Abstract
Modification of the phosphate groups of lipid A with amine-containing substituents, such as phosphoethanolamine, reduces the overall net negative charge of gram-negative bacterial lipopolysaccharide, thereby lowering its affinity to cationic antimicrobial peptides. Modification of the 1 position of Helicobacter pylori lipid A is a two-step process involving the removal of the 1-phosphate group by a lipid A phosphatase, LpxEHP (Hp0021), followed by the addition of a phosphoethanolamine residue catalyzed by EptAHP (Hp0022). To demonstrate the importance of modifying the 1 position of H. pylori lipid A, we generated LpxEHP-deficient mutants in various H. pylori strains by insertion of a chloramphenicol resistance cassette into lpxEHP and examined the significance of LpxE with respect to cationic antimicrobial peptide resistance. Using both mass spectrometry analysis and an in vitro assay system, we showed that the loss of LpxEHP activity in various H. pylori strains resulted in the loss of modification of the 1 position of H. pylori lipid A, thus confirming the function of LpxEHP. Due to its unique lipid A structure, H. pylori is highly resistant to the antimicrobial peptide polymyxin (MIC > 250 μg/ml). However, disruption of lpxEHP in H. pylori results in a dramatic decrease in polymyxin resistance (MIC, 10 μg/ml). In conclusion, we have characterized the first gram-negative LpxE-deficient mutant and have shown the importance of modifying the 1 position of H. pylori lipid A for resistance to polymyxin.
Helicobacter pylori is a prevalent human pathogen infecting the stomachs of nearly half of the world's population (43). Although most H. pylori infections are asymptomatic, chronic infection of the gastric mucosa by the bacterium causes recurrent gastroduodenal inflammatory disease (4, 55). In addition, persistent infection with H. pylori is considered a risk factor for the development of mucosa-associated lymphoid tissue lymphoma, loss of gastric glands (atrophy), and adenocarcinoma (14, 41). A unique feature of H. pylori is its ability to persist in the human gastric mucosa, an ecological niche in which no other microorganism is known to thrive. H. pylori persistence in the mucosa is dependent on a number of different virulence factors, including production of vacuolating toxin, adhesions, urease, and presumably like other mucosal pathogens, the ability to resist the action of cationic antimicrobial peptides (CAMPs) (5, 7).
CAMPs are key components of the innate immune response and play an important role in the host defense against microbial infection (21, 31, 32). Although the exact mechanisms by which CAMPs kill gram-negative bacteria are not well understood, it has been proposed that the antimicrobial activity of CAMPs is initiated predominately through binding to the lipid A moiety of lipopolysaccharide (LPS) (9, 50). LPS is the major surface molecule of gram-negative bacteria and is held in the outer membrane by its hydrophobic anchor, lipid A (39, 45). The lipid A moiety of LPS is the bioactive component of LPS associated with endotoxic shock and is required to maintain the integrity of the outer membrane barrier (45, 59). The electrostatic interaction between CAMPs and the negatively charged phosphate groups of lipid A disrupts the structure and integrity of the outer membrane, causing the membrane barrier to become more permeable. Permeabilization of the outer membrane allows CAMPs to gain access to the inner membrane, where they are thought to impose their bactericidal effects (62, 63).
Microorganisms have evolved several mechanisms to avoid being killed by CAMPs, including covalent modification of gram-negative bacterial lipid A (59). In some organisms, such as Salmonella enterica serovar Typhimurium and Pseudomonas aeruginosa, modification of the lipid A structure is under the control of the two-component regulatory systems PhoP/PhoQ and PmrA/PmrB (13, 15, 18). For example, activation of Salmonella PmrA results in the masking of the lipid A phosphate groups with 4-amino-4-deoxy-l-arabinose (l-Ara4N) and phosphoethanolamine (pEtN). Addition of these amine-containing substituents results in reduction in the overall negative charge of the lipid A domain of LPS, promoting resistance to antimicrobial peptides, including polymyxin (15, 18). Polymyxin, a cyclic lipopeptide produced by the gram-positive soil bacterium Paenibacillus polymyxa (36), binds to the phosphate groups of lipid A and kills gram-negative bacteria in a manner that shares common features with the CAMPs of the innate immune system (62, 63).
Instead of masking the negatively charged phosphate groups with positively charged moieties, some pathogenic bacteria, like Francisella tularensis (65), Porphyromonas gingivalis (28), Bacteroides fragilis (68), and H. pylori (37, 53), take an alternative approach by expressing enzymes that remove the lipid A phosphate groups from their LPSs. The first lipid A phosphatase, LpxE, was identified in the nitrogen-fixing gram-negative endosymbiont Rhizobium leguminosarum (26). Recently, LpxE homologs in both Francisella novicida and H. pylori have been cloned and characterized (56, 67). LpxE is distantly related to Escherichia coli PgpB, a phosphatidylglycerol phosphatase, and is highly selective for the 1-phosphate group of lipid A (26, 56, 67).
In H. pylori, removal of the lipid A 1-phosphate group is followed by addition of a pEtN residue to the C-1 hydroxyl, forming a phosphodiester linkage (Fig. 1). The reaction is catalyzed by the Helicobacter protein Hp0022 (EptAHP) and occurs only after removal of the 1-phosphate group (Fig. 1) (56). Hp0022 is a homolog of other described lipid A pEtN transferases, such as E. coli EptA (60), S. enterica serovar Typhimurium PmrC (30), and Neisseria meningitidis LptA (8). However, these organisms do not express lipid A phosphatases and attach a pEtN residue directly to the lipid A phosphate group in a pyrophosphate linkage (30, 60). Previously, we demonstrated that membranes of E. coli containing LpxEHP (Hp0021) catalyzed the dephosphorylation of lipid A (56). In this study, we generated the first gram-negative LpxE-deficient mutant and demonstrated that modification at the 1 position of H. pylori lipid A by LpxE plays an important role in H. pylori resistance to the antimicrobial peptide polymyxin.
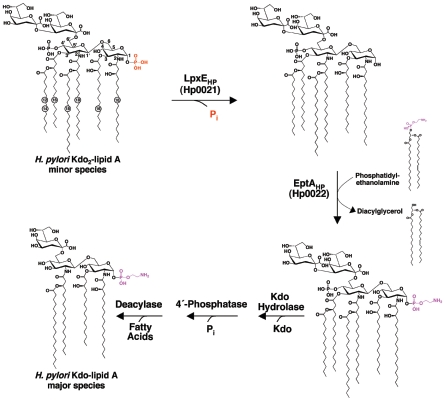
FIG. 1.
The proposed reactions catalyzed by LpxEHP (Hp0021) and EptAHP (Hp0022) of H. pylori. H. pylori synthesizes a minor lipid A species that is bis-phosphorylated and hexa-acylated, resembling E. coli Kdo2-lipid A (37, 58). Modification of the 1 position of H. pylori Kdo2-lipid A is a two-step enzymatic process that involves the removal of the 1-phosphate group by LpxEHP, followed by the addition of a pEtN unit directly to the glucosamine disaccharide backbone by EptAHP (56). Further modifications to the H. pylori lipid A structure are catalyzed by a Kdo (3-deoxy-d-manno-octulosonic acid) hydrolase (51), a 4′-phosphatase (A. X. Tran and M. S. Trent, unpublished data), and a 3′-O-deacylase (C. M. Stead and M. S. Trent, unpublished data), resulting in the major lipid A species produced by the organism (58).
MATERIALS AND METHODS
Chemicals and other materials.
[γ-32P]ATP and 32Pi were obtained from Amersham International. Silica gel 60 (0.25-mm) thin-layer plates were purchased from EM Separation Technology (Merck). Yeast extract, tryptone, and brucella broth were from Difco. Triton X-100 and bicinchoninic acid were from Pierce. All other chemicals were reagent grade and were purchased from Sigma.
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are summarized in Table 1. H. pylori strains 26695 and J99 were obtained from the American Type Culture Collection. Clinical isolates of H. pylori were obtained from human gastric biopsy specimens, as previously described (33). H. pylori was cultivated in brucella broth supplemented with 10% fetal bovine serum (HyClone) and vancomycin (10 μg/ml) or on blood agar plates (BAP) supplemented with vancomycin (10 μg/ml), chloramphenicol (8 μg/ml), and/or kanamycin (8 μg/ml) when appropriate. The cells were grown under microaerobic conditions as previously described (56). E. coli strain XL-1 Blue was routinely grown in LB broth or on LB agar (46) supplemented with ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), and/or kanamycin (30 μg/ml) when appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| Helicobacter pylori | ||
| J99 | Wild type | ATCC 700824 |
| 26695 | Wild type | ATCC 700392 |
| Hp7-91 | Clinical isolate | 51 |
| Cp20-84 | Clinical isolate | 51 |
| Hsp110-93 | Clinical isolate | 51 |
| 26695/lpxEHP::cm | 26695 with chloramphenicol resistance cassette in Hp0021 gene | This work |
| J99/lpxEHP::cm | J99 with chloramphenicol resistance cassette in Hp0021 gene | This work |
| Hp7-91/lpxEHP::cm | H. pylori clinical isolate with chloramphenicol resistance cassette in Hp0021 gene | This work |
| Hp7-91/lpxEHP::cm/pHel3-lpxEHP | Hp7-91/lpxEHP::cm; pHel3 carrying Hp0021 gene | This work |
| Cp20-84/lpxEHP::cm | H. pylori clinical isolate with chloramphenicol resistance cassette in Hp0021 gene | This work |
| Hsp110-93/lpxEHP::cm | H. pylori clinical isolate with chloramphenicol resistance cassette in Hp0021 gene | This work |
| Escherichia coli | ||
| XL-1 Blue | recA1 endA1 gyrA96thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIq ZΔM15::Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector; Ampr; lac promoter (lacZ) f1 ColE1 | Stratagene |
| pBSHp0021 | pBluescript II SK(+) containing Hp0021 gene | This work |
| pBSHp0021NX | pBHp0021 engineered with NdeI and XhoI sites | This work |
| pBSHp0021::cm | pBSHp0021NX containing a chloramphenicol resistance cassette in Hp0021 gene | This work |
| pHel2 | E. coli-H. pylori shuttle vector; Camr | 23 |
| pHel3 | E. coli-H. pylori shuttle vector; Kanr | 23 |
| pHel3-lpxEHP | pHel3 containing Hp0021 gene | This work |
Recombinant DNA techniques.
Plasmids were isolated using the QIAGEN Spin Prep kit. Custom primers were obtained from Integrated DNA Technologies. PCR reagents were purchased from Stratagene. PCR cleanup was performed using the QIAquick PCR Purification kit (QIAGEN). DNA fragments were isolated from agarose gels using the QIAquick Gel Extraction kit (QIAGEN). Restriction endonucleases, T4 DNA ligase, and shrimp alkaline phosphatase were purchased from New England Biolabs. All modifying enzymes were used according to the manufacturers' instructions.
Natural transformation of H. pylori.
Natural transformation of H. pylori strains was performed with plasmid DNA according to the procedures described by Haas et al. (19). In brief, bacteria were harvested from BAP and suspended to an A600 of 0.1 in brucella broth medium containing 10% fetal bovine serum and vancomycin (10 μg/ml). The cells were incubated for 6 to 12 h under microaerophilic conditions (85% N2, 10% CO2, 5% O2) at 37°C to allow nonmotile cells to recover. Afterward, plasmid DNA (2.5 to 5 μg) was added, and the cells were incubated for another 6 h before the suspension was plated on selective BAP. Resistant colonies were repurified on chloramphenicol (8 μg/ml)- and/or kanamycin (8 μg/ml)-containing plates.
Construction and complementation of an H. pylori lpxEHP (Hp0021 gene)-deficient mutant.
The Hp0021 gene and its flanking sequences, including 654 base pairs upstream and 807 base pairs downstream, were amplified by PCR (primers U-hp0021 and L-hp0021) (Table 2) from H. pylori 26695 genomic DNA using Pfu Turbo (Stratagene) according to the manufacturer's instructions. The flanking DNA was digested with BamHI and KpnI, gel purified, and subsequently cloned into pBluescript II SK(+) (Stratagene), resulting in the plasmid pBSHp0021. The vector pBSHp0021 was then subjected to two separate rounds of site-directed mutagenesis, using the QuikChange XL Site-Directed Mutagenesis kit (Stratagene), to create both NdeI (primers U-hp0021NdeI and L-hp0021NdeI) and XhoI (primers U-hp0021XhoI and L-hp0021XhoI) restriction sites (pBSHp0021NX). In order to disrupt the Hp0021 gene, a chloramphenicol resistance cassette (cm), obtained by PCR (primers U-hp0021CAM and L-hp0021CAM) from the E. coli-H. pylori shuttle vector (pHel2) (23), was inserted into the NdeI and XhoI sites of pBSHp0021NX. The resulting suicide plasmid, pBSHp0021::cm, containing an interrupted Hp0021 gene, was transformed into the H. pylori wild-type strains shown in Table 1 by natural transformation (23), and resistant colonies were selected on BAP containing 8 μg/ml of chloramphenicol. Resistant colonies were repurified on chloramphenicol-containing plates, and the successful insertion of the resistance cassette was verified by PCR of genomic DNA to ensure a double crossover (data not shown). Additionally, PCR primers were designed for amplification of the constructed pBluescript II SK(+) suicide vector to confirm that no vector DNA remained in the genome (data not shown).
TABLE 2.
Oligonucleotides
| Name | Sequencea |
|---|---|
| U-hp0021 | 5′-GCGCGCGGATCCAACGACGGCGAAAAGAAT-3′ |
| L-hp0021 | 5′-GCGCGCGGTACCAGGATAAACCCTCTCTAT-3′ |
| U-hp0021NdeI | 5′-GTTTGGGCGAGCGCCCATATGGAGGTAATTTCAAC-3′ |
| L-hp0021NdeI | 5′-GTTGAAATTACCTCCATATGGGCGCTCGCCCAAAC-3′ |
| U-hp0021XhoI | 5′-AACATGCCAAGCGGGCACTCGAGTATGGTGGGTTTGGCGGTG-3′ |
| L-hp0021XhoI | 5′-CACCGCCAAACCCACCATACTCGAGTGCCCGCTTGGCATGTT-3′ |
| U-hp0021CAM | 5′-GCGCGCCATATGCCGAGATTTTCAGGAGCT-3′ |
| L-hp0021CAM | 5′-GCGCGCCTCGAGTTACGCCCCGCCCTGCCA-3′ |
| U-hp0021Comp | 5′-GCGCGCCTCGAGAGCATTCCATCATTCAAA-3′ |
| L-hp0021Comp | 5′-GCGCGCGGTACCTTAAGGTTTTTTGATTAA-3′ |
Underlining indicates restriction sites.
To construct a plasmid for complementation of the lpxE mutant, the lpxE gene, including 150 base pairs upstream, was amplified by PCR (U-hp0021Comp and L-hp0021Comp) from H. pylori Hp7-91 genomic DNA using Pfu Turbo (Stratagene) according to the manufacturer's instructions. The PCR product was digested with XhoI and KpnI, gel purified, and cloned into the E. coli-H. pylori shuttle vector (pHel3) (23). The resulting plasmid, pHel3-lpxEHP, was transformed into H. pylori lpxEHP-deficient mutants by natural transformation and selected on BAP containing chloramphenicol (8 μg/ml) and kanamycin (8 μg/ml).
Isolation and analysis of lipid A and phospholipid species from 32Pi-labeled cells.
H. pylori strains were grown as described above. Typically, a 20-ml cell culture was labeled uniformly with 5 μCi/ml 32Pi until the cells reached late log phase. Bacteria were collected using a clinical centrifuge and washed with 5 ml of phosphate-buffered saline (pH 7.4). 32P-labeled lipid A and phospholipids were isolated using published protocols (70) and spotted onto a Silica Gel 60 thin-layer chromatography (TLC) plate (∼10,000 cpm/lane). Lipids were separated using the solvent chloroform, pyridine, 88% formic acid, and water (50:50:16:5 [vol/vol/vol/vol]). The TLC plates were exposed overnight to a phosphorimager screen, and product formation was detected and analyzed using a Bio-Rad Molecular Imager phosphorimager equipped with Quantity One software.
Large-scale isolation of H. pylori lipid A.
Cultures (500 ml) were grown to an A600 of 1.0 at 37°C under microaerophilic conditions for 36 to 40 h. Cells were harvested by centrifugation at 6,000 × g for 15 min, washed once with phosphate-buffered saline, and resuspended in 20 ml of phosphate-buffered saline (12). Lipid A was released from the cells and separated by DEAE anion-exchange chromatography as previously described (40, 70) and stored frozen at −20°C.
Mass spectrometry of lipid A species.
Mass spectra of the purified lipids were acquired in the negative linear mode using a matrix-assisted laser desorption-ionization-time of flight (MALDI-TOF) mass spectrometer (AXIMA-CFR; Kratos Analytical, Manchester, United Kingdom) equipped with a nitrogen laser (337 nm). The instrument was operated using 20-kV extraction voltage and time-delayed extraction, providing a mass resolution of about ±1 atomic mass unit for compounds with an Mr of ∼2,000. Each spectrum represented the average of 100 laser shots. Saturated 6-aza-2-thiothymine in 50% acetonitrile and 10% tribasic ammonium citrate (9:1 [vol/vol]) served as the matrix. The samples were dissolved in chloroform/methanol (4:1 [vol/vol]) and deposited on the sample plate, followed by an equal portion of matrix solution (0.3 μl). The sample was dried at 25°C prior to mass analysis.
Preparation of cell extracts, double-spun cytosol, and washed membrane.
Typically, 250 ml of H. pylori cultures was grown to an A600 of 1.0 at 37°C and harvested by centrifugation at 6,000 × g for 30 min. All samples were prepared at 4°C. Cell extract, membrane-free cytosol, and washed membranes were prepared as previously described (56) and stored in aliquots at −20°C. The protein concentration was determined by the bicinchoninic acid method (49), using bovine serum albumin as the standard.
Preparation of radiolabeled substrates.
The substrate [4′-32P]lipid IVA was generated from 125 μCi of [γ-32P]ATP and the tetra-acyl-disaccharide 1-phosphate lipid acceptor (a generous gift from C. R. H. Raetz), using the overexpressed 4′-kinase (LpxK) present in membranes of E. coli BLR(DE3)/pLysS/pJK2, as previously described (1, 2, 56).
H. pylori lipid A 1-phosphatase assay.
The H. pylori lipid A 1-phosphatase activity was assayed under optimized conditions based on the method of Tran et al. (56). Reaction mixtures (10 μl) contained 50 mM MES (morpholineethanesulfonic acid) (pH 6.0), 0.2% Triton X-100, and 5 μM [4′-32P]lipid IVA (∼3,000 to 5,000 cpm/nmol) as the substrate. Washed membranes (1.0 mg/ml) were employed as the enzyme source, as indicated. Dephosphorylation reaction mixtures were incubated at 30°C for the indicated times and terminated by spotting 4.5-μl portions of the mixtures onto silica gel 60 TLC plates. Reaction products were separated and visualized as described above.
Polymyxin B sensitivity assays.
Polymyxin B sulfate (Sigma) was used at concentrations of 0.1 to 250 μg/ml in both plate and broth assays. Standard MIC testing of susceptibility to polymyxin was performed as described by Gunn and Miller and Steinberg et al. (16, 52). To assay H. pylori strains for polymyxin resistance, the strains were grown to mid-log phase (optical density at 600 nm, ∼0.35) and diluted to a concentration of approximately 2,500 CFU/ml in brucella broth. The cells (200 μl) were mixed on a microtiter plate with various concentrations of polymyxin B sulfate (Sigma) and incubated at 37°C for 1 h. Then, 150 μl of polymyxin-treated cells was directly plated on BAP and incubated at 37°C under microaerobic conditions for 36 to 40 h. The colony counts of cells incubated with the various concentrations of polymyxin were compared to colony counts of cells not exposed to polymyxin. The percent survival was defined as follows: survival (%) = (CFU of peptide-exposed culture/CFU of nonexposed culture) × 100.
Transmission electron microscopy.
H. pylori cells were grown to mid-log phase (A600, ∼0.5) and diluted to a concentration of approximately 2,500 CFU/ml in brucella broth. Cells (2 ml) were exposed to 100 μg/ml of polymyxin B sulfate (Sigma) and incubated at 37°C under microaerobic conditions for 1 h. Afterward, samples were washed, fixed in 2% (vol/vol) glutaraldehyde plus 0.5% (vol/vol) paraformaldehyde, processed, embedded in Epon-Araldite resin, and stained for high-contrast morphology as described previously (25, 69). Ultrathin sections were examined with a Philips Tecnai 10 transmission electron microscope (FEI Company, Hillsboro, Ore.) operating at 80 kV.
RESULTS
MALDI-TOF mass spectrometry and 32P labeling of the lipids A from H. pylori strains 26695 and 26695/lpxEHP::cm.
In strain 26695, the Hp0021 (accession number DQ447324) and Hp0022 (accession number DQ447325) genes encode the lipid A 1-phosphatase and pEtN transferase, respectively (56) (Fig. 2), with a similar genomic organization found in strain J99. In either case, these genes are unidirectionally transcribed beginning with the Hp0022 gene and separated by only 10 bp. To determine if modification of H. pylori lipid A at the 1 position was important for resistance to antimicrobial peptides and to confirm its role in the modification of Helicobacter lipid A, we constructed an lpxEHP-deficient mutant of the sequenced strain 26695. Based upon previously reported biochemical evidence (56), it was expected that inactivation of lpxEHP would result in the loss of 1-phosphatase activity, thereby preventing the addition of pEtN catalyzed by H. pylori EptAHP (Hp0022).
FIG. 2.
Schematic representation of the H. pylori chromosomal region of strain 26695 encoding LpxEHP and EptAHP. The overall genomic organizations of H. pylori genes encoding enzymes modifying the 1 position of H. pylori lipid A are similar in strains 26695 and J99. However, the region between the Hp0022 and Hp0025 genes are not annotated as open reading frames in strain J99. As indicated in the figure, 28 bp of DNA was removed from the lpxEHP gene prior to insertion of the catGC cassette conferring chloramphenicol resistance. Based upon homology to other lipid phosphatases, such as Francisella LpxE, Rhizobium LpxE, and E. coli PgpB, the deleted 28-bp region encodes a protein domain that is predicted to function as part of the active site of H. pylori LpxE.
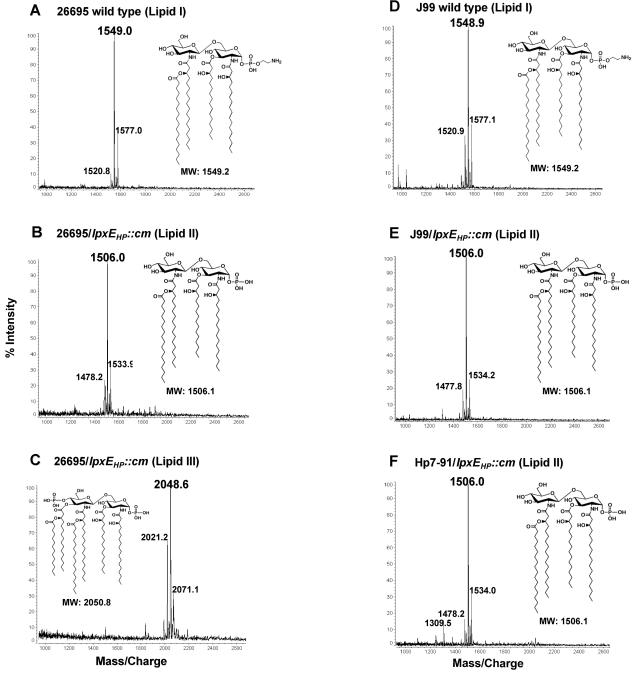
The lipid A species from both wild-type 26695 and the 26695/lpxEHP::cm mutant were isolated and fractionated based upon their charges by DEAE-cellulose column chromatography with increasing salt step elutions. Following DEAE purification, the lipids were analyzed by MALDI-TOF mass spectrometry in the negative mode. Mass spectrometry analysis of wild-type 26695 showed a major ion peak at m/z 1549.0 atomic mass units, corresponding to a tetra-acylated lipid A structure that lacks the 4′-phosphate group and is substituted at the C-1 position with a pEtN residue (lipid I) (Fig. 3A). This is the first analysis of the lipid A structure synthesized by one of the sequenced strains of H. pylori and is identical to that previously reported by Moran and coworkers for H. pylori strain NCTC 11637 (37). As predicted, inactivation of lpxEHP in H. pylori strain 26695 prevented the enzymatic activity of EptAHP and resulted in the presence of a tetra-acylated lipid A species with the 1-phosphate group still attached to the C-1 position (lipid II), as indicated by the major ion peak at m/z 1506.0 (Fig. 3B). Lipid II, bearing a single phosphate group, eluted from the DEAE column during the 120 mM salt elution. However, based upon the dry mass of lipid A fractions produced by the 26695 lpxE mutant, lipid II was <5% of the total lipid (data not shown).
FIG. 3.
Comparison of the lipid A fractions isolated from wild-type H. pylori and lpxEHP mutants. Lipids A of the indicated strains were analyzed by MALDI-TOF mass spectrometry in the negative-ion mode. Although mass spectrometry data was acquired on lipids eluting in each fraction from the DEAE column (40), only those fractions containing lipids are shown. Lipid I from wild-type strains 26695 (A) and J99 (D) were found to elute in the flowthrough and wash fractions, whereas lipid II (B, E, and F) produced by lpxEHP mutants eluted in the 120 mM salt fraction (see Materials and Methods). Lipid III (C), produced only by the 26695/lpxEHP::cm mutant, eluted in the 240 mΜ salt fraction.
The bulk of the lipid A synthesized by 26695/lpxEHP::cm eluted in the 240 mM salt fraction during DEAE chromatography, showing a major ion peak at m/z 2048.6 during mass spectrometry. The ion peak at m/z 2048.6 atomic mass units corresponds to a lipid A species retaining both the 1- and 4′-phosphate groups (lipid III), resulting in an increase in the negative charge. Furthermore, lipid III retained the 3′-ester-linked fatty acyl chains, resulting in a hexa-acylated structure (Fig. 3C). TLC analysis of 32P-labeled lipid A species isolated from 26695 and its isogenic lpxEHP mutant confirmed that 26695/lpxEHP::cm produces only one major lipid A species (Fig. 4A, lane 2) that migrates faster than wild-type lipid A (Fig. 4A, lane 1). The bis-phosphorylated and hexa-acylated lipid A structure synthesized by 26695/lpxEHP::cm, lipid III, confirms that H. pylori contains the necessary enzymatic machinery to synthesize a lipid A molecule structurally similar to that of E. coli. However, unlike E. coli lipid A, the lipid A of Helicobacter bears fatty acyl chains 16 to 18 carbons in length. Previously, Moran and coworkers and Tran et al. reported that a lipid A species identical to lipid III (Fig. 1) could be isolated from wild-type H. pylori strain NCTC 11637, but only as a minor component of the total lipid (37, 58). However, we were unable to detect any species similar to lipid III in wild-type strains grown in liquid culture. Given that LpxE is distantly related to E. coli PgpB, a phosphatidylglycerol phosphatase (11), the 32P-labeled phospholipids of wild-type strain 26695 were compared to those of 26695/lpxEHP::cm (Fig. 4B). Based upon TLC analysis, inactivation of lpxEHP had no affect on phospholipid biosynthesis.
FIG. 4.
Comparison of 32P-labeled lipid A (A) and phospholipid fractions (B) isolated from H. pylori 26695 and 26695/lpxEHP::cm. 32P-labeled lipid A species or phospholipids were isolated as described in Materials and Methods and separated by TLC using the solvent chloroform, pyridine, 88% formic acid, and water (50:50:16:5 [vol/vol/vol/vol]). The 32P-labeled lipids were visualized by phosphorimaging. The identities of the major phospholipids are based upon the major phospholipids reported for H. pylori (24) and the migration of the major phospholipids found in E. coli (72). PG, phosphatidylglycerol; PE, phosphatidylethanolamine; CL, cardiolipin.
MALDI-TOF mass spectrometry of lipid A From H. pylori strains J99, J99/lpxEHP::cm, and Hp7-91/lpxEHP::cm.
The bis-phosphorylated and hexa-acylated lipid A structure resulting from disruption of lpxEHP in 26695 (Fig. 3C) suggested that loss of 1-phosphatase activity may prevent other enzymatic modifications of H. pylori lipid A, such as removal of the 4′-phosphate or deacylation (Fig. 1) (58). Both pEtN addition and removal of the Kdo hydrolase have been shown to require prior removal of the 1-phosphate group in H. pylori (51, 56). Therefore, we were interested to determine whether inactivation of lpxEHP in strain J99, a second strain for which the genomic sequence is available, resulted in production of similar lipid A structures. Like 26695, the outer membrane of H. pylori J99 contained a tetra-acylated lipid A lacking the 4′-phosphate group and bearing a pEtN at the 1 position (Fig. 3D). Mass spectrometry analysis of lipid A isolated from J99/lpxEHP::cm revealed one major ion peak at m/z 1506.0 atomic mass units (Fig. 3E), resembling the minor tetra-acylated lipid A species (lipid II) of the 26695 lpxEHP mutant. Interestingly, the tetra-acylated species bearing a single phosphate group was the major lipid produced by the J99 mutant, and no hexa-acylated structures were detected (Fig. 4).
Further investigation of three clinical isolates, Hp7-91 (Fig. 3F), Cp20-84, and Hsp110-93 (data not shown), lacking a functional 1-phosphatase, revealed that, like J99, these strains synthesized only a tetra-acylated molecule. Taken together, these data suggest that insertional inactivation of lpxEHP results in replacement of a pEtN residue with a phosphate group at the C-1 position of H. pylori lipid A, increasing the overall negative charge of the molecule and, thus, of the bacterial surface. At present, it is unclear why the 26695 lpxEHP mutant produces a lipid A species that is bis-phosphorylated and hexa-acylated.
Complementation of the H. pylori lpxEHP-deficient mutant.
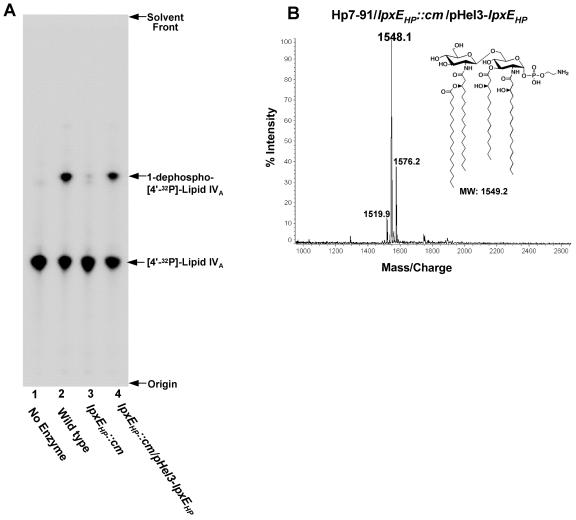
Unlike strains 26695 and J99 (48), H. pylori Hp7-91 was transformed readily with the pHel3-lpxEHP covering plasmid and therefore was selected for complementation studies. To confirm the enzymatic function of LpxEHP, membranes of wild-type H. pylori strain Hp7-91 and Hp7-91/lpxEHP::cm were isolated and assayed with [4′-32P]lipid IVA, a key lipid A precursor, for 1-phosphatase activity. Removal of the 1-phosphate group from [4′-32P]lipid IVA increases the hydrophobicity of the lipid substrate, resulting in a faster-migrating lipid species, denoted 1-dephospho-[4′-32P]lipid IVA, when analyzed by TLC in the solvent system employed (56). As shown in Fig. 5A, membranes from wild-type H. pylori Hp7-91 displayed 1-phosphatase activity (Fig. 5A, lane 2). In contrast, membranes obtained from Hp7-91/lpxEHP were essentially unable to catalyze the removal of the 1-phosphate group from [4′-32P]lipid IVA (Fig. 5A, lane 3). To demonstrate that recovery of the 1-phosphatase activity was solely dependent upon LpxE, the lpxEHP gene, along with 150 base pairs of DNA upstream of its start codon, was cloned into the E. coli/H. pylori shuttle vector, pHel3 (23). The resulting plasmid, pHel3-lpxEHP, was used to complement the lpxEHP mutant, resulting in restoration of 1-phosphatase activity (Fig. 5, lane 4). Introduction of the vector control, pHel3, into Hp7-91/lpxEHP::cm had no effect upon 1-phosphatase activity (data not shown). Furthermore, natural transformation of pHel3-lpxEHP into Hp7-91/lpxEHP::cm restored the synthesis of a lipid A structure identical to that of wild-type H. pylori showing a predominant peak at m/z 1548.1 atomic mass units during mass spectrometry (Fig. 5B).
FIG. 5.
Reconstitution of 1-phosphatase activity in membranes of an H. pylori Hp7-91/lpxEHP mutant by the introduction of an lpxEHP-complementing plasmid (A) and MALDI-TOF mass spectrometry of lipid A obtained from the complemented Hp7-91/lpxEHP::cm mutant (B). Membranes isolated from the indicated H. pylori strains were assayed for 1-phosphatase activity under conditions described in Materials and Methods. Protein concentrations were 1.0 mg/ml, and the assays were carried out for 1 h at 30°C. Each reaction mixture contained 5 μΜ of [4′-32P]lipid IVA substrate. The reaction products were separated by TLC and detected by phosphorimager analysis. Removal of the 1-phosphate group was indicated by conversion of the [4′-32P]lipid IVA substrate to a faster-migrating species (56). (B) The lipid A of Hp7-91/lpxEHP::cm transformed with pHel3-lpxEHP was isolated and fractionated by DEAE-cellulose chromatography using published protocols (40). The lipid A sample was analyzed by MALDI-TOF mass spectrometry in the negative-ion mode. The data shown are from lipid A species found to elute in the flowthrough and wash fractions.
Disruption of lpxEHP affects H. pylori resistance to polymyxin.
Polymyxin primarily interacts with the negatively charged phosphate groups of lipid A. H. pylori is intrinsically resistant to polymyxin (Table 3), presumably due in part to the low number of negative charges in its lipid A. Modification of H. pylori lipid A, including pEtN decoration of the C-1 hydroxyl group, does not appear to require additional signals when the bacterium is cultured under standard laboratory conditions. The direct attachment of pEtN to the 1 position of the disaccharide backbone of Helicobacter lipid A results in a greater reduction in the negative charge than the pyrophosphate linkage from pEtN addition in E. coli (70), S. enterica serovar Typhimurium (71), and N. meningitidis (27).
TABLE 3.
MICs of polymyxin of H. pylori strains
| Strain | Polymyxin MIC (μg/ml) |
|---|---|
| 26695 | >250 |
| 26695/lpxEHP::cm | 0.2 |
| J99 | >250 |
| J99/lpxEHP::cm | 10 |
| Hp7-91 | >250 |
| Hp7-91/lpxEHP::cm | 25 |
| Hp7-91/lpxEHP::cm/pHel3-lpxEHP | >250 |
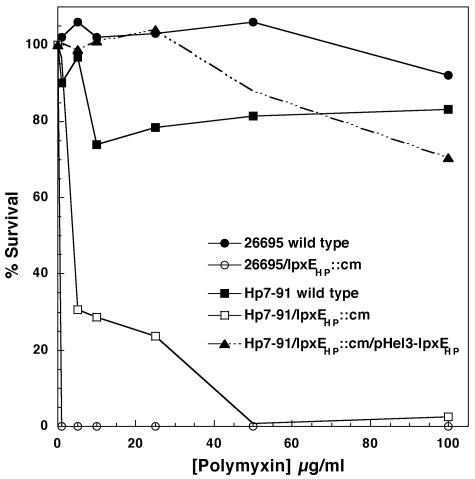
To demonstrate the importance of modifying the 1 position of H. pylori lipid A and to examine the significance of LpxEHP for polymyxin resistance, wild-type H. pylori and lpxEHP mutants were exposed to increasing concentrations of polymyxin and examined for the ability to survive in comparison to bacteria not exposed to the peptide (Fig. 6). As shown in Fig. 6, wild-type H. pylori strain 26695 demonstrated nearly 100% survival even at a polymyxin concentration of 100 μg/ml. Additionally, we determined the MICs of polymyxin for all wild-type strains to be greater than 250 μg/ml (Table 3).
FIG. 6.
Effect of lpxEHP mutation on the polymyxin resistance of H. pylori. H. pylori strains were exposed to increasing concentrations of polymyxin, and the survival percentages were determined as described in Materials and Methods. Wild-type strains 26695 and Hp7-91 served as the polymyxin-resistant controls. Survival data for strain J99 were similar to those shown for strain Hp7-91 (data not shown).
Disruption of lpxEHP in all strains resulted in a dramatic decrease in the percent survival when exposed to polymyxin (Fig. 6 and Table 3). The MICs of 26695/lpxEHP::cm and J99/lpxEHP::cm were found to be 0.2 and 10 μg/ml, respectively (Table 3). The differences in polymyxin sensitivity between the two lpxEHP mutant strains can be attributed to the number of phosphate groups on the lipid A molecule that are exposed to attack by polymyxin. The lipid A isolated from 26695/lpxEHP::cm resembles the bis-phosphorylated lipid A structure of unmodified E. coli lipid A (45), which is also highly sensitive to polymyxin with a MIC of 1 μg/ml (64). Having two phosphates attached to the lipid A domain of 26695/lpxEHP::cm dramatically increased the sensitivity to polymyxin by more than 1,000-fold compared to wild-type 26695 (Table 3). However, the sensitivity to polymyxin of strain J99/lpxEHP::cm, which has a single phosphate group attached to lipid A at the C-1 position, increased by 25-fold compared to wild-type J99 (Table 3). As shown in Table 3, 26695/lpxEHP::cm was 50 times more sensitive to polymyxin than J99/lpxEHP::cm, demonstrating the importance of modifying the lipid A phosphate groups of H. pylori LPS for resistance to CAMPs in H. pylori.
The level of polymyxin resistance was also investigated in H. pylori strains Hp7-91 and Hp7-91/lpxEHP::cm and the complemented Hp7-91 mutant. The percent survival and MIC of polymyxin for the H. pylori clinical isolate Hp7-91 were similar to those observed for wild-type 26695 and J99 (Fig. 6 and Table 3). The lipid A structures of both Hp7-91/lpxEHP::cm and J99/lpxEHP::cm were identical when analyzed by mass spectrometry and therefore displayed similar sensitivities to polymyxin (Table 3). Polymyxin resistance was fully restored in Hp7-91/lpxEHP::cm upon introduction of the lpxEHP covering plasmid (Fig. 6 and Table 3), ruling out any possible polar effects.
Transmission electron microscopy of the H. pylori 1-phosphatase mutant following exposure to polymyxin.
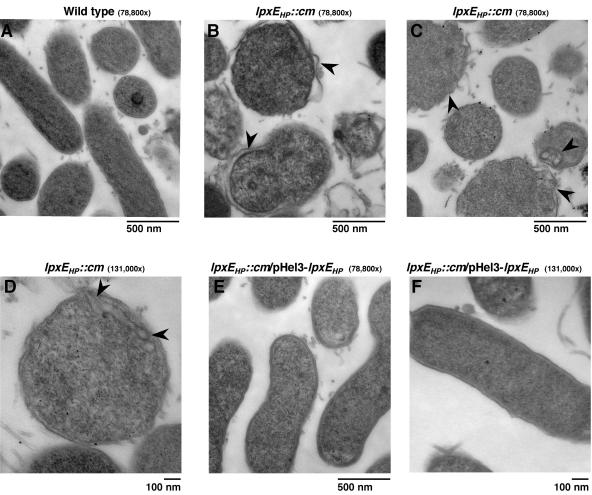
Having demonstrated that deletion of lpxEHP in H. pylori dramatically reduces resistance to polymyxin, we next examined the effect of polymyxin on the bacterial morphology of the lpxE mutant by transmission electron microscopy. Electron micrographs of both wild-type Hp7-91 and Hp7-91/lpxEHP::cm that were not exposed to polymyxin showed rods or curved rods (data not shown). Exposure to polymyxin at 100 μg/ml did not affect the cell morphology of wild-type Hp7-91 (Fig. 7A). Conversely, exposure of bacteria to polymyxin lacking a functional LpxEHP resulted in morphological alterations resembling those reported for H. pylori undergoing coccoid formation, eventually leading to cell death (Fig. 7B to D) (3, 29). The cell-damaging effect of polymyxin was characterized by ruffling of the outer membrane, membrane invaginations, and in some cases an overall increase in the periplasmic region of the bacterial cell envelope (Fig. 7B to D). Restoration of polymyxin resistance in Hp7-91/lpxEHP::cm was observed upon transformation of the plasmid pHel3-lpxEHP (Fig. 7E and F).
FIG. 7.
Transmission electron micrographs of the H. pylori 1-phosphatase mutant following exposure to polymyxin. H. pylori strains Hp7-91, Hp7-91/lpxEHP::cm, and Hp7-91/lpxEHP::cm containing pHel3-lpxEHP were exposed to 100 μg/ml of polymyxin for 1 h. The cells were processed as described in Materials and Methods. Changes to the cell envelope structure seen in the lpxEHP mutant following exposure to polymxyin are indicated by arrowheads.
DISCUSSION
H. pylori heavily modifies the lipid A moiety of its LPS, resulting in the production of a structure with a hydroxyl group at the 4′ position and a pEtN residue at the 1 position (Fig. 1) (58). The unique structure of H. pylori lipid A should help promote resistance to CAMPs during host infection, since it is well known that the lipid A phosphate groups are necessary for binding of cationic peptides (59). Here, we report the characterization of an H. pylori mutant unable to express functional LpxEHP, a lipid A 1-phosphatase. Insertional inactivation of lpxEHP resulted in the loss of 1-phosphatase activity and the subsequent transfer of pEtN. These findings corroborated our previous report that the H. pylori lipid A pEtN transferase was unique in that it functions only after the removal of the 1-phosphate group by LpxEHP (56).
Wild-type H. pylori strains were found to be inherently resistant to the antimicrobial peptide polymyxin, displaying a MIC of >250 μg/ml. This high level of polymyxin resistance has also been reported for other gram-negative organisms, including Proteus mirabilis (35), Brucella abortus (34), and N. meningitidis (61). H. pylori strain J99 and three additional clinical isolates lacking a functional LpxEHP all synthesized lipid A structures bearing a single phosphate group at the 1 position, resulting in up to a 25-fold increase in polymyxin sensitivity. Introduction of a complementing plasmid expressing the phosphatase completely restored polymyxin resistance (Fig. 6B), correlating with a lipid A structure bearing a pEtN group at the C-1 position (Fig. 5B). An unexpected but reproducible finding of these studies was the synthesis of a bis-phosphorylated species in the 26695 lpxEHP mutant. Interestingly, the 26695 lpxEHP mutant displayed complete sensitivity to polymyxin (MIC, 0.2 μg/ml) in comparison to other strains, suggesting that removal of the 4′-phosphate group of H. pylori lipid A is also critical for CAMP resistance.
It has long been known that polymyxin-resistant variants of both S. enterica serovar Typhimurium and E. coli produce lipid A species modified with both pEtN and l-Ara4N. Masking of a lipid A phosphate group with l-Ara4N has been conclusively shown to be critical for CAMP resistance (15, 17, 57). However, the importance of pEtN addition for resistance to CAMPs is not as clearly defined. For example, Salmonella pmrC (eptA) mutants show only slight increases in susceptibility to killing by polymyxin (less than fivefold) in comparison to the wild type (30, 54). On the other hand, loss of LptA function, the neisserial homolog of EptA, results in more than a 200-fold reduction in polymyxin resistance (61). Other gram-negative pathogens, such as Vibrio cholerae (6), Campylobacter jejuni (38), and P. gingivalis (28), modify the lipid A domain of their LPSs with either one or two pEtN groups. Further investigation of pEtN modification in other pathogens will be required to determine its role in CAMP resistance and pathogenesis.
During infection of the gastric mucosa, H. pylori encounters antimicrobial peptides produced by its human host. For example, expression of the human cathelicidin LL-37 (22) and human β-defensin 2 is upregulated in the gastric mucosa of H. pylori-infected patients (20, 66). Furthermore, H. pylori itself produces a cecropin-like peptide, Hp (2-20), to which it is resistant, that is derived from the amino-terminal part of its ribosomal protein L1 (44). Cecropin has been shown to bind to bis-phosphorylated lipid A (10), and phoP mutants of Salmonella unable to mask their lipid A phosphate groups show increased sensitivity to cecropin A (47). Therefore, reduction or masking of the phosphate groups of H. pylori lipid A should provide resistance to cecropin-like peptides. Indeed, the H. pylori 26695 lpxEHP mutant producing an unmodified lipid A structure displays complete sensitivity to cecropin A in comparison to the wild type (unpublished results). Since H. pylori undergoes “altruistic lysis” in vivo (42), it has been suggested that H. pylori cecropin may provide an advantage to the bacterium by inhibiting the growth of competing organisms (44). On the whole, the current work provides initial evidence supporting the importance of lipid A modifications for the survival of H. pylori upon exposure to antimicrobial peptides.
Acknowledgments
M. S. Trent thanks Anthony Moran for helpful conversations and Rainer Haas for generously providing vectors pHel2 and pHel3.
This work was supported by National Institutes of Health grants RO1-AI064184 (to M.S.T.) and RO1-GM6440 (to R.J.C.)
REFERENCES
- 1.Basu, S. S., J. D. York, and C. R. Raetz. 1999. A phosphotransferase that generates phosphatidylinositol 4-phosphate (PtdIns-4-P) from phosphatidylinositol and lipid A in Rhizobium leguminosarum. A membrane-bound enzyme linking lipid A and ptdins-4-p biosynthesis. J. Biol. Chem. 274:11139-11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belunis, C. J., and C. R. Raetz. 1992. Biosynthesis of endotoxins. Purification and catalytic properties of 3-deoxy-d-manno-octulosonic acid transferase from Escherichia coli. J. Biol. Chem. 267:9988-9997. [PubMed] [Google Scholar]
- 3.Benaissa, M., P. Babin, N. Quellard, L. Pezennec, Y. Cenatiempo, and J. L. Fauchere. 1996. Changes in Helicobacter pylori ultrastructure and antigens during conversion from the bacillary to the coccoid form. Infect. Immun. 64:2331-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaser, M. J. 1993. Helicobacter pylori: microbiology of a ‘slow’ bacterial infection. Trends Microbiol. 1:255-260. [DOI] [PubMed] [Google Scholar]
- 5.Bylund, J., T. Christophe, F. Boulay, T. Nystrom, A. Karlsson, and C. Dahlgren. 2001. Proinflammatory activity of a cecropin-like antibacterial peptide from Helicobacter pylori. Antimicrob. Agents Chemother. 45:1700-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee, S. N., and K. Chaudhuri. 2003. Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim. Biophys. Acta 1639:65-79. [DOI] [PubMed] [Google Scholar]
- 7.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 8.Cox, A. D., J. C. Wright, J. Li, D. W. Hood, E. R. Moxon, and J. C. Richards. 2003. Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide. J. Bacteriol. 185:3270-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daugelavicius, R., E. Bakiene, and D. H. Bamford. 2000. Stages of polymyxin B interaction with the Escherichia coli cell envelope. Antimicrob. Agents Chemother. 44:2969-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Lucca, A. J., T. J. Jacks, and K. A. Brogden. 1995. Binding between lipopolysaccharide and cecropin A. Mol. Cell Biochem. 151:141-148. [DOI] [PubMed] [Google Scholar]
- 11.Dillon, D. A., W. I. Wu, B. Riedel, J. B. Wissing, W. Dowhan, and G. M. Carman. 1996. The Escherichia coli pgpB gene encodes for a diacylglycerol pyrophosphate phosphatase activity. J. Biol. Chem. 271:30548-30553. [DOI] [PubMed] [Google Scholar]
- 12.Dulbecco, R., and M. Vogt. 1954. Plaque formation and isolation of pure lines with poliomyelitis viruses. J. Exp. Med. 99:167-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 14.Forman, D., D. G. Newell, F. Fullerton, J. W. Yarnell, A. R. Stacey, N. Wald, and F. Sitas. 1991. Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from a prospective investigation. BMJ 302:1302-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 16.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 19.Haas, R., T. F. Meyer, and J. P. van Putten. 1993. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol. Microbiol. 8:753-760. [DOI] [PubMed] [Google Scholar]
- 20.Hamanaka, Y., M. Nakashima, A. Wada, M. Ito, H. Kurazono, H. Hojo, Y. Nakahara, S. Kohno, T. Hirayama, and I. Sekine. 2001. Expression of human beta-defensin 2 (hBD-2) in Helicobacter pylori induced gastritis: antibacterial effect of hBD-2 against Helicobacter pylori. Gut 49:481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock, R. E., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 22.Hase, K., M. Murakami, M. Iimura, S. P. Cole, Y. Horibe, T. Ohtake, M. Obonyo, R. L. Gallo, L. Eckmann, and M. F. Kagnoff. 2003. Expression of LL-37 by human gastric epithelial cells as a potential host defense mechanism against Helicobacter pylori. Gastroenterology 125:1613-1625. [DOI] [PubMed] [Google Scholar]
- 23.Heuermann, D., and R. Haas. 1998. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257:519-528. [DOI] [PubMed] [Google Scholar]
- 24.Hirai, Y., M. Haque, T. Yoshida, K. Yokota, T. Yasuda, and K. Oguma. 1995. Unique cholesteryl glucosides in Helicobacter pylori: composition and structural analysis. J. Bacteriol. 177:5327-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodinka, R. L., and P. B. Wyrick. 1986. Ultrastructural study of mode of entry of Chlamydia psittaci into L-929 cells. Infect. Immun. 54:855-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karbarz, M. J., S. R. Kalb, R. J. Cotter, and C. R. Raetz. 2003. Expression cloning and biochemical characterization of a Rhizobium leguminosarum lipid A 1-phosphatase. J. Biol. Chem. 278:39269-39279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulshin, V. A., U. Zahringer, B. Lindner, C. E. Frasch, C. M. Tsai, B. A. Dmitriev, and E. T. Rietschel. 1992. Structural characterization of the lipid A component of pathogenic Neisseria meningitidis. J. Bacteriol. 174:1793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumada, H., Y. Haishima, T. Umemoto, and K. Tanamoto. 1995. Structural study on the free lipid A isolated from lipopolysaccharide of Porphyromonas gingivalis. J. Bacteriol. 177:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusters, J. G., M. M. Gerrits, J. A. Van Strijp, and C. M. Vandenbroucke-Grauls. 1997. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect. Immun. 65:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, H., F. F. Hsu, J. Turk, and E. A. Groisman. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186:4124-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehrer, R. I., and T. Ganz. 1999. Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol. 11:23-27. [DOI] [PubMed] [Google Scholar]
- 32.Lehrer, R. I., A. K. Lichtenstein, and T. Ganz. 1993. Defensins: antimicrobial and cytotoxic peptides of mammalian cells. Annu. Rev. Immunol. 11:105-128. [DOI] [PubMed] [Google Scholar]
- 33.Li, C., D. A. Ferguson, Jr., T. Ha, D. S. Chi, and E. Thomas. 1993. A highly specific and sensitive DNA probe derived from chromosomal DNA of Helicobacter pylori is useful for typing H. pylori isolates. J. Clin. Microbiol. 31:2157-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manterola, L., I. Moriyon, E. Moreno, A. Sola-Landa, D. S. Weiss, M. H. Koch, J. Howe, K. Brandenburg, and I. Lopez-Goni. 2005. The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J. Bacteriol. 187:5631-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy, A. J., H. Liu, T. J. Falla, and J. S. Gunn. 2001. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob. Agents Chemother. 45:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLeod, G. I., and M. P. Spector. 1996. Starvation- and stationary-phase-induced resistance to the antimicrobial peptide polymyxin B in Salmonella typhimurium is RpoS (σS) independent and occurs through both phoP-dependent and -independent pathways. J. Bacteriol. 178:3683-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moran, A. P., B. Lindner, and E. J. Walsh. 1997. Structural characterization of the lipid A component of Helicobacter pylori rough- and smooth-form lipopolysaccharides. J. Bacteriol. 179:6453-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moran, A. P., U. Zahringer, U. Seydel, D. Scholz, P. Stutz, and E. T. Rietschel. 1991. Structural analysis of the lipid A component of Campylobacter jejuni CCUG 10936 (serotype O:2) lipopolysaccharide. Description of a lipid A containing a hybrid backbone of 2-amino-2-deoxy-d-glucose and 2,3-diamino-2,3-dideoxy-d-glucose. Eur. J. Biochem. 198:459-469. [DOI] [PubMed] [Google Scholar]
- 39.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odegaard, T. J., I. A. Kaltashov, R. J. Cotter, L. Steeghs, P. van der Ley, S. Khan, D. J. Maskell, and C. R. Raetz. 1997. Shortened hydroxyacyl chains on lipid A of Escherichia coli cells expressing a foreign UDP-N-acetylglucosamine O-acyltransferase. J. Biol. Chem. 272:19688-19696. [DOI] [PubMed] [Google Scholar]
- 41.Parsonnet, J., S. Hansen, L. Rodriguez, A. B. Gelb, R. A. Warnke, E. Jellum, N. Orentreich, J. H. Vogelman, and G. D. Friedman. 1994. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 330:1267-1271. [DOI] [PubMed] [Google Scholar]
- 42.Phadnis, S. H., M. H. Parlow, M. Levy, D. Ilver, C. M. Caulkins, J. B. Connors, and B. E. Dunn. 1996. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect. Immun. 64:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pounder, R. E., and D. Ng. 1995. The prevalence of Helicobacter pylori infection in different countries. Aliment. Pharmacol. Ther. 9(Suppl. 2):33-39. [PubMed] [Google Scholar]
- 44.Putsep, K., C. I. Branden, H. G. Boman, and S. Normark. 1999. Antibacterial peptide from H. pylori. Nature 398:671-672. [DOI] [PubMed] [Google Scholar]
- 45.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Shi, Y., M. J. Cromie, F. F. Hsu, J. Turk, and E. A. Groisman. 2004. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol. Microbiol. 53:229-241. [DOI] [PubMed] [Google Scholar]
- 48.Smeets, L. C., J. J. Bijlsma, S. Y. Boomkens, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 50.Srimal, S., N. Surolia, S. Balasubramanian, and A. Surolia. 1996. Titration calorimetric studies to elucidate the specificity of the interactions of polymyxin B with lipopolysaccharides and lipid A. Biochem. J. 315:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stead, C., A. Tran, D. Ferguson, Jr., S. McGrath, R. Cotter, and S. Trent. 2005. A novel 3-deoxy-d-manno-octulosonic acid (Kdo) hydrolase that removes the outer Kdo sugar of Helicobacter pylori lipopolysaccharide. J. Bacteriol. 187:3374-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinberg, D. A., M. A. Hurst, C. A. Fujii, A. H. Kung, J. F. Ho, F. C. Cheng, D. J. Loury, and J. C. Fiddes. 1997. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob. Agents Chemother. 41:1738-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suda, Y., Y. M. Kim, T. Ogawa, N. Yasui, Y. Hasegawa, W. Kashihara, T. Shimoyama, K. Aoyama, K. Nagata, T. Tamura, and S. Kusumoto. 2001. Chemical structure and biological activity of a lipid A component from Helicobacter pylori strain 206. J. Endotoxin Res. 7:95-104. [PubMed] [Google Scholar]
- 54.Tamayo, R., B. Choudhury, A. Septer, M. Merighi, R. Carlson, and J. S. Gunn. 2005. Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar Typhimurium lipopolysaccharide core. J. Bacteriol. 187:3391-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor, D. N., and M. J. Blaser. 1991. The epidemiology of Helicobacter pylori infection. Epidemiol. Rev. 13:42-59. [DOI] [PubMed] [Google Scholar]
- 56.Tran, A. X., M. J. Karbarz, X. Wang, C. R. Raetz, S. C. McGrath, R. J. Cotter, and M. S. Trent. 2004. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. J. Biol. Chem. 279:55780-55791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran, A. X., M. E. Lester, C. M. Stead, C. R. Raetz, D. J. Maskell, S. C. McGrath, R. J. Cotter, and M. S. Trent. 2005. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J. Biol. Chem. 280:28186-28194. [DOI] [PubMed] [Google Scholar]
- 58.Tran, A. X., C. M. Stead, and M. S. Trent. 2005. Remodeling of Helicobacter pylori lipopolysaccharide. J. Endotoxin Res. 11:161-166. [DOI] [PubMed] [Google Scholar]
- 59.Trent, M. S. 2004. Biosynthesis, transport, and modification of lipid A. Biochem. Cell Biol. 82:71-86. [DOI] [PubMed] [Google Scholar]
- 60.Trent, M. S., and C. R. H. Raetz. 2002. Cloning of EptA, the lipid A phosphoethanolamine transferase associated with polymyxin resistance. J. Endotoxin Res. 8:158. [Google Scholar]
- 61.Tzeng, Y. L., K. D. Ambrose, S. Zughaier, X. Zhou, Y. K. Miller, W. M. Shafer, and D. S. Stephens. 2005. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J. Bacteriol. 187:5387-5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaara, M., and T. Vaara. 1981. Outer membrane permeability barrier disruption by polymyxin in polymyxin-susceptible and -resistant Salmonella typhimurium. Antimicrob. Agents Chemother. 19:578-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaara, M., and T. Vaara. 1983. Polycations as outer membrane-disorganizing agents. Antimicrob. Agents Chemother. 24:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vaara, M., and T. Vaara. 1983. Polycations sensitize enteric bacteria to antibiotics. Antimicrob. Agents Chemother. 24:107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vinogradov, E., M. B. Perry, and J. W. Conlan. 2002. Structural analysis of Francisella tularensis lipopolysaccharide. Eur. J. Biochem. 269:6112-6118. [DOI] [PubMed] [Google Scholar]
- 66.Wada, A., N. Mori, K. Oishi, H. Hojo, Y. Nakahara, Y. Hamanaka, M. Nagashima, I. Sekine, K. Ogushi, T. Niidome, T. Nagatake, J. Moss, and T. Hirayama. 1999. Induction of human beta-defensin-2 mRNA expression by Helicobacter pylori in human gastric cell line MKN45 cells on cag pathogenicity island. Biochem. Biophys. Res. Commun. 263:770-774. [DOI] [PubMed] [Google Scholar]
- 67.Wang, X., M. J. Karbarz, S. C. McGrath, R. J. Cotter, and C. R. Raetz. 2004. MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: topography of Francisella novicida LpxE expressed in Escherichia coli. J. Biol. Chem. 279:49470-49478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weintraub, A., U. Zahringer, H. W. Wollenweber, U. Seydel, and E. T. Rietschel. 1989. Structural characterization of the lipid A component of Bacteroides fragilis strain NCTC 9343 lipopolysaccharide. Eur. J. Biochem. 183:425-431. [DOI] [PubMed] [Google Scholar]
- 69.Wyrick, P. B., J. Choong, C. H. Davis, S. T. Knight, M. O. Royal, A. S. Maslow, and C. R. Bagnell. 1989. Entry of genital Chlamydia trachomatis into polarized human epithelial cells. Infect. Immun. 57:2378-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou, Z., S. Lin, R. J. Cotter, and C. R. Raetz. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-l-arabinose, phosphoethanolamine and palmitate. J. Biol. Chem. 274:18503-18514. [DOI] [PubMed] [Google Scholar]
- 71.Zhou, Z., A. A. Ribeiro, S. Lin, R. J. Cotter, S. I. Miller, and C. R. Raetz. 2001. Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PMRA-dependent 4-amino-4-deoxy-l-arabinose, and phosphoethanolamine incorporation. J. Biol. Chem. 276:43111-43121. [DOI] [PubMed] [Google Scholar]
- 72.Zhou, Z., K. A. White, A. Polissi, C. Georgopoulos, and C. R. Raetz. 1998. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 273:12466-12475. [DOI] [PubMed] [Google Scholar]