Abstract
GlnD of Escherichia coli is a bifunctional signal-transducing enzyme (102.4 kDa) which uridylylates the allosteric regulatory protein PII and deuridylylates PII-UMP in response to growth with nitrogen excess or limitation, respectively. GlnD catalyzes these reactions in response to high or low levels of cytoplasmic glutamine, respectively, and indirectly directs the expression of nitrogen-regulated genes, e.g., the glnK-amtB operon. We report that chromosomal mini-Tn10 insertions situated after nucleotide number 997 or 1075 of glnD partially suppressed the osmosensitive phenotype of ΔotsBA or otsA::Tn10 mutations (defective osmoregulatory trehalose synthesis). Strains carrying these glnD::mini-Tn10 mutations either completely repressed the expression of trp::(glnKp-lacZ) or induced this reporter system to nearly 60% of the wild-type glnD level in response to nitrogen availability, an essentially normal response. This was in contrast to the much-studied glnD99::Tn10 mutation, which carries its insertion in the 3′ end of the gene, causes a complete repression of glnKp-lacZ expression under all growth conditions, and also confers leaky glutamine auxotrophy. When expressed from the Pm promoter in plasmid constructs, the present glnD mutations produced proteins with an apparent mass of 39 or 42 kDa. These proteins were deduced to comprise 344 or 370 N-terminal residues, respectively, harboring the known nucleotidyltransferase domain of GlnD, plus a common C-terminal addition of 12 residues encoded by IS10. They lacked three other domains of GlnD. Apparently, the transferase domain by itself enabled the cells to catalyze the uridylylation reaction and direct nitrogen-regulated gene expression. Our data indicate that there exists a link between osmotic stress and the nitrogen response.
Escherichia coli cells grown in minimal medium with elevated osmotic strength generated with NaCl accumulate potassium glutamate and trehalose as compatible solutes to prevent osmotic dehydration of the cytoplasm (20, 38). E. coli has an osmotically inducible pathway for synthesis of trehalose consisting of trehalose-6-P synthase (OtsA), which utilizes UDP-glucose and glucose-6-P as substrates, and trehalose-6-P phosphatase (OtsB), which catalyzes the final step. The otsBA genes constitute an operon (33) with RpoS-dependent transcription (23, 32). Mutants of E. coli that are defective in trehalose synthesis tolerate less osmotic stress than isogenic otsBA+ cells, but their osmotic tolerance can be restored to wild-type levels by an external supply of glycine betaine, which is the preferred compatible solute in E. coli (21). Besides serving as a compatible solute, endogenously synthesized trehalose protects E. coli cells against environmental stress caused by heat and chill (23, 35). Trehalose can also serve as the sole energy and carbon source both in the absence and presence of osmotic stress (14, 22).
Glutamate has many functions in the cell. Together with glutamine, it is a central metabolite in ammonia assimilation of E. coli and most other bacteria. Assimilation of ammonia can be catalyzed by glutamate dehydrogenase (GDH) and glutamine synthetase (GS). Alternatively, glutamine produced by GS can be converted to glutamate by glutamate synthase (GOGAT). The GS/GOGAT cycle is best suited for assimilation of ammonia into cells growing under nitrogen limitation, because GS has a higher affinity for ammonia than GDH (Km, 1 to 5 mM) (49). GDH comes into play when the external NH4+ concentration is high. Accumulation of potassium glutamate is an early response in osmoregulation of E. coli (20). At high levels of external ammonium, both pathways can participate in this synthesis of glutamate, since a mutation in either gltB (GOGAT) or gdhA (GDH) does not influence the initial rate of K+ and glutamate accumulation in cells subjected to osmotic upshock in the presence of 16 mM NH4+ (40). However, although osmotically stressed gltB mutants of Salmonella enterica serovar Typhimurium display a high initial accumulation of glutamate and K+ (63), the lack of a functional GS/GOGAT cycle causes reduced steady-state glutamate and K+ pools in the presence of both low and high concentrations of ammonium compared with the wild type (18, 63).
To our knowledge, regulation of the nitrogen response of E. coli has been studied only with cells that are grown in the absence of osmotic stress. In such cells, the synthesis and activity of GS are regulated by the availability of nitrogen through a cascade of three cyclic reactions. GlnD is the primary sensor of the nitrogen status of the cell by sensing the glutamine concentration of the cytoplasm (26, 30, 31). In nitrogen-limited cells (e.g., cells grown with glutamine as the only source of nitrogen), GlnD uridylylates two signal transduction proteins, PII and GlnK (uridylyltransferase [UTase] activity). In cells grown with nitrogen excess, GlnD removes the UMP groups of PII-UMP and GlnK-UMP by hydrolysis (uridylyl-removing [UR] activity). PII and GlnK display 67% sequence identity (57). They form homo- and heterotrimers and have overlapping, but not identical, functions in the cell (59, 60, 61). The allosteric regulatory protein PII is expressed constitutively at a low level, and its main function is to regulate the expression of nitrogen-regulated genes and the activity of GS (42, 56). In contrast, GlnK is strongly induced by nitrogen limitation (7, 58). The glnK gene forms an operon with amtB (58), which encodes a gas channel for ammonia (64). The unuridylylated form of GlnK binds to the AmtB transporter creating a membrane-bound complex (29), but the physiological role of the complex formation remains uncertain for E. coli.
The glnALG operon encodes GS, NRII, and NRI, respectively (10, 48). NRII is a soluble histidine protein kinase which controls phosphorylation and dephosphorylation of the response regulator NRI. There is a basal σ70-dependent transcription of the glnALG operon from the glnAp1 and glnLp promoters, but NRI-P is required together with σ54 for high level of transcription from the glnAp2 promoter (44, 48). NRI-P is absolutely required for transcription of the glnK-amtB operon (8). Under growth conditions of excess nitrogen, PII stimulates the phosphatase activity of NRII, which results in a low NRI-P level and low or no transcription of nitrogen-regulated operons (34, 36). On the other hand, PII-UMP does not interact with NRII (6, 47). Therefore, under nitrogen limitation, NRII does not display its phosphatase activity and most NRI remains phosphorylated. In turn, this results in higher expression of NRI itself and other nitrogen-regulated genes and operons, e.g., glnK-amtB. Additionally, the activity of GS is regulated at the protein level by adenylylation and deadenylylation catalyzed by ATase; i.e., unadenylylated GS is active and adenylylated GS is partially inactivated and feedback inhibited (53). Under nitrogen limitation, PII-UMP stimulates ATase to deadenylylate GS, and under nitrogen excess, PII stimulates the opposite reaction (53).
Compared with the other proteins of the nitrogen regulation cascade of E. coli, relatively little is known about GlnD. In the present study, we have generated glnD transposon mutations, which partially suppress the osmosensitive phenotype of otsBA. The present mutant glnD alleles expressed N-terminal polypeptide fragments comprising only 37 or 40% of the GlnD protein. These fragments contained a nucleotidyltransferase (NT) domain but lacked the three other domains of GlnD. We used a glnKp-lacZ fusion, which has been made previously by Atkinson and Ninfa (7), to compare the nitrogen response in cells producing wild-type or mutant GlnD proteins. We concluded that the truncated GlnD proteins had unregulated UTase activity and lacked UR activity. Our data showed that the nitrogen response system is very robust, since cells which carried the glnD transposon mutations on the chromosome displayed an essentially normal regulation of the glnKp-lacZ fusion. Studies with the reporter system indicated that there exists a link between osmotic stress and nitrogen response.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phages.
The bacterial strains, plasmids, and phages used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and bacteriophages
| Strain, plasmid, or bacteriophage | Genotype or description | Reference, source, or construction |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac) U169 flbB5301 ptsF25 relA1 rpsL150 deoC1 rbsR fruA25 | CGSC6152 |
| DH5α | F− φ80dlacZΔ M15 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 λ−relA1 Δ(argF-lac)U169 | Bethesda Research Laboratories |
| Pφ | F− Δpta-ackA.zej::Tn10 trpDC700::putPA1303 [Φ(glnKp-lacZ) Knr] | 7 |
| FF4031 | MC4100 ΔotsBA | 21 |
| FF4169 | MC4100 otsA1::Tn10 (Tcr) | 21 |
| FF1005 | MC4100 recA56 srl-300::Tn10 (Tcr) | 32 |
| RB9040 | glnD99::Tn10 (Tcr) | 15 |
| SR62 | FF4031 glnD39::mini-Tn10 cam (Cmr) | This study |
| SR110 | FF4031 glnD42::mini-Tn10 kan (Knr) | This study |
| SR942 | FF4031 glnD42::mini-Tn10 cam (Cmr) | This study |
| AT1 | MC4100 trpDC700::putPA1303 [Φ(glnKp-lacZ) Knr] | P1 (Pφ) × MC4100 |
| AT2 | AT1 glnD39::mini-Tn10 cam (Knr Cmr) | P1 (SR62) × AT1 |
| AT3 | AT1 glnD99::Tn10 (Knr Tcr) | P1 (RB9040) × AT1 |
| AT4 | AT3 ΔglnD99::Tn10 (Knr Tcs) | This study |
| AT5 | AT4 recA56 srl-300::Tn10 (Knr Tcr) | P1 (FF1005) × AT4 |
| AT7 | AT2 otsA1::Tn10 (Knr Cmr Tcr) | P1 (FF4169) × AT2 |
| AT9 | AT10 glnD42::mini-Tn10 cam (Knr Tcr Cmr) | P1 (SR942) × AT10 |
| AT10 | AT1 otsA1::Tn10 (Knr Tcr) | P1 (FF4169) × AT1 |
| Plasmids | ||
| pGEM7-Zf(+) | Cloning vector; Apr | Promega |
| pJB655 | Expression vector; Apr | 12 |
| pTG7420 | Vector carrying wild-type glnD; Apr | 19 |
| pUC-4K | Vector carrying a BamHI Knr fragment from Tn903 | Pharmacia |
| pHT3 | pGEM7-Zf(+) carrying the EcoRI-SacI chromosomal fragment with mini-Tn10 kan of SR110; Apr Knr | This study |
| pHT4 | pGEM7-Zf(+) carrying the BamHI-SacI fragment from pHT3; Apr | This study |
| pHT5 | pGEM7-Zf(+) carrying the EcoRI-BamHI fragment from pHT3; Apr | This study |
| pHT6 | pHT5 was digested with EcoRI and BsgI, made blunt, and religated; Apr | This study |
| pHK1 | pGEM7-Zf(+) carrying the ClaI-SacI chromosomal fragment with mini-Tn10 cam of SR942; Apr Cmr | This study |
| pHK2 | pGEM7-Zf(+) carrying the ClaI-BamHI fragment from pHK1; Apr | This study |
| pHK3 | pGEM7-Zf(+) carrying the BamHI-SacI fragment from pHK1; Apr | This study |
| pAT7 | pGEM7-Zf(+) carrying the ClaI-SacI chromosomal fragment with mini-Tn10 cam of SR62; Apr Cmr | This study |
| pAT8 | pGEM7-Zf(+) carrying the BamHI-SacI fragment from pAT7; Apr | This study |
| pAT9 | pGEM7-Zf(+) carrying the ClaI-BamHI fragment from pAT7; Apr | This study |
| pAT14 | The ClaI-BamHI fragment of pHT6 was replaced with the corresponding fragment from pAT7; Apr | This study |
| pAT24 | glnD′ from pAT14 was cloned as an AflIII-BamHI PCR fragment into pJB655; Apr | This study |
| pAT25 | glnD′ from pHT6 was cloned as an AflIII-BamHI PCR fragment into pJB655; Apr | This study |
| pAT33 | glnD from pTG7420 was cloned as an AflIII-BamHI PCR fragment into pJB655; Apr | This study |
| Phages | ||
| P1 | vir | Laboratory collection |
| λNK1316 | Mini-Tn10 kan/Ptac-ATS transposase | 37 |
| λNK1324 | Mini-Tn10 cam/Ptac-ATS transposase | 37 |
Growth media and growth conditions.
The standard minimal growth medium was M63 with 22 mM glucose (41). The minimal medium used for measurements of nitrogen response contained the following: 100 mM KH2PO4, 75 mM KOH, 2.9 mM K2SO4, 1 mM MgCl2, 3.9 μM FeSO4, 22 mM glucose, and 0.2% (wt/vol) (NH4)2SO4 (nitrogen excess) or glutamine (nitrogen limitation). This medium is referred to as nitrogen response medium in Tables 2 and 3. When glutamine was used as the nitrogen source, the cells were grown until the optical density at 600 nm (OD600) had reached at least 1.0 (see Results). The osmotic strength of the minimal medium was increased by addition of 0.3 M, 0.4 M, or 0.55 M NaCl as stated in the text. For the nitrogen excess medium, this corresponded to a final osmolality of 730, 890, and 1,130 mosmol kg−1, respectively. The osmolality was measured by freezing point depression. pH was adjusted to 7.2 with KOH when necessary. The rich medium routinely used was LB. Triple-strength LB was used for cultivation of cells overproducing recombinant GlnD proteins. In these cells, the Pm promoter was induced by addition of 0.5 mM m-toluic acid when the OD600 of the cultures was 0.2, after which the cultivation was continued for 4 h. For selection of resistance markers, antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg ml−1; chloramphenicol (Cm), 30 μg ml−1; kanamycin (Kn), 40 μg ml−1; and tetracycline (Tc), 15 μg ml−1. Tryptophan (2 mM) was added to the minimal medium and glutamine (1 mM) was added to the LB medium when required by the cells. Nitrogen response was measured in cells grown at 30°C; otherwise, the cells were grown at 37°C (see Results).
TABLE 2.
Effect of osmotic stress, nitrogen availability, and the glnD allele on expression of glnKp-lacZ
| Strainc | glnD allele | β-Galactosidase activitya (Miller units) when grown in nitrogen response mediumb with:
|
||||
|---|---|---|---|---|---|---|
| No NaCl
|
0.3 M NaCl
|
0.55 M NaCl
|
||||
| NH4+ | Gln | NH4+ | Gln | NH4+ | ||
| AT10 | Wild type | <1 | 2,800 ± 110 | 8.3 ± 1.8 | 610 ± 170 | 6.9 ± 5.4 |
| AT9 | glnD42 | <1 | 1,700 ± 150 | 4.1 ± 1.2 | 150 ± 28 | 36 ± 11 |
| AT7 | glnD39 | 2.0 ± 0.2 | 1,800 ± 190 | 5.4 ± 1.6 | 270 ± 93 | 95 ± 14 |
Values are means of duplicate determinations from two to four separate cultures ± standard deviations.
Medium contained 0.2% (wt/vol) (NH4)2SO4 or glutamine.
All strains carried otsA1::Tn10.
TABLE 3.
Effect of overexpression of full-length and truncated GlnD proteins on expression of glnKp-lacZ in strain AT5 (ΔglnD99::Tn10 recA56)
| Plasmid | Description of GlnD | β-Galactosidase activitya (Miller units) when grown in nitrogen response mediumb with:
|
|
|---|---|---|---|
| NH4+ | Gln | ||
| None | None | 4 | |
| pAT33 | Full length | 460 ± 120 | 2,600 ± 170 |
| pAT25 | Truncated, GlnD42 | 1,700 ± 230 | 2,700 ± 220 |
| pAT24 | Truncated, GlnD39 | 1,600 ± 230 | 2,700 ± 150 |
Values are means of triplicate determinations from two to five separate cultures ± standard deviations.
Medium contained 0.2% (wt/vol) (NH4)2SO4 or glutamine.
Standard recombinant DNA techniques.
Restriction endonuclease digestions, ligations, and agarose gel electrophoresis were performed in accordance with standard protocols (50). Transformations were performed by the method of Chung et al. (16). Genomic DNA was prepared as described by Ausubel et al. (9). Plasmid DNA was prepared by a plasmid midi kit from QIAGEN or a Wizard miniprep kit from Promega. DNA was extracted from agarose gels using the Qiaex or the Qiaquick kit from QIAGEN. DNA sequencing was performed using cycle sequencing with an AmpliTaq kit from PE Applied Biosystems. PCR was performed by utilizing the Expand high-fidelity PCR system from Roche or the PfuTurbo DNA polymerase from Stratagene.
Preparation of pools of transposon mutants of FF4031.
Strain FF4031 was infected with phage λNK1316 (mini-Tn10 kan) or phage λNK1324 (mini-Tn10 cam) as described previously (37). After the transposition, the cells were spread on 50 LB agar plates containing Kn or Cm, and about 50,000 Knr or Cmr colonies representing individual transposition events were then washed off the plates, pooled, and stored at −80°C in LB containing 15% glycerol until used.
Genetic characterization of transposon-containing chromosomal fragments.
Genomic DNA from strain SR110 (Knr) was digested with selected restriction endonucleases, and Southern blot analysis was performed by probing with a 1.7-kb BamHI fragment from pUC-4K that carries the Knr marker from Tn903. The DIG system from Boehringer Mannheim was used to label the probe, and the Southern blot analysis was performed as specified by the manufacturer. The mini-Tn10 kan insertion in SR110 was found to reside on a 5.5-kb EcoRI-SacI fragment. Chromosomal DNA was cut with EcoRI and SacI, and fragments of the appropriate size were isolated from agarose gel, ligated into the vector pGEM7-Zf(+), and transformed into DH5α. After several attempts, we obtained a Knr transformant carrying pHT3 with the correct 5.5-kb insert. This insert contained only two BamHI sites that were located in each of the IS10 sequences of the mini-transposon (37). The 0.8-kb BamHI-SacI and 3.1-kb EcoRI-BamHI fragments of pHT3 were cloned into the corresponding sites of pGEM7-Zf(+), yielding plasmids pHT4 and pHT5, respectively. pHT5 was digested with EcoRI and BsgI, made blunt by treatment with Klenow polymerase, and religated to yield pHT6, which carried a remaining insert of 1.3 kb. The junctions between IS10 and chromosomal DNA in pHT4 and pHT6 were sequenced by using forward and reverse sequencing primers as specified for pGEM7-Zf(+) by Promega.
In order to clone the mini-Tn10 cam insertions of strains SR62 and SR942, genomic DNA was digested with ClaI and SacI (see Results), and DNA fragments of about 3.1 kb were isolated from agarose gels, ligated into the same sites of pGEM7-Zf(+), and transformed into DH5α. From the resulting transformants, we obtained plasmids pHK1 and pAT7, which carried the Cmr determinant of SR942 and SR62, respectively. By using the subcloning strategy which was outlined above for pHT3, the ClaI-BamHI and BamHI-SacI fragments of pHK1 and pAT7 were subcloned into pGEM7-Zf(+). The resulting plasmids, which were called pHK2 and -3 and pAT9 and -8, respectively, were used for sequencing of the junctions between chromosomal DNA and IS10. Two additional Cmr mutants were subjected to the same cloning procedures (see Results). Plasmid pAT14 was obtained by replacing the ClaI-BamHI fragment of pHT6 with the corresponding fragment of pAT7.
Construction of glnD expression vectors.
IS10-truncated glnD fragments of pAT14 and pHT6 were PCR amplified by using the following primers: GTGGCGCAACATGTATACCCTTCCAGA (forward) and AAAAAGGATCCGGGATCATATGACAAG (reverse). A full-length glnD gene derivative was amplified from pTG7420 by using the same forward primer in combination with AAAAAAGGATCCACATCACCCTTTATCG (restriction sites are underlined). This procedure introduced an AflIII site at the start codon, changed the second codon of glnD from AAT (Asn) to TAT (Tyr), and introduced a BamHI site downstream of the IS10 generated or the natural stop codons. The amplified AflIII-BamHI fragments were cloned into the same sites of expression vector pJB655 (12), leading to plasmids pAT24, pAT25, and pAT33, respectively.
Strain constructions and other methods.
P1 phage transductions were performed as described by Miller (41). The recA56 mutation was cotransduced with srl-300::Tn10, and increased sensitivity to UV light was used to screen Tcr colonies for the recA56 mutation (39). Strain AT4 (ΔglnD99::Tn10) was generated by selecting for a Tcs mutant of AT3 by using the method described by Bochner et al. (13). For analysis of GlnD proteins, overproducing strains were disrupted by sonication in 40 mM imidazol (pH 7.4), and the sonicate was centrifuged for 30 min at 6,000 × g (4°C). The soluble fractions were separated by sodium dodecyl sulfate (SDS)-8% polyacrylamide gel electrophoresis (PAGE) as described by Sambrook et al. (50). Fifteen micrograms of protein was added in each lane. The protein content of these samples was determined by the microassay procedure provided by Bio-Rad. The β-galactosidase assay was performed using the method described by Miller (41), and the cells were permeabilized with chloroform and SDS.
Domain search.
Searches for domains in GlnD were done on 30 March 2006 at the following URLs: http://pfam.wustl.edu/hmmsearch.shtml and http://myhits.isb-sib.ch/cgi-bin/motif_scan.
RESULTS
Transposon mutations which suppress the osmosensitive phenotype of ΔotsBA.
The otsBA genes for trehalose synthesis were originally identified by selecting mutants of E. coli which do not grow in M63 with 0.45 M NaCl added (21). In order to identify transposon mutations which suppress this ots phenotype, we generated two pools of FF4031 (ΔotsBA) cells which were mutagenized with mini-Tn10 kan or mini-Tn10 cam. Each pool comprised cells from about 50,000 Knr or Cmr colonies which had been formed by individual transposition events. Cells from these pools were grown to mid-exponential phase in liquid M63, and then about 107 cells were spread on agar plates with M63-0.4 M NaCl. To prevent drying, the plates were wrapped in plastic bags, incubated at 37°C, and inspected regularly during the next 4 days. The first colonies that appeared on the plates were purified, and their antibiotic resistance marker was transduced with P1 into a new FF4031 background. From the pool of cells carrying mini-Tn10 kan, we obtained the transductant SR110 for which the osmotolerant phenotype was 100% cotransducible with the Knr marker. From the other mutant pool, we obtained four Cmr transductants with similar properties. When the transposons of these cells were transduced into SR110, all Cmr transductants obtained were Kns. Since three of the Cmr mutants carried identical insertions (see below), only strains SR62 and SR942 are included in Table 1.
The mutants selected were among those that grew best on the salt agar plates, but they did not grow as well as MC4100 (otsBA+), which is the parental strain of FF4031. It should be noted that for the majority of the mutants tested, the antibiotic resistance marker was not cotransducible with their osmotolerant phenotype. Their osmotolerance was apparently caused by spontaneous mutations.
Cloning and physical mapping of Tn10 insertions.
Based on Southern blot analysis, the mini-Tn10 kan insertion of SR110 was cloned as a 5.5-kb EcoRI-SacI fragment into pGEM7-Zf(+), thereby creating plasmid pHT3 (Table 1). By subcloning of chromosomal DNA fragments with the flanking IS10 sequences and sequencing relevant parts of these subclones, we found that the transposon was inserted after nucleotide (nt) 1075 in the coding sequence of the glnD gene. From the known chromosomal DNA sequence of E. coli, we deduced that the EcoRI site was located upstream of the rpsB gene and that the SacI site was at nt 1846 of glnD. The map gene, which is transcribed in the same direction as glnD, is located between rpsB and glnD on the E. coli chromosome.
From the initial transductional mapping, we assumed that the mini-Tn10 cam insertions of the four Cmr mutants were situated in glnD. Since we had experienced that the EcoRI-SacI fragment of strain SR110 was difficult to clone, the mini-Tn10 cam insertions of the Cmr mutants were cloned into pGEM7-Zf(+) as 3.1-kb chromosomal fragments extending from a ClaI site at nt 118 in the coding sequence of glnD to the identified SacI site. These clones were easily obtained by selecting for Cmr transformants of DH5α.
Characterization of plasmid pAT7 and its subclones (Table 1) which carried chromosomal fragments of SR62 revealed that the transposon insertion was situated after nt 997 of glnD. The ClaI-SacI fragment of SR942, which was cloned into pHK1, carried the mini-Tn10 cam insertion in exactly the same location as the mini-Tn10 kan insertion of SR110. Analyses of clones obtained from the two remaining Cmr mutants revealed that they were identical with those obtained from SR942, and they are not included in the list of plasmids (Table 1).
Comparisons with the phenotype of the glnD99::Tn10(Tc) mutation.
Strain RB9040 carries the much-studied glnD99::Tn10 mutation, which confers a leaky Gln− phenotype (15). A convenient way to score the Gln− phenotype is to grow the cells on nutrient agar with or without glutamine added (11). In the presence of added glutamine (1 mM), RB9040 formed large colonies on the LB plates. Otherwise, RB9040 formed very small colonies which threw off larger colonies upon prolonged incubation of the plates, as noted previously for cells grown in minimal medium with arginine as the sole nitrogen source (5, 7). In contrast, all glnD transposon mutants selected by us displayed a Gln+ phenotype in the LB-plate assay, i.e., they formed large colonies of uniform size.
When the glnD99::Tn10 (Tcr) mutation of RB9040 was transduced into SR110, the majority of the selected Tcr transductants were Kns and displayed the same Gln− phenotype as RB9040. Selected Knr Tcr transductants displayed a Gln+ growth phenotype. Apparently, in these double mutants, the glnD::mini-Tn10 kan mutation of SR110 was phenotypically dominant over the glnD99::Tn10 mutation. The glnD99::Tn10 insertion has previously been physically mapped to a location which is downstream of the present mini-Tn10 insertions, i.e., 2,106 nt into the coding sequence (4).
Expression of truncated and full-length GlnD proteins.
The length and the mass of the wild-type GlnD protein are 890 amino acid residues and 102.4 kDa, respectively (56). The results from the genetic and phenotypic characterizations of the present mutants indicated that they produced truncated GlnD proteins with biological activity. For SR62, this protein was deduced to comprise the 332 N-terminal residues plus a C-terminal tail of 12 residues (TDESPNDFGKNH) encoded by the IS10 sequence of the transposon. Its deduced mass was 39.3 kDa. For convenience, this protein was named GlnD39 and the corresponding mutation of SR62 was named glnD39::mini-Tn10 cam. Both SR110 and SR942 were deduced to produce a corresponding GlnD derivative of 370 residues (42.2 kDa), including exactly the same 12-residue tail found in GlnD39. Correspondingly, this protein was named GlnD42, and their mutations were named glnD42::mini-Tn10 kan and glnD42::mini-Tn10 cam, respectively (Table 1).
From the chromosomal insert of pHT3, we constructed pHT6 (Table 1), which carried the coding sequence of GlnD42 with the native promoter, i.e., a 1.3-kb DNA fragment extending from a BsgI site located 98 bp upstream of the stop codon of map to a BamHI site located 34 bp downstream of the first in-frame stop codon in the IS10 sequence. Plasmid pAT14, carrying the coding sequence of GlnD39, was created by replacing the 1.0-kb ClaI-BamHI fragment of pHT6 with the corresponding fragment of pAT7. Both plasmids conferred a Gln+ phenotype to AT5 (ΔglnD99::Tn10 recA56) in the LB plate assay (see above). These data verified that biologically active GlnD39 and GlnD42 proteins were expressed from the native glnD promoter.
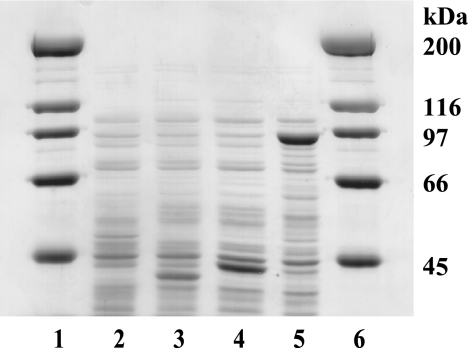
By a PCR-based cloning procedure, the coding sequences of GlnD39, GlnD42, and full-length GlnD were cloned into the expression vector pJB655 (12) under the control of the inducible Pm promoter and its XylS regulator, thereby generating plasmids pAT24, pAT25, and pAT33, respectively. When induced with m-toluic acid (0.5 mM), strain AT5 (ΔglnD99::Tn10 recA56) carrying pAT24 or pAT25 produced large amounts of soluble proteins with the expected apparent mass of 39 or 42 kDa, respectively, as judged by SDS-PAGE. AT5(pAT33) overproduced a protein with the apparent mass of 89 kDa (Fig. 1). The observed mobility of the latter protein was in accordance with the previously reported mobility of wild-type GlnD on SDS gels (56). The proteins were biologically active as judged from their ability to confer the Gln+ phenotype to the ΔglnD99::Tn10 host cells. For this complementation, it was unnecessary to induce the Pm promoter with m-toluic acid.
FIG. 1.
SDS-PAGE analysis of recombinant GlnD proteins. The production strains were AT5 (ΔglnD99::Tn10 recA56) carrying various plasmids. Lanes 1 and 6, molecular mass standards; lane 2, pJB655 (vector control); lane 3, pAT24 (GlnD39); lane 4, pAT25 (GlnD42); lane 5, pAT33 (full-length GlnD).
Effects of the glnD mutations and osmotic stress on the nitrogen response.
Atkinson and Ninfa (7) have previously constructed a glnKp-lacZ fusion which is integrated in the trp operon. This construct allows glnK expression to be tested in cells with a glnK+ background. They have shown that the β-galactosidase activity produced by the fusion responds to nitrogen limitation of E. coli cells in a GlnD-dependent manner. We constructed strains carrying trp::Φ(glnKp-lacZ) and otsA1::Tn10 together with either the wild-type or a mutant glnD allele. As observed for the ΔotsBA strains, the otsA1::Tn10 strains with the mutant glnD alleles displayed increased osmotolerance.
In the absence of osmotic stress, AT10 cells which carried the wild-type glnD gene displayed the same strong nitrogen response as previously reported for corresponding glnD+ otsA+ cells (7); i.e., the glnKp-lacZ fusion was nearly completely repressed (<1 U) in cells grown under excess nitrogen conditions [0.2% (wt/vol) (NH4)2SO4], and it produced 2,800 U of β-galactosidase in cells grown under nitrogen limitation conditions (0.2% [wt/vol] glutamine as the sole nitrogen source). Also, strains AT7 (glnD39) and AT9 (glnD42) displayed a strong nitrogen response, albeit the full production of β-galactosidase was reduced by 36 to 39% (Table 2). In comparison, nitrogen limitation-induced AT5 cells carrying ΔglnD99::Tn10 produced only 4 U of β-galactosidase (Table 3), which was similar to the value previously reported for E. coli strain DΦ carrying glnD99::Tn10 (7).
Under nitrogen starvation conditions, mild osmotic stress (0.3 M NaCl) reduced the expression of glnKp-lacZ by about 80% for AT10 cells producing wild-type GlnD and about 85 to 90% for AT7 and AT9 cells producing the truncated GlnD proteins (Table 2). However, under excess-nitrogen conditions, mild osmotic stress (0.3 M NaCl) increased the production of β-galactosidase from glnKp-lacZ slightly for cells producing either the wild-type (8 U) or the truncated versions of GlnD (4 to 5 U). Severe osmotic stress generated with 0.55 M NaCl did not elicit a further increase in β-galactosidase production of AT10 but increased the β-galactosidase production to 36 U in AT9 and to 95 U in AT7, which produced GlnD42 and GlnD39, respectively. It should be noted that cells carrying trp::Φ(glnKp-lacZ) required tryptophan (e.g., 2 mM), which stimulated slightly growth of osmotically stressed cells and reduced the growth differences between the strains. Thus, under nitrogen excess conditions, the doubling times of AT10 (wild-type GlnD), AT7 (GlnD39), and AT9 (GlnD42) were 2.3, 2.2, and 2.2 h, respectively, with 0.3 M NaCl and 5.3, 4.7, and 4.8 h, respectively, with 0.55 M NaCl.
For AT5 (ΔglnD99::Tn10 recA56) cells grown with excess nitrogen, we observed that overexpression of the truncated GlnD proteins from pAT24 and pAT25 provoked a higher level of expression of glnKp-lacZ than overexpression of full-length GlnD from pAT33. Under nitrogen starvation conditions, all three plasmids had the same strong effect (Table 3). A similar observation has been made previously for wild-type GlnD (43).
Apparent growth-phase dependent expression of glnKp-lacZ.
Atkinson et al. (8) have reported previously that glnKp-lacZ expression in nitrogen-limited E. coli (glnD+) cells (i.e., glutamine used as a nitrogen source) does not occur before the cultures reach a certain cell density. Therefore, they have warned against the practice of using mid-log cultures in investigations of the nitrogen response when glutamine is used as a nitrogen source. When grown with glutamine, we observed the same phenomenon for cells carrying glnKp-lacZ (data not shown). A likely explanation for this phenomenon is the fact that glutamine is spontaneously decomposed following first-order kinetics to pyrrolidone-carboxylic acid and ammonium. At 37°C, 10% of the glutamine present in common culture medium or phosphate buffer (pH 7.2) decomposes per day. The reaction rate can be reduced by using the common precaution of growing the cells at 30°C, but the reaction proceeds at a measurable speed even at 4°C. At this temperature, a 10% decomposition takes 9 days (55). Thus, in practice, the decomposition of glutamine in E. coli cultures is unavoidable. In our experiments, we did not record the β-galactosidase activity of nitrogen-limited cultures (30°C) before the OD600 of the cultures was at least 1.0 and the specific β-galactosidase activity of the cells was fully expressed. At this high cell density, the ammonium concentration of the medium was evidently kept below the critical value for induction of the nitrogen response. In other words, the preformed ammonium had been catabolized, and the rate of ammonium catabolism exceeded the rate of ammonium formation.
DISCUSSION
The aim of this investigation was to isolate and characterize transposon mutations that suppress the osmosensitive phenotype of an E. coli mutant with defective trehalose synthesis. This work led us to find a new class of glnD transposon mutants. Our positive selection procedure, which was colony formation of ΔotsBA cells with random mini-Tn10 insertions on solid NH4+-rich medium of elevated osmotic strength (0.4 M NaCl), was sufficiently clear to allow this type of glnD transposon mutants to be isolated repeatedly.
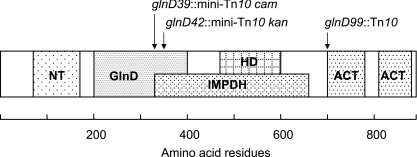
Searches in the data banks revealed that the large GlnD proteins of bacteria have a common domain structure. In GlnD of E. coli, which comprises 890 amino acid residues, residues 70 to 169 compose an NT domain, residues 468 to 602 compose a His-Asp (HD) (see below) domain, and residues 708 to 783 and 815 to 880 compose two aspartokinase-chorismate mutase-TyrA (ACT) (see below) domains (1-3) (Pfam, St. Louis database). Furthermore, the region between residues 191 and 400 contains a motif which is found in GlnD-like UR/UTases but not assigned a specific function (marked GlnD in Fig. 2). These motifs were found by searches in all databases tested. In addition, a search with the Pfam global program of the MyHits database revealed that the 332-686 region comprises an IMP dehydrogenase domain which is found in IMP dehydrogenase itself and GMP reductase. Consequently, some of the domains overlap (Fig. 2). Only the NT domain was retained in the present mutant proteins (Fig. 2), which comprised only 332 (GlnD39) or 358 (GlnD42) N-terminal residues plus a common C-terminal tail of 12 residues encoded by the fused IS10 sequence. The present GlnD′ proteins appeared to be stable in the cytoplasm in that they were biologically active and could be overexpressed in amounts similar to that of the full-length protein. The transposase of the λ phages which were used to generate the present mini-Tn10 insertions has been modified to decrease its DNA sequence specificity (37). Since two independently generated Cmr and Knr mutants carried the transposon in the same site (producing GlnD42) and a third mutant carried the transposon in a nearby location (producing GlnD39), we suspect that mutants with the present osmotolerant phenotype can be formed only by Tn10 insertions in a small region of glnD.
FIG. 2.
Schematic presentation of the domain structure of the 890-residue GlnD protein. The functions and exact locations of the domains are given in the text.
Unlike ATase (GlnE; see below), GlnD has only one identifiable NT domain (25). It has been reported that certain single-amino-acid substitutions within this sequence can cause loss or reduction of UTase activity (D107N), UR activity (G98A), or both activities (D105N), and it has therefore been proposed that the conserved transferase sequence constitutes the catalytic center of both activities (43). Kinetic data for E. coli GlnD are consistent with the existence of a single binding site for PII and PII-UMP, a single catalytic center for the UTase and UR activities, and a single binding site for glutamine from where glutamine inhibits the UTase activity and stimulates the UR activity (31). However, since the HD domain is found in many enzymes with known or deduced phosphohydrolase activity, it has been proposed previously that the UR activity of GlnD may reside in this domain (1). The ACT domain is a conserved regulatory ligand binding fold found in many enzymes that are involved in the control of amino acid metabolism, therefore pointing to a regulatory function of the two ACT domains of GlnD (2, 3).
It has been shown previously that PII is partially (8%) uridylylated in nitrogen-starved cells carrying the glnD99::Tn10 mutation (52) in which the transposon is inserted only 567 bp upstream of the stop codon (4), i.e., just upstream of the first ACT domain (Fig. 2). The leaky nature of the glnD99::Tn10 mutation allows slow growth with arginine as the only source of nitrogen (5), but the mutation represses the expression of glnKp-lacZ (7). In contrast, the present glnD transposon mutants caused a nitrogen-starvation-dependent expression of glnKp-lacZ which was more than 60% of the expression induced by wild-type GlnD. These data showed that the truncated GlnD39 and GlnD42 proteins are capable of regulating PII activity and that they enabled the cell to sense a nitrogen starvation signal. The most likely explanation is that the GlnD′ proteins had retained their UTase activity; alternatively, they may have the capacity to interact with and sequester unmodified PII (as discussed below). It is believed that glutamine is the main regulator of E. coli GlnD under physiological conditions (reviewed in reference 47). Since the truncated GlnD proteins lacked the ACT domains, it seems unlikely that they have retained their ability to sense glutamine. Thus, either the N-terminal fragment of GlnD can sense another metabolite or another regulatory mechanism came into play when the glutamine-sensing ability was defective. This mechanism could involve the interaction of PII with other metabolites, such as ATP or 2-oxoglutarate, which are required for both uridylylation of PII and deuridylylation of PII-UMP (31). Anyway, our data showed that the nitrogen response system is very robust in that the deletion of about 60% of the main sensor protein had small consequences for nitrogen regulation in cells grown at steady state under laboratory conditions.
The finding that osmotic stress increased the expression of glnKp-lacZ in cells which were grown at steady state with excess nitrogen indicated that osmotic stress elicited a nitrogen response in E. coli. The glnKp-lacZ expression was low in cells producing wild-type GlnD but rather pronounced in severely stressed (0.55 M NaCl) cells producing mutant GlnD′ proteins, correlating with their more osmotolerant phenotype. Under nitrogen starvation in E. coli, there is a sequential activation of nitrogen-regulated genes due to various affinities and arrangements of their binding sites for NRI-P, and it has been shown that glnK requires a high level of NRI-P for expression (8). Thus, the observed glnK expression may be a direct effect of increased NRI-P level or an indirect effect of increased accumulation of metabolites in stressed cells. Anyway, the apparent link between osmotic stress and nitrogen response correlates with the finding of Yan et al. (63) that the GS/GOGAT cycle is required for the maintenance of normal steady-state pools of glutamate and K+ in osmotically stressed cells of Salmonella enterica serovar Typhimurium.
Corynebacterium glutamicum produces a GlnD protein which comprises only 692 residues (28) and lacks ACT domains. This GlnD protein catalyzes adenylylation of GlnK (the only PII homolog of the organism) and deadenylylation of GlnK-AMP, but it is not the primary sensor of the nitrogen status of the cell (45, 54). It has been reported that a cloned 1.1-kb fragment of the 5′ end of the gene expresses a protein which catalyzes the adenylylation reaction but not the reverse deadenylylation reaction and that the expression causes a more severe growth defect than a glnD deletion (54). It has also been shown previously that an engineered chromosomal glnD mutation in Streptomyces coelicolor which expresses the N-terminal 482 residues of the protein causes deregulated adenylylation of GlnK in vivo. However, neither this truncation nor a GlnD null mutation causes a changed growth phenotype or changes in the adenylylation pattern of GS, and it has therefore been concluded that GlnD of S. coelicolor does not have a function analogous to that of GlnD of E. coli (24). We cannot decide from our data if the GlnD39 and GlnD42 proteins have UR activity, but with the absence of the HD domain and by analogy to the findings for the gram-positive bacteria, it seems likely that they lack UR activity. In vitro studies done by M. R. Atkinson and A. J. Ninfa with the purified N-terminal NT domain which was prepared by proteolytic cleavage of GlnD support the conclusion that the present GlnD′ proteins lack UR activity and possess UTase activity which is unregulated by glutamine (A. J. Ninfa, personal communication).
Ninfa et al. (43) have previously reported that overexpression of wild-type GlnD elicits a nitrogen response in E. coli, and they proposed that excess amount of GlnD can titrate out PII.
We found that overexpression of the truncated GlnD′ proteins caused a stronger expression of glnK-lacZ than overexpression of wild-type GlnD (Table 3). If the previous explanation (43) is correct, the truncated GlnD′ proteins may bind PII more strongly than wild-type GlnD. An additional explanation for our finding is that the lack of UR activity and the presence of unregulated UTase activity in the GlnD′ proteins resulted in increased accumulation of PII-UMP.
It has been suspected for years that the 5′ end of glnD transposon mutants of gram-negative bacteria may encode a protein fragment which is essential, at least for the diazotrophic ones (4). Besides the glnD99::Tn10 mutation, the present mutations are the only glnD transposon mutations of E. coli which have been physically mapped. None of the mapped glnD transposon mutations of other gram-negative bacteria, which have been obtained by screening for loss of a biological function, carry the transposon in the 5′ end of the gene. A mutant of Gluconacetobacter diazotrophicus has been constructed with an insertion in the 3′ end of the chromosomal glnD gene, but attempts to construct mutants with an insertion in the central region of glnD failed (46). Similarly, Rhizobium leguminosarum does not seem to tolerate an internal deletion within the 5′ end and the central region of the chromosomal glnD gene but tolerates insertions in the central region or the 3′ end of the gene (51). In Azotobacter vinelandii, a glnD null mutation is lethal unless it carries a suppressor mutation (presumably in glnE encoding ATase) which prevents adenylylation of GS or produces a mutant GS protein with an inactive adenylylation site. This bacterium lacks a glutamine uptake system and succumbs to glutamine starvation if GS is permanently adenylylated (17). Whether the explanation for the apparent lethality of certain glnD mutations is the same for other gram-negative bacteria remains to be seen.
We have not physically mapped the ΔglnD99::Tn10 mutation which we used in our complementation studies. The deletion mutant was phenotypically similar to the parental transposon mutant, and the only purpose of making the deletion was to be able to introduce the recA56 mutation by cotransduction with a nearby Tn10 mutation. Thus, the inactive chromosomal glnD gene and the truncated plasmid-borne gene could not have recombined in our complementation and overexpression experiments with strain AT5 (ΔglnD99::Tn10 recA56).
Compared with the other proteins in the nitrogen response regulation of E. coli, relatively little is known about GlnD, apparently because GlnD is difficult to handle experimentally due to its large size. The biologically active GlnD39 and GlnD42 proteins or variants of them may become useful for further characterization of the structure and enzymology of GlnD. It is noteworthy that E. coli ATase has been separated into an N-terminal fragment which catalyzes adenylylation of GS and a C-terminal fragment which catalyzes the reverse reaction; both fragments carry a separate nucleotidyltransferase domain (27). The crystal structure of the N-terminal domain has now been determined (62).
Acknowledgments
We thank H. Devold for help with isolation of transposon mutants. We thank A. J. Ninfa for providing strain PΦ and E. Degryse for providing plasmid pTG7420.
This work was supported by a grant from the Research Council of Norway.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469-472. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 1999. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 287:1023-1040. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L., and E. V. Koonin. 1999. DNA polymerase β-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 27:1609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arcondéguy, T., R. Jack, and M. Merrick. 2001. PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65:80-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkinson, M. R., and A. J. Ninfa. 1992. Characterization of Escherichia coli glnL mutations affecting nitrogen regulation. J. Bacteriol. 174:4538-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkinson, M. R., E. S. Kamberov, R. L. Weiss, and A. J. Ninfa. 1994. Reversible uridylylation of the Escherichia coli PII signal transduction protein regulates its ability to stimulate the dephosphorylation of the transcription factor nitrogen regulator I (NRI or NtrC). J. Biol. Chem. 269:28288-28293. [PubMed] [Google Scholar]
- 7.Atkinson, M. R., and A. J. Ninfa. 1998. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol. Microbiol. 29:431-447. [DOI] [PubMed] [Google Scholar]
- 8.Atkinson, M. R., T. A. Blauwkamp, V. Bondarenko, V. Studitsky, and A. J. Ninfa. 2002. Activation of the glnA, glnK, and nac promoters as Escherichia coli undergoes the transition from nitrogen excess growth to nitrogen starvation. J. Bacteriol. 184:5358-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1991. Current protocols in molecular biology. Wiley, New York, N.Y.
- 10.Backman, K., Y. M. Chen, and B. Magasanik. 1981. Physical and genetic characterization of the glnA-glnG region of the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 78:3743-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bancroft, S., S. G. Rhee, C. Neumann, and S. Kustu. 1978. Mutations that alter the covalent modification of glutamine synthetase in Salmonella typhimurium. J. Bacteriol. 134:1046-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, P. Karunakaran, and S. Valla. 1997. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 38:35-51. [DOI] [PubMed] [Google Scholar]
- 13.Bochner, B. R., H. C. Huang, G. L. Schieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boos, W., U. Ehmann, H. Forkl, W. Klein, M. Rimmele, and P. Postma. 1990. Trehalose transport and metabolism in Escherichia coli. J. Bacteriol. 172:3450-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bueno, R., G. Pahel, and B. Magasanik. 1985. Role of glnB and glnD gene products in regulation of the glnALG operon of Escherichia coli. J. Bacteriol. 164:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colnaghi, R., P. Rudnick, L. He, A. Green, D. Yan, E. Larson, and C. Kennedy. 2001. Lethality of glnD null mutations in Azotobacter vinelandii is suppressible by prevention of glutamine synthetase adenylylation. Microbiology 147:1267-1276. [DOI] [PubMed] [Google Scholar]
- 18.Csonka, L. N., T. P. Ikeda, S. A. Fletcher, and S. Kustu. 1994. The accumulation of glutamate is necessary for optimal growth of Salmonella typhimurium in media of high osmolality but not induction of the proU operon. J. Bacteriol. 176:6324-6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Degryse, E. 1991. Polymorphism in the dgt-dapD-tsf region of Escherichia coli K-12 strains. Gene 102:141-142. [DOI] [PubMed] [Google Scholar]
- 20.Dinnbier, U., E. Limpinsel, R. Schmid, and E. P. Bakker. 1988. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 150:348-357. [DOI] [PubMed] [Google Scholar]
- 21.Giæver, H. M., O. B. Styrvold, I. Kaasen, and A. R. Strøm. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170:2841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez, C., M. Ardourel, E. Bremer, A. Middendorf, W. Boos, and U. Ehmann. 1989. Analysis and DNA sequence of the osmoregulated treA gene encoding the periplasmic trehalase of Escherichia coli K-12. Mol. Gen. Genet. 217:347-354. [DOI] [PubMed] [Google Scholar]
- 23.Hengge-Aronis, R., W. Klein, R. Lange, M. Rimmele, and W. Boos. 1991. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 173:7918-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hesketh, A., D. Fink, B. Gust, H. U. Rexer, B. Scheel, K. Chater, W. Wohlleben, and A. Engels. 2002. The GlnD and GlnK homologues of Streptomyces coelicolor A3(2) are functionally dissimilar to their nitrogen regulatory system counterparts from enteric bacteria. Mol. Microbiol. 46:319-330. [DOI] [PubMed] [Google Scholar]
- 25.Holm, L., and C. Sander. 1995. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 20:345-347. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda, T. P., A. E. Shauger, and S. Kustu. 1996. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 259:589-607. [DOI] [PubMed] [Google Scholar]
- 27.Jaggi, R., W. C. van Heeswijk, H. V. Westerhoff, D. L. Ollis, and S. G. Vasudevan. 1997. The two opposing activities of adenylyl transferase reside in distinct homologous domains, with intramolecular signal transduction. EMBO J. 16:5562-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakoby, M., R. Krämer, and A. Burkovski. 1999. Nitrogen regulation in Corynebacterium glutamicum: isolation of genes involved and biochemical characterization of corresponding proteins. FEMS Microbiol. Lett. 173:303-310. [DOI] [PubMed] [Google Scholar]
- 29.Javelle, A., E. Severi, J. Thornton, and M. Merrick. 2004. Ammonium sensing in Escherichia coli. Role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J. Biol. Chem. 279:8530-8538. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, P., J. A. Peliska, and A. J. Ninfa. 1998. The regulation of Escherichia coli glutamine synthetase revisited: role of 2-ketoglutarate in the regulation of glutamine synthetase adenylylation state. Biochemistry 37:12802-12810. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, P., J. A. Peliska, and A. J. Ninfa. 1998. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry 37:12782-12794. [DOI] [PubMed] [Google Scholar]
- 32.Kaasen, I., P. Falkenberg, O. B. Styrvold, and A. R. Strøm. 1992. Molecular cloning and physical mapping of the otsBA genes, which encode the osmoregulatory trehalose pathway of Escherichia coli: evidence that transcription is activated by KatF (AppR). J. Bacteriol. 174:889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaasen, I., J. McDougall, and A. R. Strøm. 1994. Analysis of the otsBA operon for osmoregulatory trehalose synthesis in Escherichia coli and homology of the OtsA and OtsB proteins to the yeast trehalose-6-phosphate synthase/phosphatase complex. Gene 145:9-15. [DOI] [PubMed] [Google Scholar]
- 34.Kamberov, E. S., M. R. Atkinson, J. L. Feng, P. Chandran, and A. J. Ninfa. 1994. Sensory components controlling bacterial nitrogen assimilation. Cell. Mol. Biol. Res. 40:175-191. [PubMed] [Google Scholar]
- 35.Kandror, O., A. DeLeon, and A. L. Goldberg. 2002. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. USA 99:9727-9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keener, J., and S. Kustu. 1988. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc. Natl. Acad. Sci. USA 85:4976-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 38.Larsen, P. I., L. K. Sydnes, B. Landfald, and A. R. Strøm. 1987. Osmoregulation in Escherichia coli by accumulation of organic osmolytes: betaines, glutamic acid, and trehalose. Arch. Microbiol. 147:1-7. [DOI] [PubMed] [Google Scholar]
- 39.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.McLaggan, D., J. Naprstek, E. T. Buurman, and W. Epstein. 1994. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J. Biol. Chem. 269:1911-1917. [PubMed] [Google Scholar]
- 41.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Ninfa, A. J., and P. Jiang. 2005. PII signal transduction proteins: sensors of α-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 8:168-173. [DOI] [PubMed] [Google Scholar]
- 43.Ninfa, A. J., P. Jiang, M. R. Atkinson, and J. A. Peliska. 2000. Integration of antagonistic signals in the regulation of nitrogen assimilation in Escherichia coli. Curr. Top. Cell Regul. 36:31-75. [DOI] [PubMed] [Google Scholar]
- 44.Ninfa, A. J., and B. Magasanik. 1986. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc. Natl. Acad. Sci. USA 83:5909-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nolden, L., C. E. Ngouoto-Nkili, A. K. Bendt, R. Krämer, and A. Burkovski. 2001. Sensing nitrogen limitation in Corynebacterium glutamicum: the role of glnK and glnD. Mol. Microbiol. 42:1281-1295. [DOI] [PubMed] [Google Scholar]
- 46.Perlova, O., R. Nawroth, E. M. Zellermann, and D. Meletzus. 2002. Isolation and characterization of the glnD gene of Gluconacetobacter diazotrophicus, encoding a putative uridylyltransferase/uridylyl-removing enzyme. Gene 297:159-168. [DOI] [PubMed] [Google Scholar]
- 47.Reitzer, L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57:155-176. [DOI] [PubMed] [Google Scholar]
- 48.Reitzer, L. J., and B. Magasanik. 1985. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc. Natl. Acad. Sci. USA 82:1979-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakamoto, N., A. M. Kotre, and M. A. Savageau. 1975. Glutamate dehydrogenase from Escherichia coli: purification and properties. J. Bacteriol. 124:775-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 51.Schlüter, A., M. Nöhlen, M. Krämer, R. Defez, and U. B. Priefer. 2000. The Rhizobium leguminosarum bv. viciae glnD gene, encoding a uridylyltransferase/uridylyl-removing enzyme, is expressed in the root nodule but is not essential for nitrogen fixation. Microbiology 146:2987-2996. [DOI] [PubMed] [Google Scholar]
- 52.Son, H. S., and S. G. Rhee. 1987. Cascade control of Escherichia coli glutamine synthetase: purification and properties of PII protein and nucleotide sequence of its structural gene. J. Biol. Chem. 262:8690-8695. [PubMed] [Google Scholar]
- 53.Stadtman, E. R. 1990. Discovery of glutamine synthetase cascade. Methods Enzymol. 182:793-809. [DOI] [PubMed] [Google Scholar]
- 54.Strösser, J., A. Lüdke, S. Schaffer, R. Krämer, and A. Burkovski. 2004. Regulation of GlnK activity: modification, membrane sequestration and proteolysis as regulatory principles in the network of nitrogen control in Corynebacterium glutamicum. Mol. Microbiol. 54:132-147. [DOI] [PubMed] [Google Scholar]
- 55.Tritsch, G. L., and G. E. Moore. 1962. Spontaneous decomposition of glutamine in cell culture media. Exp. Cell Res. 28:360-364. [DOI] [PubMed] [Google Scholar]
- 56.van Heeswijk, W. C., M. Rabenberg, H. V. Westerhoff, and D. Kahn. 1993. The genes of glutamine synthetase adenylylation cascade are not regulated by nitrogen in Escherichia coli. Mol. Microbiol. 9:443-457. [DOI] [PubMed] [Google Scholar]
- 57.van Heeswijk, W. C., B. Stegeman, S. Hoving, D. Molneaar, D. Kahn, and H. V. Westerhoff. 1995. An additional PII in Escherichia coli: a new regulatory protein in the glutamine synthetase cascade. FEMS Microbiol. Lett. 132:153-157. [DOI] [PubMed] [Google Scholar]
- 58.van Heeswijk, W. C., S. Hoving, D. Molenaar, B. Stegeman, D. Kahn, and H. V. Westerhoff. 1996. An alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol. Microbiol. 21:133-146. [DOI] [PubMed] [Google Scholar]
- 59.van Heeswijk, W. C., D. Wen, P. Clancy, R. Jaggi, D. L. Ollis, H. V. Westerhoff, and S. G. Vasudevan. 2000. The Escherichia coli signal transducers PII (GlnB) and GlnK form heterotrimers in vivo: fine tuning the nitrogen signal cascade. Proc. Natl. Acad. Sci. USA 97:3942-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vasudevan, S. G., C. Gedye, N. E. Dixon, E. Cheah, P. D. Carr, P. M. Suffolk, P. D. Jeffrey, and D. L. Ollis. 1994. Escherichia coli PII protein: purification, crystallization and oligomeric structure. FEBS Lett. 337:255-258. [DOI] [PubMed] [Google Scholar]
- 61.Xu, Y., E. Cheah, P. D. Carr, W. C. van Heeswijk, H. V. Westerhoff, S. G. Vasudevan, and D. L. Ollis. 1998. GlnK, a PII homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J. Mol. Biol. 282:149-165. [DOI] [PubMed] [Google Scholar]
- 62.Xu, Y., R. Zhang, A. Joachimiak, P. D. Carr, T. Huber, S. G. Vasudevan, and D. L. Ollis. 2004. Structure of the N-terminal domain of Escherichia coli glutamine synthetase adenylyltransferase. Structure 12:861-869. [DOI] [PubMed] [Google Scholar]
- 63.Yan, D., T. P. Ikeda, A. E. Shauger, and S. Kustu. 1996. Glutamate is required to maintain the steady-state potassium pool in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:6527-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng, L., D. Kostrewa, S. Berneche, F. K. Winkler, and X. D. Li. 2004. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. USA 7:17090-17095. [DOI] [PMC free article] [PubMed] [Google Scholar]