2A, lanes 7 and 11), which is consistent with the swarm size and the degree of flagellin and filament production observed for cells expressing these proteins. These results suggest that the intramolecular disulfide bond in FlgI is involved in the accumulation of the FlgI protein.
FIG. 2.
Levels of the mature and the precursor forms of FlgI in the ΔflgI strain producing Cys-eliminated FlgI. The FlgI proteins from RP437 and YZ1 cells harboring pBAD24 (vector), pYZ201 (wild-type FlgI), or the derivatives of pYZ201 (C254A, C338A, and C254A/C338A substitutions) were detected by immunoblotting using anti-FlgI antibodies. Whole-cell extracts were prepared by TCA precipitation. (A) The position of FlgI is indicated on the right side of the figure. Ara, l-arabinose. (B) A culture in the exponential growth phase was supplemented with l-arabinose and incubated for 10 min. Then the aliquot was incubated for another 10 min in the absence or presence of 2 mM NaN3. The positions of the precursor (38-kDa band) and the mature (36-kDa band) forms of FlgI are indicated on the right side of the figure. Whereas the levels of the mature forms of the Cys-eliminated FlgI derivatives were lower than that of wild-type FlgI, those of the precursor forms were unchanged.
FlgI is synthesized as a precursor protein with a cleavable 19-amino-acid signal sequence at the N terminus before it is transported to the periplasmic space via the Sec apparatus. In order to determine if the FlgI bands shown in Fig. 2A was the mature form or the precursor form of the protein and how much the precursor forms can be detected, ΔflgI cells producing the Cys-eliminated FlgI variants were treated with NaN3, a SecA inhibitor (24), and examined by immunoblotting (Fig. 2B). The mature form (36-kDa band) was the only band detected in all the preparations that did not receive the NaN3 treatment. On the other hand, an additional 38-kDa band corresponding to the precursor form appeared in the wild-type and the derivative FlgI samples after the NaN3 treatment. The accumulated amounts of the precursor forms of wild-type FlgI and the FlgI derivatives were comparable. These results suggest that like wild-type FlgI, the Cys-eliminated FlgI variants were produced and translocated to the periplasmic space. The levels of the mature FlgI variants significantly decreased in the presence of NaN3 (Fig. 2B, compare the lanes with and without NaN3 treatment), whereas wild-type FlgI levels were almost unchanged (Fig. 2B, lanes 3 and 4). The levels of the mature forms after the NaN3 treatment were indicative of the protein in the periplasmic space after the inhibition of the translocation of newly synthesized FlgI. Thus, we speculate that in the periplasmic space, the Cys-eliminated FlgI variants were degraded more quickly than wild-type FlgI.
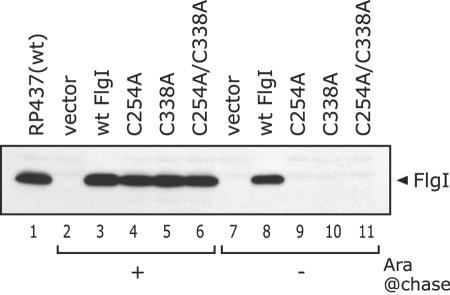
We performed a pulse-induction experiment to examine the degradation of the Cys-eliminated FlgI variants. Arabinose was added to the cell cultures at the exponential growth phase, and the cultures were incubated for 30 min to induce protein synthesis. Then the medium was exchanged to one with or without arabinose and each culture was further incubated for 20 min, followed by immunoblotting analysis (Fig. 3). To avoid complexity, the flhDC strain RP3098 was used in this experiment. Because this strain cannot express all the flagellar genes, the FlgI proteins expressed from the plasmid are not incorporated into flagella. The Cys-eliminated FlgI variants were depleted within 20 min when incubated in the absence of arabinose (Fig. 3, lanes 9 to 11), whereas wild-type FlgI remained (Fig. 3, lane 8). These lines of evidence suggest that FlgI protein that cannot form the intramolecular disulfide bond is susceptible to degradation in the periplasmic space.
FIG. 3.
Degradation of the Cys-eliminated FlgI variants in flhDC cells. The FlgI proteins from RP437 or RP3098 (flhDC) cells harboring pBAD24 (vector), pYZ201 (wild-type [wt] FlgI), or the pYZ201 derivatives (C254A, C338A, and C254A/C338A substitutions) were detected by immunoblotting using anti-FlgI antibodies. In the pulse-induction experiment, the second incubation lasted for 20 min. Whole-cell extracts from RP437 cells at the exponential growth phase were prepared as a positive control. For comparison, a fifth of a sample from cells producing wild-type FlgI was loaded on the gel (lanes 3 and 8). The position of FlgI is indicated on the right side of the figure. Ara @chase, l-arabinose in the medium for the second incubation. The Cys-eliminated FlgI derivatives were depleted when the production of the protein was arrested.
Even without the disulfide bond, the FlgI protein is stabilized when incorporated into the flagellar structures.
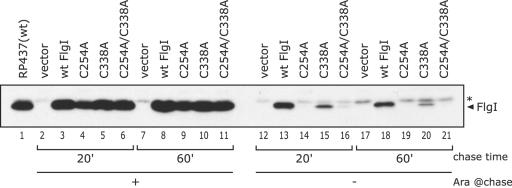
In the ΔflgI strain, some of the plasmid-derived FlgI protein is presumed to exist as the P rings incorporated into the flagella. To address whether the FlgI derivatives incorporated into the flagella were still susceptible to degradation, we performed a pulse-induction experiment with the ΔflgI strain YZ1 (Fig. 4). In the ΔflgI strain, some of the C338A protein remained after a 20-min incubation (Fig. 4, lane 15), or even after an additional 40-min incubation (Fig. 4, lane 20), although without arabinose, the Cys-eliminated FlgI variants were depleted within 20 min in the flhDC strain (Fig. 3). Consistent with these results, after a 60-min incubation, the C338A protein-producing cells were still able to swim (data not shown). Although the FlgI C338A protein was degraded in the flhDC strain, it remained in the ΔflgI strain, indicating that some of the C338A protein was not rapidly degraded, presumably due to incorporation into the flagella as the P rings. If this was the case, overproduction of the FlgI protein should suppress the motility defect of dsb cells by allowing FlgI to accumulate to sufficient levels in the periplasmic space for P-ring assembly, in spite of the absence of the normal mechanism for disulfide bond formation.
FIG. 4.
Levels of the Cys-eliminated FlgI proteins in ΔflgI cells after the production of the proteins was halted. The FlgI protein from RP437 or YZ1 (ΔflgI) cells harboring pBAD24 (vector), pYZ201 (wild-type [wt] FlgI), or the pYZ201 derivatives (C254A, C338A, and C254A/C338A substitutions) were detected by immunoblotting using anti-FlgI antibodies. The sample preparations were the same as those used for Fig. 3, with secondary incubations of 20 min (lanes 2 to 6 and 12 to 16) or 60 min (lanes 7 to 11 and 17 to 21). For comparison, a fifth of a sample from cells producing wild-type FlgI was loaded on the gel (lanes 3, 8, 13, and 18). The position of FlgI is indicated on the right side of the figure. The asterisk denotes a nonspecific band. The chase time is the length of the second incubation in minutes. Ara @chase, l-arabinose in the medium during the second incubation. Some of the FlgI C338A derivative remained even 60 min after the production of the protein was arrested.
Overproduction of the FlgI protein suppresses the motility defect of the ΔdsbB strain.
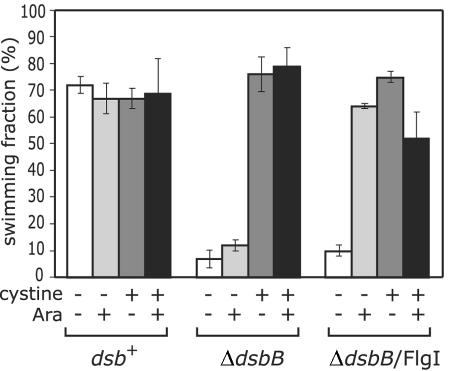
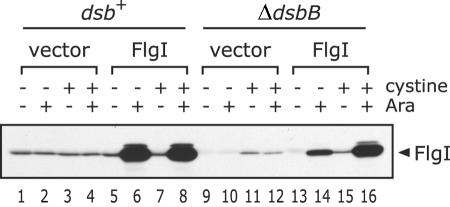
It was previously proposed that the intramolecular disulfide bond in the FlgI protein is essential for P-ring assembly. Our experimental data, however, suggest that it is instead required to prevent degradation of the protein in the periplasmic space. To investigate the possibility that the intramolecular disulfide bond in FlgI is involved in limiting the degradation of the protein, we introduced the FlgI-encoding plasmid into HK320 cells, a ΔdsbB strain that does not form disulfide bonds in periplasmic proteins, and measured the fraction of the transformants that swam in liquid M63 minimal medium (Fig. 5). As described previously (8), the ΔdsbB mutation inhibited swimming ability, and the defect was rescued by the addition of 25 μg/ml l-cystine. The level of FlgI under each set of conditions was examined by immunoblotting analysis (Fig. 6). The level of FlgI decreased in the ΔdsbB strain (Fig. 6, compare lanes 1 and 9) but increased after the addition of cystine (Fig. 6, compare lanes 9 and 11). This result indicates that the formation of the disulfide bond in the FlgI protein confers resistance to degradation.
FIG. 5.
Fraction of the ΔdsbB cells overproducing FlgI that was observed to be motile. The fractions of HK295 (dsb+) cells, HK320 (ΔdsbB) cells, and HK320 cells harboring pYZ201 (wild-type FlgI) that were observed to swim were measured. The cells were cultured in M63 glycerol minimal medium in the absence or presence of l-cystine and l-arabinose. Ara, l-arabinose. The motility defect of the ΔdsbB strain was suppressed by overproducing FlgI.
FIG. 6.
Level of FlgI in the ΔdsbB strain. The FlgI proteins from HK295 (dsb+) and HK320 (ΔdsbB) cells harboring pBAD24 (vector) or pYZ201 (wild-type FlgI) were detected by immunoblotting using anti-FlgI antibodies. The cells were cultured in M63 glycerol minimal medium in the absence or presence of l-cystine and l-arabinose. The whole-cell extracts were prepared by TCA precipitation and suspended in SDS loading buffer with β-mercaptoethanol. The position of FlgI is indicated on the right side of the figure. Ara, l-arabinose. The low level of the FlgI protein in the ΔdsbB strain increased in the presence of l-cystine.
Even in the absence of cystine, ΔdsbB cells overproducing FlgI showed a nearly wild-type level of motility (Fig. 5). For an unknown reason, the fraction of motile FlgI-overexpressing ΔdsbB cells slightly decreased upon the addition of cystine. We examined whether each strain was able to spread in M63 soft-agar plates. The ΔdsbB strain overproducing FlgI swarmed at a significantly higher rate than the ΔdsbB strain did (data not shown). These results show that overproduction of FlgI suppresses the motility defect of the ΔdsbB strain. This supports the idea that the disulfide bond in FlgI is not required for P-ring assembly. The overproduction of FlgI considerably increased the amount of FlgI in the ΔdsbB strain (Fig. 6, compare lanes 9 and 14). The increased level of the FlgI protein may account for the suppression of the motility defect of the ΔdsbB strain.
DISCUSSION
We propose that the intramolecular disulfide bond in FlgI is not absolutely required for P-ring assembly. In flgI cells, the flagellar structures are constructed up to the rivet that contains the MS ring and the rod (19). Since the FlgI chaperone FlgA is present in the ΔflgI strain, the FlgI protein derived from the plasmid is able to form a P ring around the preexisting rod. Although the FlgI C338A protein cannot form the intramolecular disulfide bond, it was able to form an apparently normal P-ring structure (Fig. 1C) and restored flagellation and motility in the ΔflgI strain when overproduced, as wild-type FlgI did (Fig. 1A and B). We therefore concluded that a P ring constructed with FlgI that did not have the intramolecular disulfide bond was structurally and functionally normal. However, the disulfide bond might somehow participate in the efficient self-assembly of FlgI into the P ring. To address such an issue, we have to develop some methods for assessing the self-assembly process.
The intramolecular disulfide bond in FlgI likely reduces the susceptibility of the protein to degradation in the periplasmic space. In the present study, we showed that the levels of the mature forms of the Cys-eliminated FlgI variants were significantly lower than that of wild-type FlgI (Fig. 2), and the Cys-eliminated FlgI variants were more susceptible to degradation than wild-type FlgI (Fig. 3 and 4). Additionally, the amount of FlgI in the ΔdsbB strain increased in the presence of l-cystine (Fig. 6). It is generally postulated that in dsb strains, most of the substrates of the Dsb proteins are not able to form disulfide bonds, resulting in unstable proteins that aggregate or are proteolyzed. For example, the PhoA protein, which is one of the most characterized substrates of DsbA, has four Cys residues that form two disulfide bonds. Under reducing conditions or in the absence of the Dsb system, the PhoA protein is unable to form the disulfide bonds and susceptible to protease in vivo and in vitro (3, 5). Additionally, the reduced form of the OmpA protein is also susceptible to protease treatment in vitro (4, 5). Recently, Hiniker and Bardwell systematically compared the proteome of the dsbA strain with that of wild-type E. coli to identify in vivo periplasmic substrates of DsbA (12). Those authors found that the levels of 10 periplasmic proteins that contained at least two Cys residues decreased in the dsbA strain. These results support the assumption that the absence of the folding catalyst DsbA causes its substrates to be unstable. Thus, we propose that the disulfide bond in FlgI is involved in forming a rigid structure that reduces the susceptibility of the protein to degradation.
Dailey and Berg proposed that the two Cys residues of FlgI form an intramolecular disulfide bond, which is essential for P-ring assembly (8). Because a pulse-labeling experiment using minicells showed that l-cystine had no effect on the level of FlgI, the authors argued against the possibility that the disulfide bond is involved in the stability of the FlgI protein. These observations appear inconsistent with the present study. Such an inconsistency might be explained by the properties of minicells. Because minicells do not have a chromosome, proteins are not produced via chromosomal expression. The levels of proteins that have a rapid turnover rate under reducing conditions (28) (e.g., DegP [HtrA], a periplasmic protease/chaperone) would decrease in minicell preparations. Therefore, it is possible that some of the factors involved in degrading incorrectly folded FlgI proteins are missing in minicells.
Spontaneous oxidation of the FlgI protein may account for the suppression of the motility defect of the ΔdsbB strain by FlgI overproduction (Fig. 5). In fact, approximately 10% of ΔdsbB cells were motile in the absence of l-cystine under our experimental conditions. It has been reported that disulfide bond formation in OmpA, an outer membrane protein, and PhoA, a periplasmic protein, takes place spontaneously but slowly in the absence of the DsbA/DsbB system (4, 5), presumably due to the oxidative environment of the periplasmic space. Therefore, to assess spontaneous oxidation in our experiments, we examined the motility of ΔdsbB cells producing the Cys-eliminated FlgI variants. Overproducing FlgI C338A restored the swimming ability of the ΔdsbB cells to almost the same level as wild-type FlgI, whereas overproduction of both FlgI C254A and FlgI C254A/C338A partially rescued the motility defect of these cells (data not shown). These results suggest that the effects of spontaneous oxidation were negligible in our overproduction experiments.
Because DsbA immediately acts on newly translocated periplasmic proteins (30), the disulfide bond forms in newly translocated FlgI as it enters the periplasmic space. With the assistance of FlgA (22), the folded monomers are assembled into the P ring around the growing rod. In ΔdsbB strains, however, disulfide bond formation and therefore the folding of the FlgI monomers should be the rate-limiting step in the P-ring formation pathway. In these strains, most of the translocated FlgI proteins likely aggregate or are degraded, with only a small fraction of FlgI folding appropriately without the disulfide bond. It is plausible that in strains that are deficient in disulfide bond formation, the chromosomal expression level of FlgI is insufficient to allow P-ring assembly due to the limited amount of the required building blocks. Overproduction from a plasmid is necessary to construct the P ring with FlgI proteins that do not have the intramolecular disulfide bond.
We found that the Cys-eliminated FlgI variants were rapidly degraded in the flhDC strain (Fig. 3), but some of the FlgI C338A protein remained in the ΔflgI strain even after FlgI production was arrested for 60 min (Fig. 4). These results indicate that the FlgI protein even without the disulfide bond is resistant to degradation in the presence of the other flagellar proteins. This resistance could be due to P-ring formation, because it can be postulated that the C338A proteins that remained after the 60-min incubation were incorporated into the flagellum as a P ring. Another possibility is that FlgA associates with the FlgI protein to protect it from degradation (22). The first possibility is supported by several lines of evidence. A previous study reported that the LP ring is fairly stable; several chemical treatments failed to disassemble the structure in vitro (2). In this study, no flagellar structures without a P ring were observed by electron microscopy in the flagellar preparations from the cells expressing the C338A protein (data not shown), suggesting that P rings made up of FlgI C338A were not disassembled during the preparation processes. Also, a P ring made with the C338A protein must be moderately stable in vivo, because ΔflgI cells with the C338A mutation retained their swimming ability after the synthesis of the C338A protein was arrested for 60 min. Thus, we suggest that once FlgI is incorporated into the flagellar basal body, the protein becomes resistant to degradation even without the intramolecular disulfide bond.
Acknowledgments
We express thanks to Kazuhiro Kutsukake (Okayama University) for kindly providing the anti-Salmonella FlgI antibodies, which were useful in the beginning of this study, and to Hiroshi Kadokura and Jon Beckwith (Harvard Medical School) for the dsbB strains and plasmids. We thank Hiroyuki Terashima, Akari Shinohara, Teppei Morita, and Toshifumi Inada (Nagoya University) for technical assistance. We are grateful to Mike Manson (Texas A&M University), Chi Aizawa (Hiroshima Prefectural University), and Hideyuki Matsunami (Osaka University) for their invaluable suggestions about the manuscript.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan; from the Japan Science and Technology Corporation (to M.H., I.K. and T.Y.); and from the Soft Nano-Machine Project of the Japan Science and Technology Agency (to T.Y. and M.H.).
REFERENCES
- 1.Aizawa, S. I., G. E. Dean, C. J. Jones, R. M. Macnab, and S. Yamaguchi. 1985. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J. Bacteriol. 161:836-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiba, T., H. Yoshimura, and K. Namba. 1991. Monolayer crystallization of flagellar L-P rings by sequential addition and depletion of lipid. Science 252:1544-1546. [DOI] [PubMed] [Google Scholar]
- 3.Akiyama, Y., and K. Ito. 1993. Folding and assembly of bacterial alkaline phosphatase in vitro and in vivo. J. Biol. Chem. 268:8146-8150. [PubMed] [Google Scholar]
- 4.Bardwell, J. C., J. O. Lee, G. Jander, N. Martin, D. Belin, and J. Beckwith. 1993. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. USA 90:1038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell, J. C., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 6.Blair, D. F. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545:86-95. [DOI] [PubMed] [Google Scholar]
- 7.Collet, J. F., and J. C. Bardwell. 2002. Oxidative protein folding in bacteria. Mol. Microbiol. 44:1-8. [DOI] [PubMed] [Google Scholar]
- 8.Dailey, F. E., and H. C. Berg. 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 90:1043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. [DOI] [PubMed] [Google Scholar]
- 11.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiniker, A., and J. C. Bardwell. 2004. In vivo substrate specificity of periplasmic disulfide oxidoreductases. J. Biol. Chem. 279:12967-12973. [DOI] [PubMed] [Google Scholar]
- 13.Homma, M., Y. Komeda, T. Iino, and R. M. Macnab. 1987. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J. Bacteriol. 169:1493-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, C. J., M. Homma, and R. M. Macnab. 1989. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J. Bacteriol. 171:3890-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, C. J., R. M. Macnab, H. Okino, and S. Aizawa. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 212:377-387. [DOI] [PubMed] [Google Scholar]
- 16.Kadokura, H., and J. Beckwith. 2002. Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J. 21:2354-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi, K., T. Saitoh, D. S. Shah, K. Ohnishi, I. G. Goodfellow, R. E. Sockett, and S. I. Aizawa. 2003. Purification and characterization of the flagellar basal body of Rhodobacter sphaeroides. J. Bacteriol. 185:5295-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima, S., Y. Asai, T. Atsumi, I. Kawagishi, and M. Homma. 1999. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations in putative channel components. J. Mol. Biol. 285:1537-1547. [DOI] [PubMed] [Google Scholar]
- 19.Kubori, T., N. Shimamoto, S. Yamaguchi, K. Namba, and S. Aizawa. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433-446. [DOI] [PubMed] [Google Scholar]
- 20.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 21.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 22.Nambu, T., and K. Kutsukake. 2000. The Salmonella FlgA protein, a putative periplasmic chaperone essential for flagellar P ring formation. Microbiology 146:1171-1178. [DOI] [PubMed] [Google Scholar]
- 23.Ohnishi, K., M. Homma, K. Kutsukake, and T. Iino. 1987. Formation of flagella lacking outer rings by flaM, flaU, and flaY mutants of Escherichia coli. J. Bacteriol. 169:1485-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliver, D. B., R. J. Cabelli, K. M. Dolan, and G. P. Jarosik. 1990. Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc. Natl. Acad. Sci. USA 87:8227-8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Skorko-Glonek, J., D. Zurawa, F. Tanfani, A. Scire, A. Wawrzynow, J. Narkiewicz, E. Bertoli, and B. Lipinska. 2003. The N-terminal region of HtrA heat shock protease from Escherichia coli is essential for stabilization of HtrA primary structure and maintaining of its oligomeric structure. Biochim. Biophys. Acta 1649:171-182. [DOI] [PubMed] [Google Scholar]
- 29.Slocum, M. K., and J. S. Parkinson. 1983. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: organization of the tar region. J. Bacteriol. 155:565-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sone, M., Y. Akiyama, and K. Ito. 1997. Differential in vivo roles played by DsbA and DsbC in the formation of protein disulfide bonds. J. Biol. Chem. 272:10349-10352. [DOI] [PubMed] [Google Scholar]
- 31.Sosinsky, G. E., N. R. Francis, D. J. DeRosier, J. S. Wall, M. N. Simon, and J. Hainfeld. 1992. Mass determination and estimation of subunit stoichiometry of the bacterial hook-basal body flagellar complex of Salmonella typhimurium by scanning transmission electron microscopy. Proc. Natl. Acad. Sci. USA 89:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studier, F. W. 1991. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219:37-44. [DOI] [PubMed] [Google Scholar]
- 33.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 34.Yorimitsu, T., K. Sato, Y. Asai, I. Kawagishi, and M. Homma. 1999. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J. Bacteriol. 181:5103-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]