Abstract
The catalytic core of Escherichia coli DNA polymerase III contains three tightly associated subunits, the α, ɛ, and θ subunits. The θ subunit is the smallest and least understood subunit. The three-dimensional structure of θ in a complex with the unlabeled N-terminal domain of the ɛ subunit, ɛ186, was determined by multidimensional nuclear magnetic resonance spectroscopy. The structure was refined using pseudocontact shifts that resulted from inserting a lanthanide ion (Dy3+, Er3+, or Ho3+) at the active site of ɛ186. The structure determination revealed a three-helix bundle fold that is similar to the solution structures of θ in a methanol-water buffer and of the bacteriophage P1 homolog, HOT, in aqueous buffer. Conserved nuclear Overhauser enhancement (NOE) patterns obtained for free and complexed θ show that most of the structure changes little upon complex formation. Discrepancies with respect to a previously published structure of free θ (Keniry et al., Protein Sci. 9:721-733, 2000) were attributed to errors in the latter structure. The present structure satisfies the pseudocontact shifts better than either the structure of θ in methanol-water buffer or the structure of HOT. satisfies these shifts. The epitope of ɛ186 on θ was mapped by NOE difference spectroscopy and was found to involve helix 1 and the C-terminal part of helix 3. The pseudocontact shifts indicated that the helices of θ are located about 15 Å or farther from the lanthanide ion in the active site of ɛ186, in agreement with the extensive biochemical data for the θ-ɛ system.
The holoenzyme DNA polymerase III (Pol III) is the main replicative polymerase in Escherichia coli (37). It is a remarkable multisubunit enzyme that is capable of extraordinary speed and fidelity of action. Pol III is composed of 10 different subunits, 7 of which act as accessory subunits for a catalytic core composed of 3 tightly bound subunits. The polymerase active site is in the large α subunit (130 kDa) of the core (33, 34). The 3′-5′ proofreading exonuclease activity is located in the ɛ subunit (28 kDa) (48), specifically, in the N-terminal domain (ɛ186) (21 kDa), which also contains the binding site of the θ subunit (9 kDa). The θ subunit has no unambiguously designated function. The three subunits are arranged linearly such that the ɛ subunit occupies the central position, binding to both α and θ (51).
The possibility of understanding the detailed function of the Pol III holoenzyme has provoked interest in the structure of this enzyme since its discovery more than 25 years ago. Unfortunately, neither the complete holoenzyme complex nor the catalytic core has been crystallized yet. Instead, there has been considerable effort to solve the structures of individual subunits by both X-ray crystallographic and nuclear magnetic resonance (NMR) methods, and there has been substantial success recently for two of the three elements of the core. The structure of ɛ186 has been determined by X-ray crystallography (20) and has been modeled from NMR data (8). This structure led to a detailed understanding of the mechanism of the exonuclease activity (19, 20). NMR studies showed that the structure of the θ subunit in aqueous buffer contains three helices, a short stretch of β structure, and large segments of flexible polypeptide chains (26). Preliminary spectral data suggested that some of the poorly structured regions attain structure on binding to ɛ186. The refined structure of θ in a mixed alcohol-water buffer solution showed that this subunit is a three-helix bundle (39). The bacteriophage P1 homolog of θ, HOT, has the same three-helix bundle fold with minor structural variations (7). No atomic resolution structure for the α subunit or any related polymerase belonging to the PolC family is known.
The role of θ in Pol III function beyond stabilizing the structure of ɛ is unclear. θ is not required for the polymerase or exonuclease activities of the core or the holoenzyme (35, 50). It does, however, stimulate the exonuclease activity of ɛ two- to fourfold (44, 51), enhances the interaction of ɛ with α (52), and stabilizes ɛ186 under thermal inactivation (19) and chemical denaturation (17) conditions. The biochemical evidence for a direct interaction of θ with the DNA substrate or template strand is ambiguous. HOT plays a similar role when it is substituted for θ, and there has been a report that it has an even greater stabilizing effect on ɛ (3). The structure of ɛ186 changes little when it binds to θ (6), whereas some of the poorly structured regions of θ become more ordered (26).
The θ-binding epitope on ɛ186 was defined using NMR chemical shift mapping experiments and was localized to a mainly hydrophobic surface encompassing strands β2 and β3, helices α1 and α2, and the N terminus of helix α7 (6). This places the θ-binding site on a face of ɛ186 far (approximately 15 to 20 Å) from the active site. Identification of the ɛ186-binding epitope on θ by similar experiments is more ambiguous because chemical shift changes in θ occur for most of the molecule when it binds to ɛ186 (26).
One of the goals of structural genomics is to assign functions based on studies of the three-dimensional structures of proteins. With this in mind, and knowing that θ has a modest effect on the activity of ɛ, we have performed structural studies with the θ-ɛ186 complex to clarify the source of the effect on ɛ. This study is the latest in a series of studies directed at elucidating the interactions between θ, ɛ, and the DNA substrate. Here, we report the three-dimensional structure of θ when it is complexed with ɛ186; the results were refined using distance and angular restraints supplemented with restraints derived from pseudocontact shifts (PCS) that were induced by inserting a single lanthanide ion into the metal-binding site of ɛ186 (45). We compared the structure of θ in complex with ɛ186 with the structure of θ in mixed alcohol-water buffer and with the structure of HOT in aqueous buffer. Below we also report the results of nuclear Overhauser enhancement (NOE) difference spectroscopy experiments that defined the contact surface of θ on ɛ186 and compare the results with a model of the θ-ɛ complex that was assembled using only the PCS data (46).
MATERIALS AND METHODS
Proteins.
Methods for overproduction and purification of θ and ɛ186 and for production and purification of the θ-ɛ186 complex have been described in detail previously (19, 20). 13C/15N-labeled θ and 15N-labeled θ were produced using minimal media supplemented with l-(+)-[6-13C]glucose and 15NH4Cl (Cambridge Isotope Laboratories) as described previously (26). Concentrations of θ-ɛ186 were determined spectrophotometrically using an ɛ280 value of 14,650 M−1 cm−1.
NMR spectroscopy.
NMR experiments with complexes of 13C/15N- or 15N-labeled θ and unlabeled ɛ186 were carried out at 25 or 30°C using a Varian 600-MHz INOVA spectrometer equipped with a 5-mm PENTA probe, a Varian 800-MHz INOVA spectrometer equipped with a 5-mm triple-resonance probe, and a Bruker Avance 800 spectrometer equipped with a 5-mm TXI probe. Each probe was equipped with triple-axis actively shielded gradients. Each protein sample was extensively dialyzed and then concentrated by ultrafiltration to obtain a concentration of ∼1 mM in 20 mM sodium phosphate (pH 7.0) buffer containing 100 mM NaCl and 0.1 mM dithiothreitol, and D2O was added to a concentration of 10% (vol/vol) before NMR spectra were acquired. Sequential backbone and Cβ resonances were assigned by combined analysis of the results of the following experiments: HNCA (55), HN(CO)CA (55), Cβ-decoupled HNCA (36), Cβ-decoupled HN(CO)CA (36), HNCO (41), HCACO (15), HN(CA)CO (11), HNCACB (55), CBCA(CO)NH (14), three-dimensional (3D) sensitivity-enhanced 15N-separated total correlation spectroscopy (TOCSY) (56) with a mixing time of 70 ms, and 3D sensitivity-enhanced 15N-separated nuclear Overhauser enhancement spectroscopy (NOESY) (56) and 13C-separated NOESY (40) with mixing times of 80 ms. Side chain 13C and 1H resonances were assigned based on the results of HCCH-TOCSY (25), H(CCO)NH (13), and C(CO)NH (13) experiments. The results of all the experiments mentioned above were recorded at a 1H frequency of 600 MHz. The results of an additional 3D sensitivity-enhanced 15N-separated NOESY experiment and a 13C-separated NOESY experiment, each with a mixing time of 80 ms, were recorded at a 1H frequency of 800 MHz. The results of 13C/15N-half-filtered and double-half-filtered NOESY experiments (43) were recorded (at a 1H frequency of 800 MHz) to establish the residues in 13C/15N-labeled θ that have NOEs to residues in unlabeled ɛ186. The acquisition and processing parameters were similar to those described previously (26).
The ɛ186 epitope on θ was determined by NOE difference spectroscopy by using the method of Eichmüller et al. (10), except that the experiment was not performed in the constant-time mode. The experiment created parallel (Σ mode) and antiparallel (Δ mode) alignments of the longitudinal magnetization of unlabeled ɛ186 and 13C/15N-labeled θ. Two separate experiments were performed at a 1H frequency of 600 MHz, and in each experiment the Σ and Δ modes were acquired in an interleaved fashion as described previously (10), using mixing times of 300 and 0 ms. The cross-peak intensities of the two-dimensional maps for the four data sets (Σ300, Δ300, Σ0, and Δ0) were quantified using SPARKY (12), and the ratio of the intensities from these data sets, Γ = (Δ300/Σ300)/(Δ0/Σ0), was used to verify the proximity of each cross-peak to the interface. A ratio of <0.5 was taken to indicate close proximity to the interface, and a ratio of >0.8 was taken to indicate that a side chain was not at the interface.
13C/15N-labeled θ-ɛ186 with a single lanthanide ion (Dy3+, Er3+, or Ho3+) in the active site of ɛ186 was prepared as described previously (45). For these experiments, the buffer was exchanged with 20 mM Tris and 100 mM NaCl (pH 7.0) by ultrafiltration because of the limited solubility of lanthanide phosphates. Pseudocontact shifts induced when the lanthanide ion bound to the θ-ɛ186 complex were measured and evaluated by using 15N and 13C heteronuclear single quantum correlation (HSQC) spectra. Well-resolved shifted cross-peaks in the 15N-HSQC spectra of lanthanide-labeled θ-ɛ186 were initially assigned by inspection since they were displaced from their diamagnetic counterparts along approximately diagonal lines. The remaining cross-peaks were assigned using predicted PCS calculated from a preliminary model of θ that was derived from NOE and angular restraints. The assignments were verified by an iterative process that involved comparing the calculated PCS following structure refinement with the measured PCS derived from inspection of cross-peak positions in the diamagnetic and paramagnetic two-dimensional HSQC spectra. In addition, diamagnetic and paramagnetic 3D HNCO spectra were recorded for the θ-ɛ186-Dy3+ complex, and the PCS were observed in all three dimensions. This facilitated assignment of PCS that were larger in this complex than in the complexes with other lanthanide ions.
Spectral analysis and experimental restraints.
Spectral weighting followed by multidimensional Fourier transformation employing linear prediction was carried out with Varian VNMR version 6.1 software (Varian Associates, Palo Alto, CA) or NMRPipe (5) as described previously (26). After transformation, the processed data from all experiments were converted into the XEASY (1) and/or SPARKY (12) format. Peak picking, spectral analysis, and semiautomated resonance assignment were performed with XEASY, SPARKY, and CANDID (22). Three-dimensional NOESY cross-peaks were integrated using the program SPSCAN v1.0.53 (R. W. Glaser and K. Wüthrich, http://www.mol.biol.ethz.ch/wuthrich/software/spscan/). CALIBA (part of the CYANA suite of programs) (16) was used to convert NOESY cross-peak volumes into upper distance bounds. A total of 106 chemical shift index (CSI)-derived φ and ψ dihedral angle restraints from the program SHIFTY (42) were used. In addition to the NOE restraints, 48 hydrogen bond restraints (rNH-O = 1.5 to 2.4 Å, rN-O = 2.4 to 3.4 Å) were included based on the CSI and other indicators of α-helical structure and where a single hydrogen bond acceptor was identified in preliminary structure calculations. A total of 162 pseudocontact shifts of amide protons derived from separate experiments with 13C/15N-labeled θ-ɛ186-Dy3+, 13C/15N-labeled θ-ɛ186-Er3+, and 13C/15N-labeled θ-ɛ186-Ho3+ were used in the final structural refinement with the program XPLOR-NIH (49).
Structure calculations.
Initial structures were calculated using CYANA (16) and CANDID (22) with distance and dihedral angle restraints. Typically, each CYANA run was composed of 100 steps of minimization, followed by 4,000 molecular dynamics steps and then by 1,000 energy minimization steps. An iterative process involving several rounds of calculations was employed to assign previously ambiguous NOE cross-peaks using the program module CANDID (22). During final structure refinement with the program XPLOR-NIH (49), the 1H-1H NOEs and angular restraints were supplemented with 162 PCS estimated from the chemical shifts of backbone resonances in 15N-HSQC experiments with 13C/15N-labeled θ-ɛ186 recorded with and without a lanthanide ion (Dy3+, Er3+, or Ho3+). The parameters of the magnetic susceptibility anisotropy tensor (Δχ) were determined using the PCS values obtained for samples of θ-ɛ186 complexes when each leucine and phenylalanine in ɛ186 had been selectively 15N labeled as described previously (45). The values for the tensor anisotropy parameter Δχax for Dy3+, Er3+, and Ho3+ used in the calculations were 40.3, −10.9, and 14.7 M3 × 10−32, respectively, and the values for the tensor anisotropy parameter Δχrh used in the calculations were 4.5, −5.1, and 3.2 m3 × 10−32, respectively. Each XPLOR-NIH run was composed of 18,000 molecular dynamics steps, followed by 5,000 energy minimization steps. The quality of the structures was evaluated with PROCHECK-NMR (28). Experimental restraints were analyzed using the programs XPLOR-NIH and AQUA (28). Figures 2, 3, and 6 were prepared with MOLMOL (27).
FIG. 2.
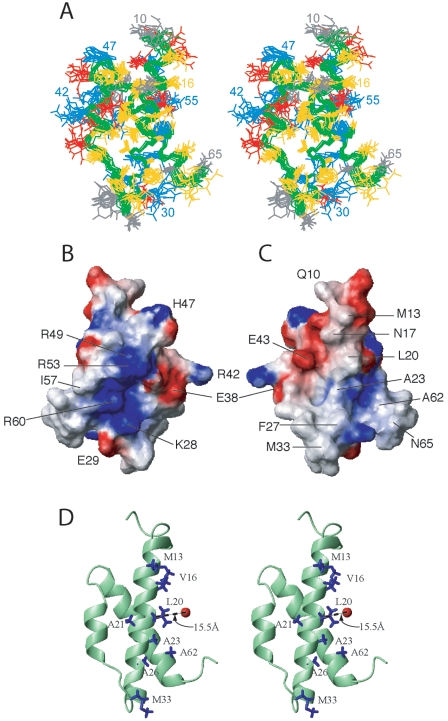
NMR-determined structures of the θ subunit in complex with ɛ186. (A) Stereo view of superposition of the backbone heavy atoms from the final ensemble of the 12 lowest-energy structures refined with distance, torsion angle, and pseudocontact shift restraints. Superposition was accomplished using the backbone atoms of Q10 to L66. (B and C) Calculated charge distribution mapped onto the surface of the θ subunit (residues 10 to 66). The regions with basic potential are indicated by blue, the regions with acidic potential are indicated by red, and white regions are uncharged regions of the θ surface. The mean structure is displayed, and the molecule in panel B has the same orientation as the stereo views in panels A and D, with helix 3 on the left, helix 1 in the center, and helix 2 on the right, whereas the molecule in panel C is rotated 180° about the vertical axis. (D) Stereo view of ɛ186-bound θ, showing some methyl-bearing side chains and the calculated position of the lanthanide ion in the active site of ɛ186. The side chains of methyl-bearing residues that showed substantial differential attenuation in the NOE difference experiment (>0.5) are blue. The approximate point of closest approach of the metal ion to the backbone is indicated. The coordinates are available from the PDB (PDB code 2AXD).
FIG. 3.
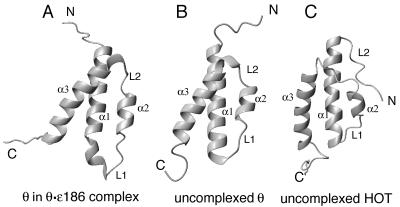
Comparison of the structures of the lowest-energy structure of ɛ186-bound θ (A), the lowest-energy uncomplexed θ structure (B), and the lowest-energy HOT structure (C). All structures have the same overall fold, but the angles between helices 1 and 3 are different. The ribbons trace the backbones of ɛ186-bound θ, free θ (PDB code 2AE9), and HOT (PDB code 1SE7), respectively.
FIG. 6.
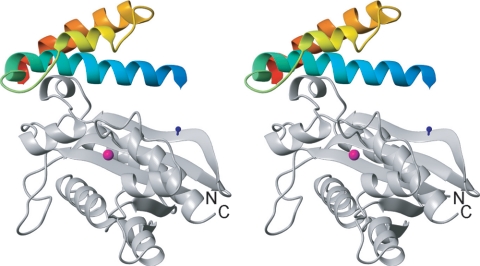
Stereo representation of the structure of the θ-ɛ186 complex as determined from pseudocontact shifts induced by Dy3+ and Er3+ ions (46). Residues 10 to 66 are included in the ribbon representation of θ, and the amino and carboxy termini are indicated by blue and red, respectively. The position of the lanthanide ion is indicated by the magenta sphere. The amide proton of H49 in ɛ186, which makes an NOE with the amide proton of A6 in θ (46), is indicated by a blue ball-and-stick diagram. The N and C termini of ɛ186 are labeled.
RESULTS
Resonance assignment and NMR spectroscopy.
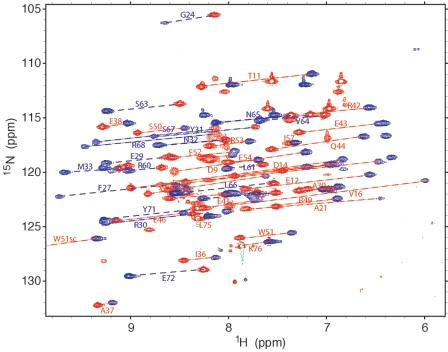
The optimum temperature range for acquisition of the spectra of the θ-ɛ186 complex was 25 to 30°C. At temperatures above 30°C, the sample aggregated, and at temperatures below 25°C, spectral resolution was not sufficient for analysis. Figure 1 shows superposition of the 15N-HSQC spectra recorded with Dy3+ and in the absence of a paramagnetic ion, from which the PCS were estimated. The 15N-HSQC and 15N-separated NOESY spectra of the 13C/15N-labeled θ subunit bound to ɛ186 showed a chemical shift dispersion characteristic of a folded, mostly helical protein. Narrow linewidths observed for the amide protons of the residues following P70 indicated that there were mobile residues at the C terminus. Nearly complete assignment of the backbone resonances (N, HN, Cα, C′) was achieved for the residues from D9 to K76, and partial assignments (N, HN) were obtained for backbone resonances for L2 to L8. The amide cross-peaks for residues L2 to L8 were broad, and their intensities in the 15N-HSQC and 15N-separated NOESY and TOCSY spectra were low. All the observable backbone amide cross-peaks (70 of an expected 71 cross-peaks) in the 15N-HSQC spectrum were assigned. Only the amide resonance of H47 could not be assigned. In contrast to the corresponding cross-peaks in the 15N-HSQC spectrum of uncomplexed θ, in which the amide resonances in the N-terminal section of helix 1 (L8 to V18) were narrow (26), more uniform signal intensities were observed for these resonances in the θ-ɛ186 complex, indicating that there was reduced mobility. Similarly, amide resonances from the C-terminal part of helix 1 (D19 to K28) and the C-terminal section of helix 3 (E54 to A62), which were either broad or not observed in uncomplexed θ (26), were easily detected and well resolved for the θ-ɛ186 complex, indicating that complex formation with ɛ186 abolished a chemical exchange process on the millisecond timescale that broadened the resonances for free θ. In contrast, the resonances of S67 to L69 were broad for the θ-ɛ186 complex, although they were narrow for free θ, which may be explained by close proximity to ɛ186. Interestingly, DeRose et al. (7) reported low-intensity amide peaks for A22 to S24 for HOT, but the equivalent peaks for ɛ186-bound θ, A21 to A23, were intense. Backbone nonexchangeable 1H and 13C resonances were assigned by standard procedures using HNCA, HN(CO)CA, and HNCO spectra, which resulted in assignment of 73 Hα (96%), 70 Cα (92%), and 61 C′ (80%) resonances. Most of the unassigned nonexchangeable resonances were from residues near the N terminus. The absence of any intense exchange peaks between the water and the amide protons of these residues in the 15N-separated NOESY spectrum indicated that the broadening of the amide signals was due to conformational exchange rather than H exchange with the water.
FIG. 1.
Effect of Dy3+ on the 15N-HSQC spectrum of the 13C/15N-labeled θ subunit complexed with ɛ186. Spectra were recorded in 90% H2O-10% D2O and 100 mM NaCl-20 mM Tris buffer (pH 7.0) at 25°C. Spectra recorded with Dy3+ (blue peaks) and in the absence of paramagnetic ion (red peaks) are superimposed. Lines connect pairs of diamagnetic and paramagnetic cross-peaks. Red lines indicate a negative pseudocontact shift, and blue lines indicate a positive pseudocontact shift. Cross-peaks are labeled with the residue assignment. For clarity, not all peaks are connected or labeled.
Secondary structure.
The NOE data were obtained from 80-ms-mixing-time 15N-separated NOESY and 13C-separated NOESY spectra at 600 and 800 MHz. The presence of medium to strong dNN and weak dαN(i,i + 1) and dαN(i,i + 3) NOEs and the consensus CSI (53, 54) identified three regions of helicity between Q10 and Y31 (helix 1), between A37 and R42 (helix 2), and between L48 and L66 (helix 3). Analysis using the programs TALOS (4) (Hα, Cα, Cβ, and C′ chemical shifts) and SHIFTY (42) (Hα, Cα, Cβ, C′, N, and HN chemical shifts) placed the helices at Q10 to Y31, A37 to R42, and R49 to R68. Helical wheel representations showed that helix 1 is mostly hydrophobic but helix 2 is amphipathic, with a predominance of acidic residues on one of its surfaces. On the other hand, helix 3 is amphipathic with mostly basic residues covering one face. The helical regions are broken by a sharp turn between N32 and P34, a short section of extended structure from V35 to A37, and a break between helices 2 and 3 that occurs near P45. The segment from P34 to I36 is characterized by weak dNN and strong dαN(i,i + 1) NOEs and an inversion of the CSI, indicating the presence of an extended conformation. Similarly, the pattern of strong dNN NOEs in the segment from E38 to Q44 is broken at P45 but resumes again at L48. The CSI again inverts at E46, but values characteristic of a helix resume at L48. Strong sequential dαδ(i,i + 1) NOEs observed for all proline Hδ resonances indicated that they are all in the trans conformation. Glycine is frequently a helix breaker (47), but all the markers for helical structure continue past G24 in the middle of helix 1. This may be due to a sequence of three alanine residues that precede G24; alanine strongly favors helix formation (47). The presence of dαN(i,i + 2) NOEs and the lack of dαN(i,i + 4) NOEs suggest that helix 1 terminates with a short stretch of a 310 helix.
Structure calculations.
The input data for structure calculations included 1H NOEs from 15N- and 13C-separated NOESY spectra, angular data from chemical shifts, and PCS as described in Materials and Methods. The PCS data were particularly valuable as the number of long-range NOEs was relatively small (Table 1), in part because most long-range contacts involved poorly resolved side chain resonances and most of the resonances from the aromatic rings of F27, Y31, and F52 were broad.
TABLE 1.
Structural statistics and root mean square deviations for the 12 NMR structure conformers representing the θ subunit of DNA polymerase III in complex with ɛ186
| Parameter | Value |
|---|---|
| Structural restraints | |
| Distance restraints | |
| Meaningful NOEs | 584 |
| Intraresidue | 152 |
| Medium range, i-j ≤ 5 | 343 |
| Long range, i-j ≥ 5 | 41 |
| Dihedral angle restraints | 111 |
| Hydrogen bond restraints | 48 |
| Pseudocontact shifts | 162 |
| Statistics for structure calculations | |
| Average restraint violations | |
| NOE violation (Å) | 0.19 ± 0.05 |
| Largest φ violation (°) | 12 |
| Largest ψ violation (°) | 10 |
| Atomic RMS deviations, residues 10 to 66 | |
| Backbone (Å) | 1.03 ± 0.30 |
| All heavy atoms (Å) | 2.11 ± 0.35 |
| Ramanchandran analysis, residues 10 to 66 | |
| Residues in the most favored regions (%) | 92.8 |
| Residues in additionally allowed regions (%) | 7.2 |
| Residues in generously allowed regions (%) | 0 |
| Residues in disallowed regions (%) | 0 |
| Energetics | |
| CHARMm energy (kcal/mol) | −881 |
Several rounds of structure calculations with CYANA (16) and CANDID (22) were used to refine the structure and assign cross-peaks. The final refinement was performed with the program XPLOR-NIH, with which the θ structure was refined against the complete set of distance, angular, and PCS (Dy3+, Er3+, and Ho3+) restraints. After the final round of calculations, the 12 structures with the lowest energies were retained from the 200 structures calculated (Fig. 2A). The backbone conformation is well defined except for nine residues at the N terminus and 10 residues at the C terminus. The structural statistics and residual violations of the experimental restraints are shown in Table 1. Inclusion of the PCS restraints reduced the root mean square (RMS) deviation values and slightly changed the relative orientations of the helices. Refinement against the distance and angular restraints yielded backbone RMS deviations of about 1.5 Å (residues 10 to 66), whereas the RMS deviations decreased to about 1.0 Å after inclusion of the PCS restraints. No well-defined structure could be calculated for residues at the N terminus (M1 to L8) and the C terminus (P70 to K76) because of the lack of meaningful intraprotein medium- and long-range NOEs in these regions of θ. Narrow cross-peaks for residues in the region from P70 to K76 indicate that the C terminus of θ is highly mobile, whereas intermolecular NOEs between A6 of θ and residues of ɛ186 and conformational exchange broadening of resonances indicate that there may be some structure at the N terminus.
The quality of the structure was supported by the appearance of the Ramanchandran plot, generated by PROCHECK-NMR (28), which showed that all the residues in the generously allowed and disallowed regions are in disordered parts of the protein (mostly the C terminus), for which there were no meaningful NOE data to restrict the protein backbone to any region of conformational space.
Description of the solution structure and comparison with uncomplexed θ and HOT.
The NMR-derived structure of θ bound to ɛ186 is a three-helix bundle (Fig. 2 and 3). Helix 1 is approximately antiparallel to helix 2 and parallel to helix 3, making angles of 170° and 50°, respectively. Helix 2 makes an angle of about 130° with helix 3. A hydrophobic core between the helices is populated mostly by short- and long-chain aliphatic residues. Several side chain resonances of K15 are shifted upfield, in agreement with the location of K15 between helices 1 and 3 and packed above W51. D19 and R55 are adjacent and partially protected from the solvent, and they may form a salt bridge. Several basic residues in helices 2 and 3 are oriented away from the protein core into the solution and are clustered on one side of θ. These residues are arginine residues at positions 42, 49, 53, 60, and 68.
The overall fold of the structure (Fig. 3A) is different from the fold described previously for free θ in aqueous buffer (26) and is more similar to the fold of free θ in a mixed methanol-H2O buffer (39) and the fold of HOT in aqueous buffer (7). Although the number of long-range NOEs for θ complexed with ɛ186 is fairly small, the NOEs are more uniformly distributed over the sequence than they were for the calculation of free θ in aqueous buffer (26). Since several of the characteristically ring current shifted resonances and the patterns of NOEs are very similar in complexed and free θ in aqueous buffer, we concluded that the structure described here and the structure of θ in the mixed solvent (39) reflect the structure of free θ in aqueous buffer more closely than the previously published structure (26). The most significant difference between free and ɛ186-bound θ appears to be in different mobilities that cause line broadening for different segments of the polypeptide chain, particularly at the interface between helix 1 and helix 3. Binding to ɛ186 stabilizes the θ structure rather than inducing any large changes.
HOT, which exhibits 53% sequence identity with θ (29), also folds into a three-helix bundle (7). Ribbon diagrams of ɛ186-bound θ, uncomplexed θ in mixed buffer (39), and uncomplexed HOT are shown in Fig. 3. Each structure has helices that are approximately the same length, and each structure has loop regions that reverse the direction of the peptide chain. The orientations of helices 1 and 3 differ by about 10° for complexed and uncomplexed θ (Fig. 3A and B) and by 20° for complexed θ and HOT (Fig. 3A and C). A DALI (23) search of the Protein Data Bank (PDB) revealed that of the proteins in the PDB, HOT is the protein most closely related to θ, with a Z score of 5.8 for the structured regions.
The structures of complexed θ and uncomplexed θ in the mixed-solvent buffer are very similar, and the greatest differences are the orientation of helix 1 with respect to the other two helices and the conformation of loop L1. These differences may be due to structural changes on complexation with ɛ or to the use of long-range PCS restraints. The latter better define the orientation of helices and loops, especially where there are few long-range distance restraints. Mueller et al. (39) commented on the paucity of long-range restraints in the vicinity of helix 2 and loop L2 of uncomplexed θ.
Electrostatic surface of θ.
Figure 2B and C show opposing faces of the electrostatic surface of the folded core of θ (residues 10 to 66) in the complex with ɛ186. Figure 2C has the same orientation as the stereo views shown in Fig. 2A and D, and Fig. 2B shows the opposing face of the surface shown in Fig. 2C. The surface shown in Fig. 2B comprising mostly residues from helices 2 and 3 is highly positively charged with several arginine and lysine side chains in close proximity. The functional significance of the cluster of arginine residues oriented outward on helix 3 is unknown. There are also small pockets of hydrophobic residues at the C terminus of helix 3 and the turn structure between helices 1 and 2. Hydrophobic residues from helix 1 and helix 3 dominate the opposing face, which can be segregated into distinct regions (Fig. 2C). The surface of helix 1 is hydrophobic, resulting from the predominance of hydrophobic residues in the central part of the helix (V16 to F27). This hydrophobic surface extends to the base of the turn structure encompassing N32, M33, and P34. On either side of the central hydrophobic spine are positively and negatively charged residues.
θ-ɛ interface.
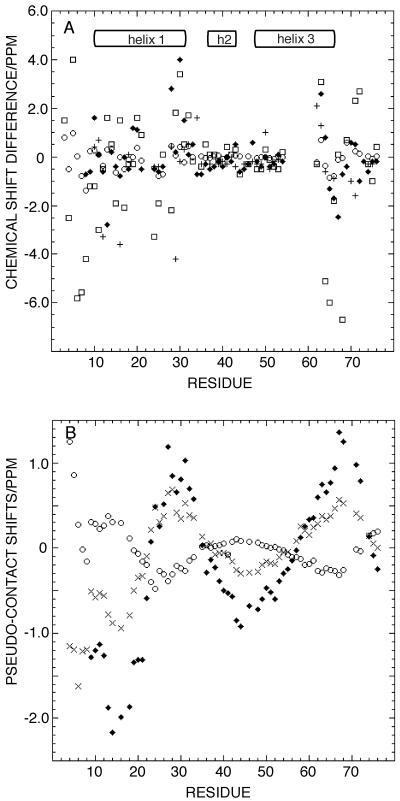
A conventional method for defining the interface between two proteins is to map the perturbation of chemical shifts and NMR signal linewidths of one protein component caused by complexation of the other. Clustering of large changes in chemical shifts in a particular region is indicative of a potential binding surface. The θ epitope on ɛ186 was identified in this way (6). Figure 4A shows the changes in the chemical shift of the amide proton, amide nitrogen, backbone carbonyl, and α carbons of θ when the complex is formed. Most of the residues show significant changes in chemical shifts; the exception is the region between M33 and W51. These results suggest that the N-terminal segment of helix 1 and the C terminus of helix 3 are involved in contacts with ɛ186. The previously identified (26) sections of uncomplexed θ that are affected by conformational or chemical exchange phenomena include the C-terminal sections of helices 1 and 3 (D19 to Y31 and R53 to S67). Thus, it is difficult to distinguish the effects of the direct contact with ɛ186 from indirect effects due to a more stable secondary structure.
FIG. 4.
Changes in chemical shifts of θ upon binding to ɛ186 and 1HN pseudocontact shifts of θ. (A) Changes in chemical shifts of 1HN (○) and 15N[amide] (□) and 13Cα (⧫) and 13C′ (+) spins. There are significant changes in regions where contact between θ and ɛ186 is also suggested by the results shown in Fig. 5. Little or no change is observed in the region from V35 to R53, suggesting that this region is not in contact with ɛ186. The resonance assignments in free θ were obtained from reference 9. (B) Plot of pseudocontact shifts of the backbone HN induced by Dy3+ (⧫), Er3+ (○), and Ho3+ (×) versus residue number.
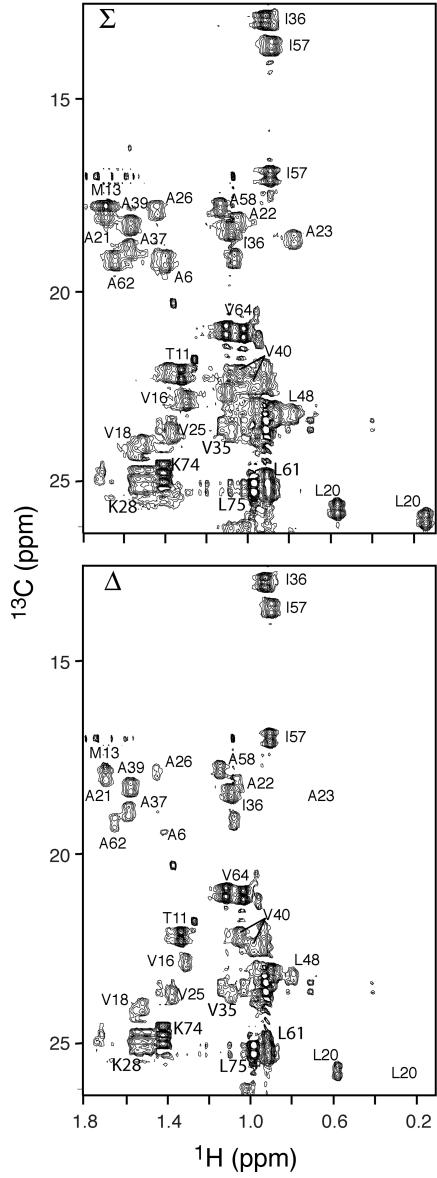
NOE difference spectroscopy (10) is an alternative method for mapping the binding epitope on a protein when the target protein is uniformly 13C/15N labeled and the other protein component is not labeled. We used this method to identify the side chains of methyl-bearing residues of 13C/15N-labeled θ that are at the interface with ɛ186. Figure 5 shows the Σ and Δ modes of part of the 1H-13C correlation map of the complex recorded with a mixing time of 300 ms. Substantial differential relaxation (>50%) was observed for the side chain resonances of A6, M13, V16, L20, A21, A23, A26, and M33 (not shown), which reside in or just precede helix 1. A62 and L66, which reside in helix 3 but face helix 1, also showed substantial differential relaxation. Figure 2D shows the locations of all of these residues in the 3D structure. Very little differential relaxation was observed for V35, I36, A37, A39, I57, A58, and V64, which reside either in the turn structure or in helix 2 or 3. A6 and L20 displayed several NOEs to the protons of unlabeled ɛ186 in half-filtered and double-half-filtered NOESY experiments (46). Furthermore, the methyl resonances of L20 were shifted upfield, indicating the presence of a nearby aromatic residue, which could only come from ɛ186 since there are no nearby aromatic rings in the θ structure. In the structure of HOT, L21 (the corresponding residue) forms part of the hydrophobic core (7), but in θ in the mixed-solvent buffer L20 is fully exposed to the solvent. The interaction interface of θ with ɛ186 clearly involves the hydrophobic surface of helix 1. The data are consistent with the chemical shift perturbation data (Fig. 4A), in that the residues with the smallest chemical shift perturbation show the smallest net differential relaxation effects.
FIG. 5.
1H-13C correlation maps obtained by the technique of Eichmüller et al. (10), showing the differential decay of θ 1H magnetization after preparation of the Σ and Δ spin modes of the 13C-HSQC spectra of ɛ186-bound 13C/15N-labeled θ. The two spectra were recorded with an NOE mixing period of 300 ms. A reference pair of spectra obtained by using a 0-ms NOESY mixing period (not shown) was also recorded to account for differences in one-bond heteronuclear scalar coupling constants.
An interface aligning helix 1 of θ with ɛ186 is also qualitatively consistent with the results of experiments with lanthanide ions at the ɛ186 active site, since residues in the N-terminal half of helix 1 had larger average PCS than residues in helices 2 and 3 (Fig. 4B). This indicates that helix 1 is closer to the active site of ɛ186 than are other regions of θ. The position of the lanthanide ion with respect to the structure of θ is shown in Fig. 2D. The closest distance of approach of the lanthanide and helix 1 of θ, ∼15 Å, is consistent with the presence of an interaction site near a hydrophobic patch on ɛ186 encompassing helix α1, strands β2 and β3, and the N terminus of helix α7 (6). Chemical shift mapping of the binding epitope of θ on ɛ186 demonstrated that the interface between ɛ and θ is primarily hydrophobic (6), which is in good agreement with the results of the experiments described above. This places the ɛ186 epitope on θ primarily along helix 1, as shown by the hydrophobic surface in Fig. 2C. Preliminary chemical shift mapping experiments of the HOT-ɛ186 complex placed the interaction site near the N terminus of helix 1 and the C terminus of helix 3 (7), in good agreement with the results of the NOE difference experiments.
Notably, 15N-HSQC cross-peaks could be assigned for residues 4 to 9 preceding helix 1 in the samples with Er3+ and Ho3+. Some of these cross-peaks could also be observed for the θ-ɛ186-Dy3+ complex, but they were more difficult to assign. The fact that cross-peaks could be observed at all for these residues in the paramagnetic samples indicates that their amide protons must be located more than 14 Å from the paramagnetic ion (2). Cross-peaks of residues 2 and 3 were already weak in the diamagnetic complex and were therefore less likely to be observed in the paramagnetic complexes. The magnitude of the PCS observed for these residues thus indicates that they are closer to the active site of ɛ186 than any other residue of θ is but may not reach all the way to this site.
DISCUSSION
The small θ subunit of DNA polymerase III forms an isolable complex with the catalytic N-terminal domain (ɛ186) of the ɛ proofreading 3′-5′ exonuclease subunit (19, 44). The high-resolution crystal structure of ɛ186 (20) has provided impetus to modeling the structure of the θ-ɛ186 complex, both to explain its stability and to obtain insight into the roles of θ in DNA replication. Although θ is not necessary for the exonuclease activity of ɛ, it has been shown to protect ɛ186 against thermal inactivation, as measured by the activity remaining after treatment for 10 min at an elevated temperature; the temperature at which 50% of the activity remained was about 14°C higher for θ-ɛ186 than for free ɛ186 (19). Free ɛ186 was also observed to precipitate irreversibly in a few hours under conditions used here for extensive NMR studies of the complex (at 25 or 30°C); NMR spectra of free ɛ186 could be obtained only at 10°C (18). Furthermore, as judged from the stability of the intact Pol III core (α-ɛ-θ complex) during 28,000-fold purification from plasmid-free E. coli (38), the θ-ɛ complex is probably extremely stable to dissociation. Direct measurement of the dissociation constant of the complex has not been reported, but an upper limit for the dissociation constant of 20 nM has been obtained by electrospray ionization-mass spectrometry (17).
Our determination of the three-dimensional structure of θ complexed with ɛ186 is the next step for understanding the basis for this intimate and stable protein-protein interaction, and our findings elucidated the binding interface. Remarkably, the NMR spectra obtained for θ in the 30-kDa complex with ɛ186 were easier to interpret than the NMR data obtained for free θ, yet the number of long-range NOEs that could be observed between the helices in θ was still small for the complex. In this situation the global PCS restraints made a significant contribution to the quality of the structure.
Overall, the structure obtained for ɛ186-bound θ is similar to the structure of free θ in mixed alcohol-water solvent (39) and to the structure of the close homolog HOT in aqueous buffer (7). The differences among these structures are either due to structural changes in the free and complexed proteins or due to limitations of the structure determination protocols. Since PCS restraints were used in the structure calculation, the present NMR structure of θ satisfies the PCS restraints better than the structure of either free θ (39) or HOT (7). We noted that the side chain of L21 is barely solvent exposed in the structure of HOT (7), so it would not be expected to engage in intermolecular NOEs with ɛ186. In contrast, the corresponding residue in θ, L20, is much more solvent exposed in the complexed and free forms of θ (39). Since the methyl groups of this leucine have intermolecular NOEs with ɛ186 and experience strong ring currents in the complex with ɛ186 but not in free θ, the two structures of θ provide plausible models of the structure in the complex. The small chemical shift changes observed for helix 2 and most of helix 3 in θ suggest that any changes in the global fold of θ upon complexation with ɛ186 are small.
The intermolecular NOEs observed for L20 and the NOE difference experiments (10) resulted in identification of a hydrophobic surface along helix 1, as shown in Fig. 2C, as an important part of the interaction surface with ɛ. The result is in agreement with the chemical shift mapping of the binding epitope of θ on ɛ186 that indicated that the interface between θ and ɛ is primarily hydrophobic (6). Electrospray ionization-mass spectrometry studies of the dissociation of θ-ɛ186 also suggested that the θ-ɛ interface is hydrophobic (17). Furthermore, preliminary chemical shift mapping experiments with the HOT-ɛ186 complex placed the interaction site near the N terminus of helix 1 and the C terminus of helix 3 (7), in agreement with our results.
The intermolecular NOEs and reduced mobility observed for A6 show that the N-terminal segment of θ is involved in specific contacts with ɛ186, whereas it is highly mobile in uncomplexed θ in aqueous (26) and mixed-solvent buffers (39). Thus, it is clear that the N-terminal segment of θ folds upon complex formation with ɛ186, although most interactions of this segment may be intermolecular rather than intramolecular. Although the absence of intraprotein long-range NOEs precluded structure determination for the first nine residues of ɛ-bound θ, PCS values and paramagnetic relaxation enhancements indicated that the N-terminal residues of θ are closer to the metal ion at the active site of ɛ186 than are any of the other residues of θ.
We used a novel NMR strategy (46) based on the PCS induced by inserting lanthanides at the ɛ186 metal-binding site to dock the NMR-derived structure of θ described above with the X-ray-derived structure of ɛ186 (20). The PCS-based routine for rigid docking of θ and ɛ186 is described in detail elsewhere (46). Briefly, the PCS detected for the amide protons of [15N]Leu- and [15N]Phe-labeled ɛ186 (with unlabeled θ) were used to derive the Δχ tensor of Dy3+ with respect to the crystal structure of ɛ186. The PCS of the amide protons of 15N-labeled θ (with unlabeled ɛ186) were subsequently used to position and orient the Δχ tensor with respect to the solution structure of θ described here. The common coordinate system defined in this way by the principal axes of the Δχ tensor of the metal ion positioned and oriented the structures of ɛ186 and θ with respect to each other. Er3+ data recorded in the same manner defined a second coordinate system that was equally centered about the metal ion but oriented somewhat differently. The combination of Dy3+ and Er3+ data provided a unique solution for the ɛ186-θ complex (46). A stereo representation of the model is shown in Fig. 6. The overall structure agrees with the results of NOE difference spectroscopy, intermolecular NOEs, and chemical shift changes, placing the hydrophobic surface at the C terminus of helix 1 (Fig. 6) and part of the C terminus of helix 3 in contact with the short helix α2 in ɛ186. The N terminus of helix 1 of θ is approximately parallel to one of the strands of the β-sheet in ɛ186. The closest approach of the backbone of θ is ∼15 Å from the active site of ɛ186, and thus the backbone of the helices of θ is too far away to have a direct role in the proofreading process. Although there is no direct interaction with the catalytically important helix 7 of ɛ186, θ may have a role in stabilizing the position of this helix by restraining the adjacent β strand. This stabilizing role is consistent with biological evidence showing that θ is not required for the proofreading function of ɛ but rather has a modest effect on the activity of ɛ and the fidelity of replication (52).
Exonuclease I, a structural relative of ɛ, has an additional C-terminal helical domain that we speculated previously may be an analog of θ (20). However, when the location of this domain in exonuclease I was compared with the location of θ on ɛ186, there was little overlap; the C-terminal domain of exonuclease I does not overlay the β-sheet and only partially overlays helix α2. In addition, there is very little structural homology between θ and the C-terminal domain of exonuclease I, and its function is probably unrelated (46).
The interface between θ and ɛ186 is large and relatively flat and is populated mostly by long-chain and aromatic side chains. The minimum contact surface between θ and ɛ186 as shown in Fig. 6 was calculated to be 1,400 Å2 per monomer by the program DSSP (24). The actual contact surface is almost certainly larger because the structure determination of θ did not take into account the interaction between ɛ186 and the N terminus of θ shown by the intermolecular NOEs between A6 of θ and ɛ186. This interface area corresponds to the standard-size protein interface as defined by Lo Conte et al. (30) and is typical of protein-protein complexes with nanomolar affinity (30), particularly those with hydrophobic interfaces.
Although there is no evidence that θ interacts with either single- or double-stranded DNA, it is intriguing that it has two motifs that figure prominently in DNA-binding proteins. The helix-loop-helix motif encompasses helix 1, loop 1, and helix 2 of θ, and the helix-hairpin-helix motif encompasses helix 2, loop 2, and helix 3 of θ. The helix-hairpin-helix motif has been proposed to be a nonspecific, single-stranded DNA-binding motif (9, 32), and the helix-loop-helix motif is also common in proteins that bind to DNA (21, 31). The solvent-facing surface of helix 3 of θ has several closely spaced arginine residues (Fig. 2B) that could also function as a DNA-binding region.
Acknowledgments
We are grateful to Varian Associates (Palo Alto, CA) for generously providing time on the INOVA 800 spectrometer. We thank Esther Bulloch and Tanya Ronson for assistance with sample preparation, Gabrielle Cavallaro for assistance with the PARA restraints module of XPLOR-NIH, and Robert London for making the coordinates of θ and HOT available prior to publication. We benefited from the use of NMR facilities at the Australian National University NMR Center.
This research was supported by grants from the Australian Research Council, including a Federation Fellowship to G.O.
REFERENCES
- 1.Bartels, C., T.-H. Xia, M. Billeter, P. Güntert, and K. Wüthrich. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6:1-10. [DOI] [PubMed] [Google Scholar]
- 2.Bertini, L., C. Luchinat, and G. Parigi. 2002. Magnetic susceptibility in paramagnetic NMR. Prog. NMR Spectrosc. 40:249-273. [Google Scholar]
- 3.Chikova, A. K., and R. M. Schaaper. 2005. Bacteriophage P1 hot gene product can substitute for the E. coli DNA polymerase III θ subunit. J. Bacteriol. 187:5528-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornilescu, G., F. Delaglio, and A. Bax. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289-302. [DOI] [PubMed] [Google Scholar]
- 5.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 6.DeRose, E. F., T. Darden, S. Harvey, S., Gabel, F. W. Perrino, R. M. Schaaper, and R. E. London. 2003. Elucidation of the ɛ-θ interface of Escherichia coli DNA polymerase III by NMR spectroscopy. Biochemistry 42:3635-3644. [DOI] [PubMed] [Google Scholar]
- 7.DeRose, E. F., T. W. Kirby, G. A. Mueller, A. K. Chikova, R. M. Schaaper, and R. E. London. 2004. Phage like it HOT: solution structure of the bacteriophage P1-encoded HOT protein, a homolog of the θ subunit of E. coli DNA polymerase III. Structure 12:2221-2231. [DOI] [PubMed] [Google Scholar]
- 8.DeRose, E. F., D. W. Li, T. Darden, S. Harvey, F. W. Perrino, R. M. Schaaper, and R. E. London. 2002. Model for the catalytic domain of the proofreading ɛ subunit of Escherichia coli DNA polymerase III based on NMR structural data. Biochemistry 41:94-110. [DOI] [PubMed] [Google Scholar]
- 9.Doherty, A. J., L. C. Serpell, and C. P. Ponting. 1996. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 24:2488-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichmüller, C., M. Tollinger, B. Kräutler, and R. Konrat. 2001. Mapping the ligand binding site at protein side-chains in protein-ligand complexes through NOE difference spectroscopy. J. Biomol. NMR 20:195-202. [DOI] [PubMed] [Google Scholar]
- 11.Engelke, J., and H. Rüterjans. 1995. Sequential protein backbone resonance assignments using an improved 3D-HN(CA)CO pulse scheme. J. Magn. Reson. B 109:318-322. [Google Scholar]
- 12.Goddard, T. D., and D. G. Kneller. 2003. Sparky, University of California, San Francisco.
- 13.Grzesiek, S., J. Anglister, and A. Bax. 1993. Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. B 101:114-119. [Google Scholar]
- 14.Grzesiek, S., and A. Bax. 1992. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114:6291-6293. [Google Scholar]
- 15.Grzesiek, S., and A. Bax. 1993. The origin and removal of artifacts in 3D HCACO spectra of proteins uniformly enriched with 13C. J. Magn. Reson. B 102:103-106. [Google Scholar]
- 16.Güntert, P., C. Mumenthaler, and K. Wüthrich. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273:283-298. [DOI] [PubMed] [Google Scholar]
- 17.Gupta, R., S. M. Hamdan, N. E. Dixon, M. M. Sheil, and J. L. Beck. 2004. Application of electrospray ionization mass spectrometry to study the hydrophobic interaction between then ɛ and θ subunits of DNA polymerase III. Protein Sci. 13:2878-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamdan, S., S. E. Brown, P. R. Thompson, J. Y. Yang, P. D. Carr, D. L. Ollis, G. Otting, and N. E. Dixon. 2000. Preliminary X-ray crystallographic and NMR studies on the exonuclease domain of the ɛ subunit of Escherichia coli DNA polymerase III. J. Struct. Biol. 131:164-169. [DOI] [PubMed] [Google Scholar]
- 19.Hamdan, S., E. M. Bulloch, P. R. Thompson, J. L. Beck, J.-Y. Yang, J. A. Crowther, P. E. Lilley, P. D. Carr, D. L. Ollis, S. E. Brown, and N. E. Dixon. 2002. Hydrolysis of the 5′-p-nitrophenyl ester of TMP by the proofreading exonuclease (ɛ) subunit of Escherichia coli DNA polymerase III. Biochemistry 41:5266-5275. [DOI] [PubMed] [Google Scholar]
- 20.Hamdan, S., P. D. Carr, S. E. Brown, D. L. Ollis, and N. E. Dixon. 2002. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure 10:535-546. [DOI] [PubMed] [Google Scholar]
- 21.Harrison, S. C. 1991. A structure taxonomy of DNA-binding domains. Nature 353:715-719. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann, T., P. Güntert, and K. Wüthrich. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319:209-227. [DOI] [PubMed] [Google Scholar]
- 23.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure—pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577-2637. [DOI] [PubMed] [Google Scholar]
- 25.Kay, L. E., G. Y. Xu, A. U. Singer, D. R. Muhandiram, and J. D. Forman-Kay. 1993. A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J. Magn. Reson. B 101:333-337. [Google Scholar]
- 26.Keniry, M. A., H. A. Berthon, J. Y. Yang, C. S. Miles, and N. E. Dixon. 2000. NMR solution structure of the θ subunit of DNA polymerase III from Escherichia coli. Protein Sci. 9:721-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koradi, R., M. Billeter, and K. Wüthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 14:51-55. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski, R. A., J. A. C. Rullmann, M. W. MacArthur, R. Kaptein, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 29.Lobocka, M. B., D. Rose, M. Rusin, A. Samojedny, H. Lehnherr, M. B. Yarmolinsky, and F. R. Blattner. 2004. The genome of bacteriophage P1. J. Bacteriol. 186:7032-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo Conte, L., C. Chothia, and J. Janin. 1999. The atomic structures of protein-protein recognition sites. J. Mol. Biol. 285:2177-2198. [DOI] [PubMed] [Google Scholar]
- 31.Luscombe, N. M., S. E. Austin, H. M. Berman, and J. M. Thornton. 2000. An overview of the structures of protein-DNA complexes. Genome Biol. 1:1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maciejewski, M. W., D. Liu, R. Prasad, S. H. Wilson, and G. P. Mullen. 2000. Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase. Correlation with DNA binding and dRP lyase activity. J. Mol. Biol. 296:229-253. [DOI] [PubMed] [Google Scholar]
- 33.Maki, H., T. Horiuchi, and A. Kornberg. 1985. The polymerase subunit of DNA polymerase III of Escherichia coli. I. Amplification of the dnaE gene product and polymerase activity of the α subunit. J. Biol. Chem. 260:12982-12986. [PubMed] [Google Scholar]
- 34.Maki, H., and A. Kornberg. 1985. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the α subunit, devoid of nuclease activities. J. Biol. Chem. 260:12987-12992. [PubMed] [Google Scholar]
- 35.Maki, H., and A. Kornberg. 1987. Proofreading by DNA polymerase III of Escherichia coli depends on cooperative interaction of the polymerase and exonuclease subunits. Proc. Natl. Acad. Sci. USA 84:4389-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuo, H., E. Kupce, H. J. Li, and G. Wagner. 1996. Increased sensitivity in HNCA and HN(CO)CA experiments by selective Cβ decoupling. J. Magn. Reson. B 113:91-96. [DOI] [PubMed] [Google Scholar]
- 37.McHenry, C. S. 2003. Chromosomal replicases as asymmetric dimers: studies of subunit arrangement and functional consequences. Mol. Microbiol. 49:1157-1165. [DOI] [PubMed] [Google Scholar]
- 38.McHenry, C. S., and W. Crow. 1979. DNA polymerase III of Escherichia coli. Purification and identification of subunits. J. Biol. Chem. 254:1748-1753. [PubMed] [Google Scholar]
- 39.Mueller, G. A., T. W. Kirby, E. F. DeRose, D. Li, R. M. Schaaper, and R. E. London. 2005. Nuclear magnetic resonance solution structure of the Escherichia coli DNA polymerase III θ subunit. J. Bacteriol. 187:7081-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhandiram, D. R., N. A. Farrow, G.-Y. Xu, S. H. Smallcombe, and L. E. Kay. 1993. A gradient 13C NOESY-HSQC experiment for recording NOESY spectra of 13C-labeled proteins dissolved in H2O. J. Magn. Reson. B 102:317-321. [Google Scholar]
- 41.Muhandiram, D. R., and L. E. Kay. 1994. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. B 103:203-216. [Google Scholar]
- 42.Neal, S., A. M. Nip, H. Zhang, and D. S. Wishart. 2003. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J. Biomol. NMR 26:215-240. [DOI] [PubMed] [Google Scholar]
- 43.Otting, G., and K. Wüthrich. 1990. Heteronuclear filters in two-dimensional [1H, 1H]-NMR spectroscopy: combined use with isotope labeling for studies of macromolecular conformation and intermolecular interactions. Q. Rev. Biophys. 23:39-96. [DOI] [PubMed] [Google Scholar]
- 44.Perrino, F. W., S. Harvey, and S. M. McNeill. 1999. Two functional domains of the ɛ subunit of DNA polymerase III. Biochemistry 38:16001-16009. [DOI] [PubMed] [Google Scholar]
- 45.Pintacuda, G., M. A. Keniry, T. Huber, A. Y. Park, N. E. Dixon, and G. Otting. 2004. Fast structure-based assignment of 15N HSQC spectra of selectively 15N-labeled paramagnetic proteins. J. Am. Chem. Soc. 126:2963-2970. [DOI] [PubMed] [Google Scholar]
- 46.Pintacuda, G., A. Y. Park, M. A. Keniry, N. E. Dixon, and G. Otting. 2006. Lanthanide labeling offers fast NMR approach to 3D structure determinations of protein-protein complexes. J. Am. Chem. Soc. 128:3696-3702. [DOI] [PubMed] [Google Scholar]
- 47.Richardson, J. S., and D. C. Richardson. 1988. Amino acid preferences for specific locations at the ends of α helices. Science 240:1648-1652. [DOI] [PubMed] [Google Scholar]
- 48.Scheuermann, R. H., S. Tam, P. M. J. Burgers, and H. Echols. 1983. Identification of the ɛ-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc. Natl. Acad. Sci. USA 80:7085-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwieters, C. D., N. Tjandra, J. Kuszewski, and G. M. Clore. 2003. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160:65-73. [DOI] [PubMed] [Google Scholar]
- 50.Studwell, P. S., and M. O'Donnell. 1990. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J. Biol. Chem. 265:1171-1178. [PubMed] [Google Scholar]
- 51.Studwell-Vaughan, P. S., and M. O'Donnell. 1993. DNA polymerase III accessory proteins. V. θ encoded by holE J. Biol. Chem. 268:11785-11791. [PubMed] [Google Scholar]
- 52.Taft-Benz, S. A., and R. M. Schaaper. 2004. The θ subunit of Escherichia coli DNA polymerase III: a role in stabilizing the ɛ proofreading subunit. J. Bacteriol. 186:2774-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wishart, D. S., C. G. Bigam, A. Holm, R. S. Hodges, and B. D. Sykes. 1995. 1H, 13C and 15N random coil chemical shifts of common amino acids. I. Investigation of nearest-neighbor effects. J. Biomol. NMR 5:67-81. [DOI] [PubMed] [Google Scholar]
- 54.Wishart, D. S., and B. D. Sykes. 1994. Chemical shifts as a tool for structure determination. Methods Enzymol. 239:363-392. [DOI] [PubMed] [Google Scholar]
- 55.Yamazaki, T., W. Lee, C. H. Arrowsmith, D. R. Muhandiram, and L. E. Kay. 1994. A suite of triple resonance NMR experiments for the backbone assignment of 15N, 13C, 2H labeled proteins with high sensitivity. J. Am. Chem. Soc. 116:11655-11666. [Google Scholar]
- 56.Zhang, O., L. E. Kay, J. P. Olivier, and J. D. Forman-Kay. 1994. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR 4:845-858. [DOI] [PubMed] [Google Scholar]