Abstract
CheY-like phosphoacceptor (or receiver [REC]) domain is a common module in a variety of response regulators of the bacterial signal transduction systems. In this work, 4,610 response regulators, encoded in complete genomes of 200 bacterial and archaeal species, were identified and classified by their domain architectures. Previously uncharacterized output domains were analyzed and, in some cases, assigned to known domain families. Transcriptional regulators of the OmpR, NarL, and NtrC families were found to comprise almost 60% of all response regulators; transcriptional regulators with other DNA-binding domains (LytTR, AraC, Spo0A, Fis, YcbB, RpoE, and MerR) account for an additional 6%. The remaining one-third is represented by the stand-alone REC domain (∼14%) and its combinations with a variety of enzymatic (GGDEF, EAL, HD-GYP, CheB, CheC, PP2C, and HisK), RNA-binding (ANTAR and CsrA), protein- or ligand-binding (PAS, GAF, TPR, CAP_ED, and HPt) domains, or newly described domains of unknown function. The diversity of domain architectures and the abundance of alternative domain combinations suggest that fusions between the REC domain and various output domains is a widespread evolutionary mechanism that allows bacterial cells to regulate transcription, enzyme activity, and/or protein-protein interactions in response to environmental challenges. The complete list of response regulators encoded in each of the 200 analyzed genomes is available online at http://www.ncbi.nlm.nih.gov/Complete_Genomes/RRcensus.html.
It has been 20 years since the first bacterial response regulator genes were sequenced (35, 39, 89) and the CheY-like receiver (phosphoacceptor) domain was identified as their common regulatory module (95, 124, 138; see references 55, 57, 127, and 150 for reviews and reference 43 for a historical perspective). Structural characterization of the CheY-like receiver domain (hereafter, the REC domain) from Escherichia coli and other organisms confirmed that it is an autonomously folding, evolutionarily stable, compact structural unit (125, 126, 147) whose conformation undergoes a distinctive change upon phosphorylation (65, 75). These conformational changes, which increase the propensity of the REC domain to form dimers, are used by bacterial and archaeal cells to transmit and propagate a wide variety of environmental and intracellular signals (29, 127, 150).
The typical scheme of bacterial two-component signal transduction involves signal sensing (ligand binding or other conformational change) by a sensory histidine kinase that leads to its autophosphorylation, followed by phosphoryl transfer to the Asp residue in the N-terminal REC domain of the cognate response regulator, which affects the properties of the C-terminal DNA-binding domain (127, 150). Structural characterization of the DNA-binding domains of various response regulators revealed several variations on the common helix-turn-helix (HTH) theme, exemplified by the NarL-type, OmpR-type “winged helix,” Spo0A-type, and Fis-like structures (12, 78, 79, 87, 88, 100). Sequence comparisons revealed additional types of DNA-binding domains in certain response regulators, some of which have been experimentally verified (94, 113).
In addition to forming associations with various DNA-binding domains, the REC domain can function as a stand-alone module. Its propensity to protein-protein interactions plays a key role in the chemotaxis machinery, as well as in some other regulatory pathways (10, 130). It also forms other types of response regulators by associating with certain enzymatic or protein-binding domains, e.g., with the methylesterase (CheB) and chemotaxis modulator (CheW) domains, both of which participate in the adaptation to attractants during chemotaxis (61). In the CheB-type response regulator, the N-terminal REC domain packs against the active site of the C-terminal methylesterase domain and inhibits methylesterase activity by restricting access to the active site (34). The conformational change occurring upon phosphorylation of the REC domain apparently disrupts this interaction and relieves the methylesterase domain from inhibition by REC (34). In the Bacillus subtilis CheV protein, the presence of the N-terminal CheW-like domain was shown to stabilize the phosphorylated state of the C-terminal REC domain (40, 61).
The REC domain can also form combinations with other signaling domains, e.g., in the Caulobacter crescentus PleD response regulator, which controls flagellum ejection and stalk formation during the C. crescentus life cycle (1, 2, 52). In PleD, the N-terminal REC domain is fused to an inactivated REC domain and a C-terminal GGDEF domain, which has diguanylate cyclase activity that produces bis-(3′→5′)-cyclic diguanosine monophosphate (c-di-GMP), a secondary messenger in bacteria (99, 112; see references 58 and 109 for reviews). In Vibrio cholerae response regulator VieA, which controls biofilm formation by this organism (137), the REC domain is associated with the EAL domain, a c-di-GMP-specific phosphodiesterase (22, 30, 114, 131). A combination of the REC domain with another c-di-GMP-specific phosphodiesterase, the HD-GYP domain, controls biosynthesis of extracellular polysaccharide in Xanthomonas campestris (45, 111, 121).
The response regulator sets encoded in E. coli, B. subtilis, Synechococystis sp., Streptococcus pneumoniae, and several other bacteria have been tabulated and classified (48, 51, 74, 90, 91, 97, 146, 148). There have been several attempts to compile comprehensive lists of response regulators in all sequenced microbial genomes (11, 66, 70, 141), as well as to list response regulators in microbial genome databases, such as KEGG, SENTRA, and COG (60, 84, 134). However, the extreme diversity of REC domain combinations encoded in recently sequenced microbial genomes continues to generate problems during genome annotation (43, 157). We have already discussed the most common errors in annotating signaling proteins and suggested that, unless exact function of the newly described protein is known, its annotation should be based on its domain composition, rather than on sequence similarity of any given domain (38, 109). To assist in this process, I present here a comprehensive domain-based classification of the response regulators encoded in completely sequenced genomes of 200 bacterial and archaeal species and describe domain architectures that involve the REC domain and known DNA-binding, enzymatic, and ligand-binding domains, as well as several new domains, not described previously.
MATERIALS AND METHODS
Data sources.
The analyzed set included complete genome sequences of 200 bacterial and archaeal species, sequenced by 20 September 2005, as listed on the NCBI's Entrez Genomes website (151). Only one representative genome per species was chosen, typically the first publicly released one, according to the NCBI Genomes database listing. As an exception, two strains of E. coli, K12 and O157:H7, and three serovars of Salmonella enterica, Paratyphi, Typhi, and Typhimurium, were included in the list, as described earlier (42). The genome sequences were downloaded from the NCBI's Genomes database or searched directly on the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/) using the PSI-BLAST (3) tool.
Identification of REC-containing proteins.
The complete list of REC-containing proteins encoded in each of the 200 analyzed genomes was compiled separately for each bacterial phylum based on the results of iterative PSI-BLAST searches (3) of the NCBI's Reference Sequence (RefSeq) database (103). In each case, the search was initiated with the 122-amino-acid sequence of the Thermotoga maritima CheY protein TM0468 (GenBank accession no. AAD35552; UniProt entry Q9WYT9) and a precomputed position-specific scoring matrix. The query sequence and the position-specific scoring matrix are both available in the supplemental material. The database search space was limited to the given phylum (e.g., Firmicutes[orgn]); sequences of incomplete genomes were excluded using the srcdb_refseq_provisional[prop] modifier. The PSI-BLAST searches used strict inclusion threshold E values of 10−4 to 10−5, adjusting as necessary, and were repeated until convergence. The phylogenetic representation and the total numbers of REC-containing proteins encoded in each given genome were estimated using the Taxonomy Report option in the BLAST output. Potential false-positive hits were checked at every step of PSI-BLAST using the CDD Domain viewer (86) and manually removed (unselected) from the hit list for each iteration of PSI-BLAST. Low-scoring hits (potential false negatives) were inspected on a case-by-case basis by comparing them against the CDD and Pfam domain databases (14, 85) using the CD search tool (86) with relaxed cutoff values.
Classification of REC-containing proteins.
The lists of REC-containing proteins from each complete genome were checked against the CDD, COG, and Pfam domain databases (14, 85, 134), using the CDD Domain viewer (86), and grouped according to their domain composition. The sequences were also matched against UniProt (13), using the UniProt batch retrieval tool, and linked to the corresponding entries in the Pfam database (14). The families of response regulators were named after their experimentally characterized representatives, where available (e.g., OmpR-like and Spo0A-like). In the absence of experimentally characterized representatives, families of response regulators were named based on their domain architectures (e.g., REC-AraC and RpoE-REC). To account for the cases where different species (e.g., three species of Brucella) had merged RefSeq entries, PSI-BLAST searches were also run against selected genomes using the NCBI's Genomic BLAST tool (31), followed by manual analysis of the outputs. Whenever possible, the protein counts were compared to the published data, and discrepancies were resolved on a case-by-case basis. In most instances, discrepancies could be attributed to different treatments of highly diverged and truncated sequences. In this work, such sequences were defined as ones showing up in the PSI-BLAST searches but not recognized as containing the REC domain by RPS-BLAST comparison against CDD, COG, or Pfam databases with default or relaxed (E value of 1.0) parameters. These were typically excluded from the total count but still listed (marked with asterisks) in Table S1 in the supplemental material. All N-terminally truncated sequences were manually inspected to see if the apparent truncation could be due to the incorrect selection of the start codon during automated annotation. Corrected open reading frames for nine genes, for which this proved to be the case (Bacillus cereus BC5352; Bacteroides thetaiotaomicron BT3258; Bradyrhizobium japonicum bsl1713, bsl2179, and bsr2863; Haloarcula marismortui rrnAC1630 and rrnAC3385; and Mesorhizobium loti msl3517 and mlr3700) (see the file RRseqUpdate in the supplemental material), have been submitted to the NCBI RefSeq database. The domain architecture of the Mlr3700 protein is discussed in the Results.
RESULTS
Response regulators encoded in prokaryotic genomes show a great variety of output domains and domain combinations. Unfortunately, owing to the relatively high sequence conservation of the REC domains, sequence database searches for relatives of certain response regulators often retrieve nonhomologous protein sequences that share only the REC domain (43). Hence, classification of response regulators has to be based primarily on their domain architectures and structures of the constituent domains. To achieve proper granularity, HTH domains of the response regulators have been grouped here by sequence similarity, following the approach utilized previously in the COG database (135). Response regulators that contained RNA-binding, protein-binding, or enzymatic output domains have been put into separate categories (Table 1). Any sequence fragments longer than 90 amino acid residues, not assigned to any known protein domain, were analyzed separately and, if appropriate, identified as members of existing or novel domain families. The results of this analysis are shown in Tables 1 and 2.
TABLE 1.
Widespread types of bacterial response regulatorsa
| Response regulator
|
Output domain
|
|||||
|---|---|---|---|---|---|---|
| Typeb | COG no. | Size (aa) | No. in the set (% of total) | Name | Pfam entry | Reference(s) |
| Stand-alone REC | 0784 | 120 | 659 (14.3%) | REC | PF00072 | 125, 146 |
| DNA-binding | ||||||
| OmpR-like | 0745 | 240 | 1526 (33.1%) | wHTH | PF00486 | 88 |
| NarL-like | 2197 | 240 | 862 (18.7%) | HTH | PF00196 | 12 |
| NtrC-like | 2204 | 450 | 393 (8.5%) | AAA-FIS | PF00158 PF02954 | 76, 95 |
| LytR-like | 3279c | 250 | 139 (3.0%) | LytTR | PF04397 | 94 |
| PrrA-like | 4567 | 170 | 48 (1.0%) | FIS | PF02954 | 73 |
| REC-AraC (YesN-like) | 4753 | 260 | 48 (1.0%) | HTH_AraC | PF00165 | 106 |
| RNA-binding, AmiR/NasR-like | 3707 | 180 | 40 (0.9%) | ANTAR | PF03861 | 119 |
| Enzymatic | ||||||
| Methylesterase (CheB-like) | 2201 | 300 | 107 (2.3%) | CheB | PF01339 | 34 |
| Diguanylate cyclase (PleD-like) | 3706 | 310 | 154 (3.4%)d | GGDEF | PF00990 | 28 |
| c-di-GMP phosphodiesterase (VieA-like) | 2200c | 370 | 83 (1.8%)d | EAL | PF00563 | 46 |
| c-di-GMP phosphodiesterase (RpfG-like) | 3437 | 360 | 72 (1.6%) | HD-GYP | 45, 111 | |
| Protein phosphatase (RsbU-like) | 2208 | 350 | 46 (1.0%) | PP2C (SpoIIE) | PF07228 | 116 |
| Histidine kinase | 0642 | 350 | 88 (1.9%) | HisKA-HATPase | PF00512 PF02518 | 19 |
| Protein-binding | ||||||
| CheW-REC (CheV-like) | 0835c | 300 | 72 (1.6%) | CheW | PF01584 | 110 |
The table lists response regulators with at least 40 representatives in the analyzed set, found in at least two bacterial phyla.
Description of the domain structure, if available, or sequence alignment.
This COG contains only the output domain of this response regulator.
Includes 50 response regulators that contain both GGDEF and EAL domains.
TABLE 2.
Response regulators with unusual domain organizations
| Family name, domain organization | Total no. | Example
|
Output domain
|
|||
|---|---|---|---|---|---|---|
| Organism name | Gene name (accession no.) | Pfam entry | COG no. | Reference(s) | ||
| DNA-binding, REC-HTH | ||||||
| Spo0A-like | 15 | Bacillus subtilis | BSU24220 (CAB14353) | PB005154 | 78 | |
| YcbB (GlnL)-like | 14 | Bacillus subtilis | BSU02450 (CAB12039) | PF08664 | 113 | |
| REC-SARP (REC-HTH-BTAD) | 5 | Deinococcus radiodurans | DR2556 (AAF12095) | PF00486 PF03704 | 3947 | 152, 154 |
| REC-CRP | 2 | Gloeobacter violaceus | glr1768 (BAC89709) | PF00325 | 0664 | 49 |
| DNA-binding, HTH-REC | ||||||
| RpoE-REC | 15 | Caulobacter crescentus | CC3477 (AAK25439) | PF04542 PF04545 | 1595 | 26 |
| MerR-REC | 6 | Treponema denticola | TDE0855 (AAS11346) | PF00376 | 2452 | 25 |
| BetR-REC | 5 | Chromobacterium violaceum | CV0537 (AAQ58214) | PF08667 | This work | |
| AlpA-REC | 2 | Dehalococcoides ethenogenes | DET1294 (AAW39407) | PF05930 | 3311 | 139 |
| HlxR-REC | 1 | Haloarcula marismortui | rrnB0301 (AAV48466) | PF01638 | 1733 | 155 |
| LysR-REC | 1 | Silicibacter pomeroyi | SPO3037 (AAV96273) | PF00126 | 0583 | 93 |
| RNA-binding, CsrA-REC | 1 | Rhodopirellula baltica | RB8820 (CAD75938) | PF02599 | 50 | |
| Enzymatic | ||||||
| Ser/Thr protein kinase REC-RsbW or REC-RsbU-RsbW | 14 | Leptospira interrogans | LA2951 (AAN50149) | 2172 | 153 | |
| Phosphoaspartate phosphatase REC-CheC | 13 | Vibrio cholerae | VCA0189 (AAF96102) | PF04509 | 1776 | 98, 129 |
| Adenylate cyclase REC-ACyc | 6 | Bradyrhizobium japonicum | Blr2288 (BAC47553) | PF00211 | 2114 | 156 |
| ParA family ATPase REC-Soj | 5 | Ralstonia solanacearum | RSc0653 (CAD14183) | PF00991 | 0455 | 77 |
| NAD(P)H:disulfide oxidoreductase REC-TrxB | 3 | Nocardia farcinica | Nfa52850 (BAD60137) | PF00070 | 2345 | 72 |
| Poly-N-acetylglucos amine deacetylase REC-LmbE | 1 | Leifsonia xyli | Lxx09320 (AAT88820) | PF02585 | 2120 | 132 |
| Glycosyltransferase REC-WcaA | 1 | Methanobacterium thermoauto-trophicum | MTH548 (AAB85054) | PF00535 | 1215 | 133 |
| PilB-like ATPase REC-PulE | 2 | Dechloromonas aromatica | Daro_0966 (AAZ45722) | PF00437 | 2804 | 102 |
| Predicted phosphatase REC-PglZ | 1 | Porphyromonas gingivalis | PG0928 (AAQ66063) | PF08665 | This work | |
| Predicted phosphodiesterase REC-HDOD | 4 | Vibrio cholerae | VC1081 (AAF94240) | PF08668 | 1639 | This work |
| Ser/Thr protein kinase REC-STYK | 1 | Rhodopirellula baltica | RB10329 (CAD78826) | PF00069 | 0515 | 117 |
| Protein-protein interaction | ||||||
| REC-TPR (VieB-like) | 20 | Desulfovibrio vulgaris | DVU2937 (AAS97409) | PF00515 | 0457 | 21 |
| Hnr-like | 18 | Escherichia coli | RssB (AAC74317) | 15 | ||
| REC-HPt | 9 | Xylella fastidiosa | XF2359 (AAF85158) | PF01627 | 2198 | 63 |
| REC-PAS | 4 | Haloarcula marismortui | RrnAC0674 (AAV45676) | PF01627 | 2202 | 136 |
| REC-GAF | 4 | Geobacter sulfurreducens | GSU1316 (AAR34692) | PF01590 | 2203 | 54 |
| Ligand-binding | ||||||
| REC-PilZ | 2 | Geobacter sulfurreducens | GSU0877 (AAR34207) | PF07238 | 3215 | 4 |
| REC-cNMPbinding | 2 | Bdellovibrio bacteriovorus | Bd3065 (CAE80821) | PF00027 | 0664 | 33 |
| Uncharacterized | ||||||
| PatA-type | 21 | Nostoc sp. PCC7120 | all0521 (BAB72479) | 80, 83 | ||
| Ycf55-type | 7 | Nostoc sp. PCC7120 | alr0960 (BAB72917) | This work | ||
| REC-HalX | 9 | Halobacterium sp. NRC-1 | VNG2036G (AAG20197) | PF08663 | This work | |
| OsaB-type | 2 | Streptomyces coelicolor | SCO5749 (CAA19850) | 20 | ||
Response regulators with DNA-binding output domains.
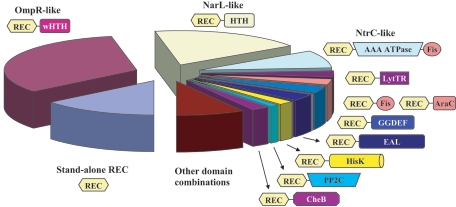
The great majority of all response regulators encoded in complete prokaryotic genomes combines the REC domain with a DNA binding HTH domain, typically either of NarL-type or of OmpR-type winged HTH family (Fig. 1). These domains are well studied and have known three-dimensional structures (12, 107). A factor of inversion stimulation (Fis)-type DNA binding domain (71, 73) is also commonly found in response regulators, either directly fused to the REC domain or containing an additional AAA+-type ATPase domain (Fig. 1). Response regulators of the latter type are referred to here as the NtrC family, after its best-studied representative (65, 76, 95, 145). The former type of domain architecture was first described in the RegA and PrrA response regulators from Rhodobacter sphaeroides (36, 73, 101, 115) and is referred to here as the PrrA family. Other widespread DNA-binding domains in response regulators include the AraC-type HTH domain (106) and the LytTR domain (94), whose structure is still unknown (Table 1).
FIG. 1.
Domain architectures of the most common types of bacterial response regulators. The sectors in the chart indicate the respective share of each domain combination and are colored similarly to the corresponding output domains. The sector marked as REC-Fis REC-AraC represents all rare DNA-binding response regulators listed in Table 2.
This work revealed a number of additional combinations of the REC domain with previously described DNA-binding domains, including cases where the HTH domain occupies the N-terminal part of the protein (Table 2). In some instances, newly identified output domains could be linked to known HTH domains by PSI-BLAST or CD searches. Thus, a previously uncharacterized N-terminal output domain in response regulators of the CV0537 family, encoded in Chromobacterium violaceum and Burkholderia spp. (beta-proteobacterial transcriptional regulator, BetR, deposited in the Pfam database as domain PF08667), was shown to be related to the XRE-type HTH domain (PF01381), found in Cro and cI repressors (see Fig. S1 in the supplemental material). In some cases, however, response regulators that were originally assumed to be DNA-binding showed no statistically significant similarity to any known DNA-binding domain. For example, nine haloarchaeal response regulators annotated as Hlx1 through Hlx6 (Table 2) contain a new output domain (renamed here HalX) (see Fig. S2 in the supplemental material), whose similarity, if any, to the homeodomain-containing animal Hlx proteins is totally spurious. All such response regulators have been classified as “uncharacterized.” Most of these newly described domain combinations are relatively rare and have a narrow phylogenetic distribution (Table 2).
Response regulators with RNA-binding output domains.
Of all the numerous RNA-binding domains described in the past several years (7), only one, ANTAR (119), has been commonly found in response regulators (Table 1). In transcriptional regulators of the AmiR and NasT type, this domain stimulates transcription by preventing transcription termination at rho-independent terminators (96, 119). A single instance of a response regulator formed by another RNA-binding domain, a fusion of the REC domain with an N-terminal CsrA domain (50, 108), was found in Rhodopirellula baltica protein RB8820 (Table 2).
Response regulators with enzymatic output domains.
Recognition of the REC domain as a universal regulatory domain was prompted by sequencing of the chemotaxis protein CheB, which combines an N-terminal REC domain with a C-terminal methylesterase domain. The latter domain retained catalytic activity in the absence of REC or when the REC domain was phosphorylated (81, 120, 124). Subsequent studies revealed response regulators that contain the REC domain in association with other enzymatic domains. PleD-like response regulators (1, 2, 52) have a GGDEF-type output domain, which has diguanylate cyclase activity (28, 112), whereas VieA-type response regulators combine the REC domain with the EAL domain, which has c-di-GMP-specific phosphodiesterase activity, and a DNA-binding domain (131, 137). Counting the response regulators containing any of these domains shows that each of them comprises more than 1% of the total set and can be found in various phylogenetic lineages (Fig. 1 and Table 1).
Two more common enzymatic output domains in response regulators are the protein phosphatase of PP2C type (RsbU-like) and the phosphodiesterase HD-GYP, a variant of the widespread HD-type phosphohydrolase (PF01966). The former domain dephosphorylates Ser∼P and Thr∼P residues in a variety of proteins (64, 116) and is often linked to the regulation of RNA polymerase sigma factors (143, 153). The HD-GYP domain has been proposed to function as a c-di-GMP hydrolase, based on its peculiar phyletic pattern, complementing that of the EAL domain (45, 46). Although this suggestion was consistent with participation of the HD-GYP-containing response regulator RpfG in regulation of extracellular polysaccharide production in Xanthomonas campestris (121) and with the wide distribution of the RpfG-type response regulators (Table 1), it has long remained unverified. Recently, the c-di-GMP-specific phosphodiesterase activity of the HD-GYP domain in RpfG has been demonstrated in a direct biochemical assay (111).
Response regulators whose output domain is a protein phosphatase of CheC type, which dephosphorylates the Asp∼P residue of REC (98, 129), are far less common and found mostly in gamma-proteobacteria. The PglZ domain, found in response regulators from Porphyromonas gingivalis and Cytophaga hutchinsonii, is another likely phosphatase. The pglZ (phage growth limitation) gene of Streptomyces coelicolor A3 (2) is part of a four-gene operon that somehow controls phage propagation in this organism (128). Its homologs, which include Salmonella enterica serovar Typhimurium protein STM4492, are often encoded on integrative plasmids (23). While its exact function is unknown, the PglZ protein and its close homologs have all the hallmarks of the alkaline phosphatase protein superfamily (see Fig. S3 in the supplemental material). Given that members of this superfamily typically function as phosphatases, phosphomutases, or phosphodiesterases (44), PglZ is likely to have a similar enzymatic activity.
Hybrid histidine kinases and REC-HisK response regulators.
In addition to response regulators, the REC domain is often found at the C terminus of sensory histidine kinases, which are then referred to as “hybrid” kinases. The search method used in this work retrieved hybrid histidine kinases as well as response regulators; these were subsequently separated based on their domain architectures. In E. coli, five histidine kinases (ArcB, BarA, EvgS, RcsC, and TorS) out of 30 are hybrid; in B. subtilis, there are no hybrid kinases with the exception of the YwpD protein that combines a truncated REC domain with a HisK domain (for simplicity, two separate subdomains of histidine kinases, the dimerization/phosphoacceptor one, HisKA, represented by Pfam entries PF00512, PF07568, PF07730, and PF02518, and the catalytic one, HATPase_c or PF02518, are treated here as a single HisK domain). However, the YwpD protein differs from typical hybrid kinases in that its REC domain is located at the N terminus, and there is no (other) sensory domain (Fig. 2). This means that the only signal that could be sensed by the YwpD protein is the phosphorylation (or other modification) of its N-terminal REC domain. Therefore, YwpD-like histidine kinases can be considered yet another form of response regulators with enzymatic output domains whose activity is modulated by appropriate sensory histidine kinases. Alternatively, the REC and HisK domains of such proteins might not be regulated at all and function solely as sinks for the phosphoryl residues. In any case, similarly to the response regulators of the PleD, VieA, and RpfG families, YwpD-like histidine kinases are members of complex signal transduction cascades operating inside the bacterial cell (41).
FIG. 2.
Domain architectures of response regulators with the histidine kinase (HisK) output domain. Domain symbols are drawn approximately to scale. Some of the domains are highly diverged and could be recognized only by searching the CDD database using relaxed stringency parameters. Source organisms, gene names, and NCBI protein database accession numbers for the domain architectures shown above are as follows (domain architecture number and accession number) B. subtilis ywpD (1; CAB05945) or Nostoc sp. PCC 7120 all3764 (1; BAB75463); Nostoc sp. PCC 7120 all1279 (2; BAB73236), alr1968 (16; BAB73667), and all4097 (17; BAB75796); Bradyrhizobium japonicum bll1030 (3; BAC46295); Haloarcula marismortui rrnAC0574 (4; AAV45588), rrnAC3379 (5; AAV48063), rrnAC2533 (6; AAV47335), rrnAC0075 (7; AAV45155), rrnAC0848 (8; AAV45829), rrnAC1626 (9; AAV46539), rrnAC3361 (10; AAV48050), rrnAC1495 (12; AAV46418), and pNG7156 (13; AAV44864); Synechocystis sp. PCC 6803 slr0222 (11; BAA10222); Anabaena variabilis Ava_B0191 (14; ABA24903); Gloeobacter violaceus gll0634 (15; BAC88575); Desulfovibrio vulgaris DVU2677 (18; AAS97149); Dechloromonas aromatica Daro_2847 (19; AAZ47577). Proteins with domain architecture numbers 1, 14, and 18 were considered bona fide response regulators.
Unfortunately, the separation between YwpD-like response regulators and hybrid histidine kinases is not clear-cut. Genome analysis reveals numerous intermediate forms with additional signaling domains, typically PAS and/or GAF, which could affect the enzymatic activity of the output domain (Fig. 2). For the purposes of this work, only YwpD-like histidine kinases without any discernible signaling domains intervening between REC and HisK were counted as bona fide response regulators.
The search for REC-containing proteins also revealed hybrid histidine kinases which, in addition to the REC domains, contain DNA-binding domains at their C-termini. These “one-component” signal transduction systems (141), exemplified by B. cereus protein BC3207, look like fusions of classical histidine kinases with full-length response regulators. Such proteins are encoded in a variety of bacterial genomes, sometimes in significant numbers. In Bacteroides thetaiotaomicron, for example, 36 out of 44 hybrid histidine kinases have tandem AraC-like DNA-binding domains at their C-termini (species-specific listings of hybrid histidine kinases are available in the files that are linked to Table S1 in the supplemental material). In Bacteroides fragilis strain YCH46, this domain organization is seen in 11 out of 17 hybrid histidine kinases. Some of such fusion proteins contain periplasmic, extracellular, or integral membrane sensory domains, so that they can only act as transcriptional regulators while their binding sites on the chromosomal DNA are positioned in the vicinity of the membrane.
Response regulators with protein-binding domains.
In some response regulators, output domains are devoid of (known) enzymatic activity and exert their effects through protein-protein interactions. These include chemotaxis response regulators of the CheV type, combining REC with the protein-binding CheW-like domain (Table 1), and response regulators of the Hnr-type (see below), as well as response regulators that contain PAS, GAF, TPR, or HPt domains as the only output domains (Table 2). While PAS and GAF domains are known to bind ligands, signal transduction likely involves their interactions with other protein domains, not ligand binding per se. The same could be true for other ligand binding domains, such as CAP_ED, cNMP-binding, and PilZ domains (Table 2).
Clade-specific response regulators.
While response regulators of OmpR, NarL, and many other families are widespread in Bacteria, some response regulators are found only in representatives of certain phylogenetic lineages. Some of them can be considered true synapomorphies—shared derived characteristics of the representatives of a particular group to the exclusion of other groups—whereas others are encoded only in organisms with larger genomes and could have been lost in streamlined genomes of parasites and other small-genome organisms.
Spo0A.
The master regulator of sporulation, Spo0A, is shared by all spore-forming representatives of Firmicutes, namely, the bacteria belonging to the orders Bacillales, Clostridiales, and Thermoanaerobacteriales (122, 123). Remarkably, in the genome sequence of Clostridium perfringens strain 13, the spo0A gene is inactivated by what authors claim is a verified frameshift (118). If so, the sequenced strain should be unable to sporulate and produce toxins, which would make it a convenient laboratory strain but hardly a representative of the naturally occurring clostridia. In any case, Spo0A combines the REC domain with an unusual HTH domain that has additional α-helices (78). Outside Firmicutes, this HTH domain has been seen so far only in the spore-forming high-GC gram-positive bacterium Symbiobacterium thermophilum, which is currently assigned to the Actinobacteria but is apparently a member of the Firmicutes (140). Thus, Spo0A is a typical synapomorphy, common to the spore-forming bacteria of the Firmicute lineage.
YcbB.
B. subtilis response regulator YcbB has been originally described as one of the genes whose disruption increased cell resistance to protonophores (105). This effect remained enigmatic until a recent study showed that YcbB controls the glutamine utilization operon (113). YcbB has been renamed GlnL, which is bound to cause confusion, as it is unrelated to the E. coli GlnL (NtrC) response regulator. In fact, the C-terminal domain of YcbB does not show obvious relation to any previously described HTH domain (see Fig. S4 in the supplemental material). Since DNA binding by YcbB has now been experimentally demonstrated (113), its C-terminal domain can be considered a new type of the DNA-binding HTH domain.
PatA.
Response regulators of the PatA type, which control heterocyst development, phototaxis, and chromatic adaptation in cyanobacteria (18, 80), combine the C-terminal REC domain with N-terminal PATAN (PatA N terminus) domain and an HTH domain in the middle (83). Although the PATAN domain itself is found in a variety of bacteria, the PATAN-REC combination appears to be limited to cyanobacteria and δ-proteobacteria (83).
RpoE-REC.
Rhizobia, brucellae, bartonellae, and other members of the alpha-subdivision of proteobacteria encode an unusual response regulator that combines a C-terminal REC domain with an N-terminal sigma-24 subunit of RNA-polymerase (σE or RpoE) (Table 2). With the sole exception of the marine bacterium Silicibacter pomeroyi, this response regulator is encoded in every alpha-proteobacterial genome that is larger than 1.6 Mb, often adjacent to the gene coding for a histidine kinase of the recently described HWE family (62). Sequence comparisons identified the RpoE-REC domain combination in the genome of Mesorhizobium loti (the mlr3700 gene product), where the original genome annotation included only its C-terminal REC domain. Response regulators with the RpoE-REC domain combination have not been described in the available literature, and the only indication of their biological function(s) so far is the observation of increased expression of the C. crescentus response regulator CC3477 in the minimal medium compared to the rich medium (56). Given the DNA-binding properties of σE, these proteins can be predicted to serve as environmentally modulated transcriptional regulators. It remains to be seen whether their RpoE-like components could function as genuine sigma subunits of the RNA polymerase.
RssB.
The Hnr-type response regulator, also referred to as RssB or SprE, regulates turnover of the stress sigma factor RpoS in E. coli, Salmonella, and several other enterobacteria (53). Due to the high degree of sequence conservation and narrow phyletic distribution, its output domain was previously considered to be unique (92). However, sequence analysis showed that this output domain is a degraded version of the PP2C-type Ser/Thr protein phosphatase that is shorter than the typical PP2C domain, has lost some of its Mn2+-binding Asp residues (Fig. 3), and is apparently devoid of the protein phosphatase activity. These observations are in line with the earlier suggestion (15) that RssB is a former anti-sigma factor that during evolution was recruited to serve as a recognition factor for proteolysis. However, RssB has apparently evolved from a protein phosphatase, not a protein kinase (an anti-sigma). As noted previously (69), phylogenetic distribution of this response regulator is limited to the gamma-proteobacterial families Enterobacteriaceae and Vibrionaceae, indicating that Hnr-dependent regulation of RpoS turnover is a peculiar feature of E. coli and its closest relatives. In pseudomonads, the degradation of the PP2C domain is much less pronounced, the key metal-binding residues are conserved, and the Hnr-related response regulators are likely to have the protein phosphatase activity (Fig. 3).
FIG. 3.
Multiple sequence alignment of the Hnr-type output domain and its comparison with the protein phosphatase domains of the PP2C family. The top sequence is that of E. coli protein RssB (GenBank accession no. AAC74317). Conserved hydrophobic amino acid residues are shaded yellow; other conserved residues are shown in bold typeface. Metal-binding residues of PP2C-type protein phosphatases are shown in white letters on dark background. The numbers show the positions of aligned residues and the lengths of gaps between them. The numbers in parentheses signify the total length of each protein. The overlap between the secondary structure prediction by PSIPRED (59) and the known structures of PP2C-type protein phosphatases (32, 104) are shown above the alignment; cylinders indicate the α-helices, and arrows indicate the β-strands.
REC-HDOD.
Several proteobacteria, including Vibrio cholerae and Pseudomonas aeruginosa, encode a response regulator with an unusual output domain, assigned to the COG1639 (41). In addition, this output domain is often encoded in the genomes of the same bacteria in a stand-alone form (see Fig. S5 in the supplemental material). The structure of one such protein, Campylobacter jejuni CJ0248, has been solved by the Joint Center for structural Genomics and deposited in the Protein Data Bank under accession code 1VQR. Sequence and structural analysis of this output domain (deposited in the Pfam database as domain PF08668) showed its distant relationship to the widespread HD phosphohydrolase (PF01966) domain (9). This domain has been named the HD-related output domain, or HDOD, but the lack of conservation of the key metal-binding residues of the typical HD domain did not allow us to predict whether it still has a phosphatase or phosphodiesterase activity and, accordingly, what its role is, if any, in the signal transduction phosphorelay.
Other response regulators.
Comparative analysis revealed several other clade-specific response regulators that are found exclusively in cyanobacteria (for example, the Slr2024 family), halobacteria (REC-HalX family), alpha-proteobacteria (CC0440 family), or epsilon-proteobacteria (CJ1608 family). A cyanobacteria-specific response regulator, Sll1879, combines the REC-domain with the Ycf55 domain, found in a stand-alone form in chloroplasts of green plants and red algae. No mention of experimental studies of these response regulators could be found in the available literature, and their functions remain unknown.
Avoidance of certain response regulator types.
The availability of a complete genome sequence allows one to determine not only which proteins are encoded there but also which proteins (protein families) are missing. Absence of certain types of response regulators in all representatives of a given phylum could be an important feature of that phylum that might help in understanding mechanisms of signal transduction in its members.
Actinobacteria, for example, encode numerous transcriptional regulators of the SARP family (152), which combine an HTH domain with the BTAD domain, whose function is unknown (154). However, the current set of 390 actinobacterial response regulators does not include a single instance of a REC-SARP domain combination. The reasons for that remain unclear; other organisms, such as Deinococcus radiodurans and Clostridium acetobutylicum, do encode response regulators with this domain combination (Table 2). None of the available actinobacterial genomes encodes response regulators of the NtrC type, which is consistent with the absence of σ54 in these genomes. The only exception is Symbiobacterium thermophilum, which is not really an actinobacterium (140) (see above). As noted earlier (42), not even the largest actinobacterial genomes encode any chemotaxis proteins; this includes the absence of the CheB, CheC, and CheV family proteins, as well as a relatively low number of stand-alone REC domains. Although actinobacteria encode signaling proteins with HisK, GGDEF, EAL, HD-GYP, and ACyc output domains, none of them is found in actinobacterial response regulators. Finally, only a small fraction of actinobacterial histidine kinases are hybrids. These features illuminate the peculiarity of the signal transduction in actinobacteria and suggest that representatives of this phylum utilize a rather straightforward mechanism of signal transduction that includes tight pairing of each HisK with a cognate DNA-binding (or RNA-binding) response regulator.
Archaeal transcriptional regulators and signal transducers are generally similar to those in bacteria (16, 47). However, archaeal response regulators differ dramatically from bacterial ones: no sequenced archaeal genome encodes OmpR, NarL, NtrC, LytR, or PrrA family response regulators (see Table S1 in the supplemental material; see also reference 141). The only response regulators that are common for bacteria and archaea are the stand-alone REC domain and CheB-like and REC-HisK combinations. Other archaeal response regulators include REC-PAS and REC-GAF domain combinations, as well as combinations of the REC domain with uncharacterized lineage-specific domains, found, for example, only in halobacteria or only in metanosarcinas.
Statistics of response regulators.
The number of response regulators encoded in each complete microbial genome shows strong positive correlation with the genome size. In the logarithmic coordinates, this dependence shows a slope of approximately 1.8, meaning that it is close to the quadratic function (Fig. 4) (see also reference 141). There are certain deviations from the general trend: some representatives of beta-, delta- and epsilon-proteobacteria, such as Geobacter sulfurreducens, Wolinella succinogenes, and Thiobacillus denitrificans, encode substantially more response regulators than other organisms with the same genome sizes. At the same time, other organisms encode relatively few response regulators or none at all (e.g., Sulfolobus spp.) (Fig. 4).
FIG. 4.
Correlation between the genome size and total number of encoded response regulators. Each symbol indicates the logarithm of the number of response regulators encoded in a single bacterial genome with the following color scheme: actinobacteria, black circles; cyanobacteria, cyan circles; alpha-proteobacteria, brown diamonds; beta-, delta-, and epsilon-proteobacteria, yellow diamonds; gamma-proteobacteria, blue squares; firmicutes, open squares; members of other bacterial phyla (Aquificales, Bacteroidetes, Chlamydiae, Deinococcus-Thermus, Planctomycetes, Spirochetes, and Thermotogales), red triangles. Archaeal response regulators are indicated by yellow triangles. Organisms that encode no response regulators are indicated by symbols on the x axis (logN = 0); these, however, were ignored for the calculation of the slope and the correlation coefficient of the regression line. The numbers in the circles indicate the data points corresponding to the following organisms: 1, Dechloromonas aromatica; 2, Geobacter sulfurreducens; 3, Wolinella succinogenes; 4, Dehalococcoides sp. strain CBDB1; 5, Sulfolobus solfataricus.
DISCUSSION
The sequence analysis of REC domain-containing response regulators presented in this work shows that functions of these proteins are not limited to transcriptional regulation and chemotaxis. While transcriptional regulators with DNA-binding output domains comprise about two-thirds of all response regulators, the remaining third consists of stand-alone REC domains, response regulators with enzymatic or protein-binding output domains, and response regulators whose output domains have not been characterized and whose functions remain unknown. In a somewhat similar analysis by Ulrich and colleagues (141), stand-alone REC domains and response regulators with unknown output domains were not considered, resulting in a much higher fraction of DNA-binding response regulators in their set.
Mechanisms of REC-mediated signaling.
Stand-alone REC domains, such as chemotaxis regulator CheY and sporulation regulator Spo0F, have two principal modes of transmitting regulatory signals. Both depend on protein-protein interactions of the phosphorylated form REC∼P with its various targets but differ in the fate of the phosphoryl group, which can stay on the REC domain or be transferred to another protein with REC∼P functioning as an intermediate in a multicomponent phosphorelay (37). In some cases, REC∼P might function just as a sink for phosphoryl groups (10, 130). In most bacteria, chemotactic signaling by CheY involves direct binding of its phosphorylated form, CheY∼P, to the flagellar switch protein, FliM (24, 149). In archaea, which have an entirely different flagellar machinery that does not include FliM, CheY-like proteins interact with a different, still unidentified, protein (130). In cyanobacteria, whose motility apparatus is related to the type IV pili, CheY∼P interacts with yet another type of switch complex (18). This is also the case for type IV pili-mediated social motility of Myxococcus xanthus (144). In addition to their role in chemotaxis, stand-alone REC domains participate in developmental processes, including regulation of sporulation in B. subtilis (Spo0F), heterocyst formation in Nostoc sp. (DevR), and cyst cell development in Rhodospirillum centenum (17, 27, 37). Furthermore, separately expressed REC and methylesterase domains of the CheB protein have been shown to form a complex even in the absence of a linker (6). These data suggest that REC domains could participate in a variety of protein-protein interactions with yet unknown target proteins.
Another big group of response regulators combines the REC domain with various enzymatic domains that are involved in signal transduction. Besides chemotactic methylesterase CheB and protein phosphatase CheC, these include the diguanylate cyclase (GGDEF), c-di-GMP phosphodiesterase (EAL and HD-GYP), adenylate cyclase, Ser/Thr-specific protein phosphatase, and histidine kinase output domains. In some cases, phosphorylation of the REC domain has been shown to activate the attached enzymatic domains (34, 99). While this activation could be due to the relief of enzyme inhibition by nonphosphorylated REC (34), it appears that in the case of CheB, both relief of inhibition and genuine activation take place (5, 6).
Another mechanism of REC-mediated signaling is the well-documented propensity of phosphorylated REC domains to form dimers. Phosphorylation-induced dimerization plays a key role in the regulation of DNA-binding response regulators, dramatically improving their binding to the palindromic or direct-repeat chromosomal binding sites. This mechanism has been demonstrated for the response regulators of the OmpR, NarL, LytR, and PrrA families (12, 73, 88, 94). In the response regulators of the NtrC family, all three domains appear to participate in oligomerization, which regulates the activity of its AAA+ ATPase domain (67, 76).
The interplay of dimerization and relief-of-inhibition mechanisms could be particularly important for response regulators involved in c-di-GMP turnover. In response regulators of the REC-GGDEF type, phosphorylation of the REC domain activates the diguanylate cyclase by enabling formation of a GGDEF dimer, the active form of the enzyme (99, 112). In contrast, the c-di-GMP-specific phosphodiesterase (the EAL domain) is active as a monomer (114). Therefore, activation of EAL phosphodiesterase by REC phosphorylation in response regulators of the REC-EAL type (131, 137) appears to be due to the relief-of-inhibition mechanism. This distinction is particularly important for the response regulators of REC-GGDEF-EAL domain architecture, which comprise 1.1% of all response regulators in the analyzed set. Although in many such proteins either the GGDEF or EAL domain appears to be inactivated (114), there are some without obvious deviations from the consensus (109). Such response regulators should have the only the phosphodiesterase activity in their monomeric form but might acquire diguanylate cyclase activity upon dimerization. It is entirely possible that upon phosphorylation of their REC domains, the overall activity of such response regulators changes from c-di-GMP-hydrolase to c-di-GMP synthetase and vice versa.
In addition to dimerization, relief-of-inhibition, direct activation, and protein-protein interactions with or without phosphoryl transfer, REC-mediated signaling might involve other mechanisms, such as, for example, allosteric regulation of the output domains. This versatility of the REC domain is consistent with the diversity of signal output domains that interact with it.
New types of response regulators.
The same regulatory mechanisms mentioned above—dimerization, protein-protein interaction, and activation/relief-of-inhibition—can be expected to work in the newly described response regulators. Among the new types of DNA-binding response regulators (Table 2), DNA-binding domains of LysR, MerR, and HxlR types are known to function as dimers (25, 93, 155), as is the RNA-binding CsrA domain (50). DNA-binding domains of the XRE-family, which includes cI and Cro regulators, also function as dimers, suggesting that the BetR domain, a newly described distant member of this family, would function the same way. However, the RpoE-like domain of the CC3477-type (RpoE-REC) response regulators is not known to form dimers, so they most likely act through relief of inhibition and/or protein-protein interactions of their C-terminal REC domains.
Among enzymatic output domains of response regulators (Tables 1 and 2), adenylate and diguanylate cyclases, histidine kinases, and RsbW-type Ser/Thr protein kinases (anti-sigma factors) are known to function as dimers (19, 28, 82, 156). In these response regulators, both phosphorylation-induced dimerization of the REC domains and relief-of-inhibition could contribute to the activity of the output domains. In all other cases, a nonphosphorylated REC domain can be expected to block enzyme activity the same way it does that in the CheB structure (34). However, just as in CheB, a direct enzyme activation by REC∼P, as well as an allosteric regulation, also remains a possibility.
Phylogenetic distribution of response regulators.
The census of response regulators confirmed several trends seen previously in the census of signal transduction proteins (42). In accordance with previous observations (46, 68, 141, 142), the total number of response regulators encoded in completely sequenced prokaryotic genomes positively correlates with the genome size (Fig. 4) and the total number of encoded proteins. The largest deviation from the general trend is seen in the same representatives of beta-, delta-, and epsilon-proteobacteria that have been previously found to encode more signaling proteins than others with the same genome size (42). Given only a minimal overlap between the two data sets, these observations show that certain environmental bacteria encode a disproportionately higher number of sensor, signal transduction, and response regulator proteins than other prokaryotes with the same genome size. On the other end of the spectrum are bacteria and archaea that encode fewer response regulators than the average bacterium of the same genome size. These are typically obligate parasites, such as Yersinia pestis, or organisms such as Sulfolobus spp. or Thermoplasma spp. that inhabit relatively stable and unique environments where few parameters, if any, ever change. These data validate the use of the term “bacterial IQ” (42) to describe the fraction of the bacterial proteome dedicated to environmental sensing and signal transduction.
Origin and function of diverse domain architectures.
The diversity of output domains in response regulators (Tables 1 and 2) suggests that virtually any DNA-binding or enzymatic domain could be fused to the REC domain to form a response regulator. For DNA-binding domains, this is probably correct, as representatives of all major classes of the HTH domain (8) could be found in response regulators (Table 2). As discussed above, the propensity of the REC domain to dimerize upon phosphorylation offers an effective mechanism of transcriptional regulation. Remarkably, HTH domains such as LysR or MerR that are typically localized at the N-termini of the corresponding transcriptional regulators have the same position in response regulators (Table 2), a clear deviation from the standard REC-HTH architecture. This diversity makes it even more difficult to explain the predominance of OmpR-type and NarL-type response regulators, far fewer numbers of the LytR-type, PrrA-type, and YesN-type response regulators, and only sporadic appearance of the response regulators with other types of the HTH domain (Fig. 1 and Tables 1 and 2).
Most enzymatic output domains found in response regulators (Tables 1 and 2) are signal transduction domains themselves, often found in transmembrane receptors (42). Such fusions add yet another layer of regulation in the complex intracellular signal transduction network, allowing modulation of the cellular levels of cAMP and c-di-GMP, as well as the activity of Ser/Thr protein kinases and phosphatases, by sensory histidine kinases in response to specific environmental cues (41). Fusions with other enzymes typically have a narrow phylogenetic distribution, and it is not known if they are ever expressed. Such combinations must have arisen from random gene translocation and fusion events and were later fixed in evolution because they allowed tight regulation of the enzyme activity. In one such case, a fusion of the REC and thioredoxin reductase domains could play an important role in environmental regulation of the cellular dithiol-disulfide ratio. This domain combination has been found in several actinobacteria and cyanobacteria and in Solibacter usitatus, a recently sequenced representative of the phylum Acidobacteria. Plant pathogen Leifsonia xyli encodes a unique response regulator with the LmbE output domain, recently shown to have N-acetylglucosamine deacetylase activity (Table 2). Such domain combination could allow this bacterium to regulate degradation of N-acetylglucosamine-containing plant polymers, such as lignin and chitosan, depending on the environmental conditions.
In general, it appears that fusing an existing metabolic enzyme to the REC domain is an evolutionarily harmless procedure: the resulting fused protein is inactive unless its REC domain is phosphorylated by a cognate histidine kinase. The same logic could be applied to the fusions between REC domains and other signaling, ligand-binding, or protein-binding domains. Indeed, the large number of rare domain fusions (Table 2; see Table S1 in the supplemental material) supports the view that signaling domain fusions follow the Lego principle: almost any fusion can occur but only some of them remain fixed in the evolution (43). If this logic is correct, many more types of response regulators can be expected to be found in microbial genomes. Several relatively common, previously uncharacterized rare or unique domains identified in this study are listed in the supplemental material and will soon be available in the public domain databases. An experimental study of these novel response regulators and analysis of their potential functions will remain the task for future work, which will provide further understanding of the organization and evolution of the prokaryotic signal transduction systems.
.
Supplementary Material
Acknowledgments
I thank Andrei Gabrielian, Bill Klimke, Darren Natale, Tao Tao, and Yuri Wolf for helpful advice; Mark Gomelsky, Igor Zhulin, and two reviewers for critical comments; and many other colleagues for suggestions.
This study was supported by the Intramural Research Program of the National Library of Medicine at the U.S. National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aldridge, P., and U. Jenal. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32:379-391. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695-1708. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zheng, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST—a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3-6. [DOI] [PubMed] [Google Scholar]
- 5.Anand, G. S., P. N. Goudreau, and A. M. Stock. 1998. Activation of methylesterase CheB: evidence of a dual role for the regulatory domain. Biochemistry 37:14038-14047. [DOI] [PubMed] [Google Scholar]
- 6.Anand, G. S., and A. M. Stock. 2002. Kinetic basis for the stimulatory effect of phosphorylation on the methylesterase activity of CheB. Biochemistry 41:6752-6760. [DOI] [PubMed] [Google Scholar]
- 7.Anantharaman, V., E. V. Koonin, and L. Aravind. 2002. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 30:1427-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravind, L., V. Anantharaman, S. Balaji, M. M. Babu, and L. M. Iyer. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231-262. [DOI] [PubMed] [Google Scholar]
- 9.Aravind, L., and E. V. Koonin. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469-472. [DOI] [PubMed] [Google Scholar]
- 10.Armitage, J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229-289. [DOI] [PubMed] [Google Scholar]
- 11.Ashby, M. K. 2004. Survey of the number of two-component response regulator genes in the complete and annotated genome sequences of prokaryotes. FEMS Microbiol. Lett. 231:277-281. [DOI] [PubMed] [Google Scholar]
- 12.Baikalov, I., I. Schroder, M. Kaczor-Grzeskowiak, K. Grzeskowiak, R. P. Gunsalus, and R. E. Dickerson. 1996. Structure of the Escherichia coli response regulator NarL. Biochemistry 35:11053-110561. [DOI] [PubMed] [Google Scholar]
- 13.Bairoch, A., R. Apweiler, C. H. Wu, W. C. Barker, B. Boeckmann, S. Ferro, E. Gasteiger, H. Huang, R. Lopez, M. Magrane, M. J. Martin, D. A. Natale, C. O'Donovan, N. Redaschi, and L. S. Yeh. 2005. The Universal Protein Resource (UniProt). Nucleic Acids Res. 33:D154-D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker, G., E. Klauck, and R. Hengge-Aronis. 2000. The response regulator RssB, a recognition factor for σS proteolysis in Escherichia coli, can act like an anti-σS factor. Mol. Microbiol. 35:657-666. [DOI] [PubMed] [Google Scholar]
- 16.Bell, S. D. 2005. Archaeal transcriptional regulation—variation on a bacterial theme? Trends Microbiol. 13:262-265. [DOI] [PubMed] [Google Scholar]
- 17.Berleman, J. E., and C. E. Bauer. 2005. Involvement of a Che-like signal transduction cascade in regulating cyst cell development in Rhodospirillum centenum. Mol. Microbiol. 56:1457-1466. [DOI] [PubMed] [Google Scholar]
- 18.Bhaya, D., A. Takahashi, and A. R. Grossman. 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA 98:7540-7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilwes, A. M., L. A. Alex, B. R. Crane, and M. I. Simon. 1999. Structure of CheA, a signal-transducing histidine kinase. Cell 96:131-141. [DOI] [PubMed] [Google Scholar]
- 20.Bishop, A., S. Fielding, P. Dyson, and P. Herron. 2004. Systematic insertional mutagenesis of a streptomycete genome: a link between osmoadaptation and antibiotic production. Genome Res. 14:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 22.Bobrov, A. G., O. Kirillina, and R. D. Perry. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247:123-130. [DOI] [PubMed] [Google Scholar]
- 23.Boltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bren, A., and M. Eisenbach. 1998. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J. Mol. Biol. 278:507-514. [DOI] [PubMed] [Google Scholar]
- 25.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 26.Campbell, E. A., J. L. Tupy, T. M. Gruber, S. Wang, M. M. Sharp, C. A. Gross, and S. A. Darst. 2003. Crystal structure of Escherichia coli σE with the cytoplasmic domain of its anti-s RseA. Mol. Cell 11:1067-1078. [DOI] [PubMed] [Google Scholar]
- 27.Campbell, E. L., K. D. Hagen, M. F. Cohen, M. L. Summers, and J. C. Meeks. 1996. The devR gene product is characteristic of receivers of two-component regulatory systems and is essential for heterocyst development in the filamentous cyanobacterium Nostoc sp. strain ATCC 29133. J. Bacteriol. 178:2037-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan, C., R. Paul, D. Samoray, N. C. Amiot, B. Giese, U. Jenal, and T. Schirmer. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA 101:17084-17089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho, H. S., J. G. Pelton, D. Yan, S. Kustu, and D. E. Wemmer. 2001. Phosphoaspartates in bacterial signal transduction. Curr. Opin. Struct. Biol. 11:679-684.11751048 [Google Scholar]
- 30.Christen, M., B. Christen, M. Folcher, A. Schauerte, and U. Jenal. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280:30829-30837. [DOI] [PubMed] [Google Scholar]
- 31.Cummings, L., L. Riley, L. Black, A. Souvorov, S. Resenchuk, I. Dondoshansky, and T. Tatusova. 2002. Genomic BLAST: custom-defined virtual databases for complete and unfinished genomes. FEMS Microbiol. Lett. 216:133-138. [DOI] [PubMed] [Google Scholar]
- 32.Das, A. K., N. R. Helps, P. T. Cohen, and D. Barford. 1996. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 Å resolution. EMBO J. 15:6798-6809. [PMC free article] [PubMed] [Google Scholar]
- 33.Diller, T. C., Madhusudan, N. H. Xuong, and S. S. Taylor. 2001. Molecular basis for regulatory subunit diversity in cAMP-dependent protein kinase: crystal structure of the type II beta regulatory subunit. Structure 9:73-82. [DOI] [PubMed] [Google Scholar]
- 34.Djordjevic, S., P. N. Goudreau, Q. Xu, A. M. Stock, and A. H. West. 1998. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc. Natl. Acad. Sci. USA 95:1381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drury, L. S., and R. S. Buxton. 1985. DNA sequence analysis of the dye gene of Escherichia coli reveals amino acid homology between the dye and OmpR proteins. J. Biol. Chem. 260:4236-4242. [PubMed] [Google Scholar]
- 36.Eraso, J. M., and S. Kaplan. 1994. PrrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J. Bacteriol. 176:32-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fedorova, N. D., A. N. Nikolskaya, and M. Y. Galperin. 2003. Using genomic context in the analysis of multi-component systems, p. 167-194. In M. Y. Galperin and E. V. Koonin (ed.), Frontiers in computational genomics, vol. 3. Caister Academic Press, Wymondham, United Kingdom. [Google Scholar]
- 39.Ferrari, F. A., K. Trach, D. LeCoq, J. Spence, E. Ferrari, and J. A. Hoch. 1985. Characterization of the spo0A locus and its deduced product. Proc. Natl. Acad. Sci. USA 82:2647-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fredrick, K. L., and J. D. Helmann. 1994. Dual chemotaxis signaling pathways in Bacillus subtilis: a σD-dependent gene encodes a novel protein with both CheW and CheY homologous domains. J. Bacteriol. 176:2727-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galperin, M. Y. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 5:35. [Online.] http://www.biomedcentral.com/1471-2180/5/35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galperin, M. Y., and M. Gomelsky. 2005. Bacterial signal transduction modules: from genomics to biology. ASM News 71:326-333. [Google Scholar]
- 44.Galperin, M. Y., and M. J. Jedrzejas. 2001. Conserved core structure and active site residues in alkaline phosphatase superfamily enzymes. Proteins 45:318-324. [DOI] [PubMed] [Google Scholar]
- 45.Galperin, M. Y., D. A. Natale, L. Aravind, and E. V. Koonin. 1999. A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol. 1:303-305. [PMC free article] [PubMed] [Google Scholar]
- 46.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 47.Geiduschek, E. P., and M. Ouhammouch. 2005. Archaeal transcription and its regulators. Mol. Microbiol. 56:1397-1407. [DOI] [PubMed] [Google Scholar]
- 48.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41:139-227. [DOI] [PubMed] [Google Scholar]
- 49.Green, J., C. Scott, and J. R. Guest. 2001. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Physiol. 44:1-34. [DOI] [PubMed] [Google Scholar]
- 50.Gutierrez, P., Y. Li, M. J. Osborne, E. Pomerantseva, Q. Liu, and K. Gehring. 2005. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J. Bacteriol. 187:3496-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagiwara, D., T. Yamashino, and T. Mizuno. 2004. Genome-wide comparison of the His-to-Asp phosphorelay signaling components of three symbiotic genera of Rhizobia. DNA Res. 11:57-65. [DOI] [PubMed] [Google Scholar]
- 52.Hecht, G. B., and A. Newton. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho, Y. S., L. M. Burden, and J. H. Hurley. 2000. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 19:5288-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoch, J. A., and T. J. Silhavy (ed.). 1995. Two-component signal transduction. ASM Press, Washington, D.C.
- 56.Hottes, A. K., M. Meewan, D. Yang, N. Arana, P. Romero, H. H. McAdams, and C. Stephens. 2004. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 186:1448-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inouye, M., and R. Dutta (ed.). 2003. Histidine kinases in signal transduction. Academic Press, San Diego, Calif.
- 58.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 59.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 60.Kanehisa, M., S. Goto, S. Kawashima, Y. Okuno, and M. Hattori. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32:D277-D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karatan, E., M. M. Saulmon, M. W. Bunn, and G. W. Ordal. 2001. Phosphorylation of the response regulator CheV is required for adaptation to attractants during Bacillus subtilis chemotaxis. J. Biol. Chem. 276:43618-43626. [DOI] [PubMed] [Google Scholar]
- 62.Karniol, B., and R. D. Vierstra. 2004. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J. Bacteriol. 186:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kato, M., T. Mizuno, T. Shimizu, and T. Hakoshima. 1997. Insights into multistep phosphorelay from the crystal structure of the C-terminal HPt domain of ArcB. Cell 88:717-723. [DOI] [PubMed] [Google Scholar]
- 64.Kennelly, P. J. 2002. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol. Lett. 206:1-8. [DOI] [PubMed] [Google Scholar]
- 65.Kern, D., B. F. Volkman, P. Luginbuhl, M. J. Nohaile, S. Kustu, and D. E. Wemmer. 1999. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature 402:894-898. [DOI] [PubMed] [Google Scholar]
- 66.Kim, D., and S. Forst. 2001. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology 147:1197-1212. [DOI] [PubMed] [Google Scholar]
- 67.Klose, K. E., A. K. North, K. M. Stedman, and S. Kustu. 1994. The major dimerization determinants of the nitrogen regulatory protein NtrC from enteric bacteria lie in its carboxy-terminal domain. J. Mol. Biol. 241:233-245. [DOI] [PubMed] [Google Scholar]
- 68.Konstantinidis, K. T., and J. M. Tiedje. 2004. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA 101:3160-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koonin, E. V., L. Aravind, and M. Y. Galperin. 2000. A comparative-genomic view of the microbial stress response, p. 417-444. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 70.Koretke, K. K., A. N. Lupas, P. V. Warren, M. Rosenberg, and J. R. Brown. 2000. Evolution of two-component signal transduction. Mol. Biol. Evol. 17:1956-1970. [DOI] [PubMed] [Google Scholar]
- 71.Kostrewa, D., J. Granzin, C. Koch, H. W. Choe, S. Raghunathan, W. Wolf, J. Labahn, R. Kahmann, and W. Saenger. 1991. Three-dimensional structure of the E. coli DNA-binding protein FIS. Nature 349:178-180. [DOI] [PubMed] [Google Scholar]
- 72.Kuriyan, J., T. S. Krishna, L. Wong, B. Guenther, A. Pahler, C. H. Williams, Jr., and P. Model. 1991. Convergent evolution of similar function in two structurally divergent enzymes. Nature 352:172-174. [DOI] [PubMed] [Google Scholar]
- 73.Laguri, C., M. K. Phillips-Jones, and M. P. Williamson. 2003. Solution structure and DNA binding of the effector domain from the global regulator PrrA (RegA) from Rhodobacter sphaeroides: insights into DNA binding specificity. Nucleic Acids Res. 31:6778-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 75.Lee, S. Y., H. S. Cho, J. G. Pelton, D. Yan, E. A. Berry, and D. E. Wemmer. 2001. Crystal structure of activated CheY. Comparison with other activated receiver domains. J. Biol. Chem. 276:16425-16431. [DOI] [PubMed] [Google Scholar]
- 76.Lee, S. Y., A. De La Torre, D. Yan, S. Kustu, B. T. Nixon, and D. E. Wemmer. 2003. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 17:2552-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leonard, T. A., P. J. Butler, and J. Lowe. 2005. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer—a conserved biological switch. EMBO J. 24:270-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewis, R. J., S. Krzywda, J. A. Brannigan, J. P. Turkenburg, K. Muchova, E. J. Dodson, I. Barak, and A. J. Wilkinson. 2000. The trans-activation domain of the sporulation response regulator Spo0A revealed by X-ray crystallography. Mol. Microbiol. 38:198-212. [DOI] [PubMed] [Google Scholar]
- 79.Lewis, R. J., D. J. Scott, J. A. Brannigan, J. C. Ladds, M. A. Cervin, G. B. Spiegelman, J. G. Hoggett, I. Barak, and A. J. Wilkinson. 2002. Dimer formation and transcription activation in the sporulation response regulator Spo0A. J. Mol. Biol. 316:235-245. [DOI] [PubMed] [Google Scholar]
- 80.Liang, J., L. Scappino, and R. Haselkorn. 1992. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc. Natl. Acad. Sci. USA 89:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lupas, A., and J. Stock. 1989. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J. Biol. Chem. 264:17337-17342. [PubMed] [Google Scholar]
- 82.Madec, E., A. Laszkiewicz, A. Iwanicki, M. Obuchowski, and S. Seror. 2002. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46:571-586. [DOI] [PubMed] [Google Scholar]
- 83.Makarova, K. S., E. V. Koonin, R. Haselkorn, and M. Y. Galperin. 16 March 2006, posting date. Cyanobacterial response regulator PatA contains a conserved N-terminal domain (PATAN) with an alpha-helical insertion. Bioinformatics [Online.] doi: 10.1093/bioinformatics/btl096. [DOI] [PubMed]
- 84.Maltsev, N., E. Marland, G. X. Yu, S. Bhatnagar, and R. Lusk. 2002. Sentra, a database of signal transduction proteins. Nucleic Acids Res. 30:349-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martinez-Hackert, E., and A. M. Stock. 1997. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5:109-124. [DOI] [PubMed] [Google Scholar]
- 88.Martinez-Hackert, E., and A. M. Stock. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301-312. [DOI] [PubMed] [Google Scholar]
- 89.Matsumura, P., J. J. Rydel, R. Linzmeier, and D. Vacante. 1984. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J. Bacteriol. 160:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mizuno, T. 1997. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 4:161-168. [DOI] [PubMed] [Google Scholar]
- 91.Mizuno, T., T. Kaneko, and S. Tabata. 1996. Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium, Synechocystis sp. strain PCC 6803. DNA Res. 3:407-414. [DOI] [PubMed] [Google Scholar]
- 92.Muffler, A., D. Fischer, S. Altuvia, G. Storz, and R. Hengge-Aronis. 1996. The response regulator RssB controls stability of the σS subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333-1339. [PMC free article] [PubMed] [Google Scholar]
- 93.Muraoka, S., R. Okumura, N. Ogawa, T. Nonaka, K. Miyashita, and T. Senda. 2003. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J. Mol. Biol. 328:555-566. [DOI] [PubMed] [Google Scholar]
- 94.Nikolskaya, A. N., and M. Y. Galperin. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 30:2453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nixon, B. T., C. W. Ronson, and F. M. Ausubel. 1986. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc. Natl. Acad. Sci. USA 83:7850-7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O'Hara, B. P., R. A. Norman, P. T. Wan, S. M. Roe, T. E. Barrett, R. E. Drew, and L. H. Pearl. 1999. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J. 18:5175-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ohmori, M., M. Ikeuchi, N. Sato, P. Wolk, T. Kaneko, T. Ogawa, M. Kanehisa, S. Goto, S. Kawashima, S. Okamoto, H. Yoshimura, H. Katoh, T. Fujisawa, S. Ehira, A. Kamei, S. Yoshihara, R. Narikawa, and S. Tabata. 2001. Characterization of genes encoding multi-domain proteins in the genome of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:271-284. [DOI] [PubMed] [Google Scholar]
- 98.Park, S. Y., X. Chao, G. Gonzalez-Bonet, B. D. Beel, A. M. Bilwes, and B. R. Crane. 2004. Structure and function of an unusual family of protein phosphatases: the bacterial chemotaxis proteins CheC and CheX. Mol. Cell 16:563-574. [DOI] [PubMed] [Google Scholar]
- 99.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pelton, J. G., S. Kustu, and D. E. Wemmer. 1999. Solution structure of the DNA-binding domain of NtrC with three alanine substitutions. J. Mol. Biol. 292:1095-1110. [DOI] [PubMed] [Google Scholar]
- 101.Phillips-Jones, M. K., and C. N. Hunter. 1994. Cloning and nucleotide sequence of regA, a putative response regulator gene of Rhodobacter sphaeroides. FEMS Microbiol. Lett. 116:269-275. [DOI] [PubMed] [Google Scholar]
- 102.Planet, P. J., S. C. Kachlany, R. DeSalle, and D. H. Figurski. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA 98:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pruitt, K. D., T. Tatusova, and D. R. Maglott. 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33:D501-D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pullen, K. E., H. L. Ng, P. Y. Sung, M. C. Good, S. M. Smith, and T. Alber. 2004. An alternate conformation and a third metal in PstP/Ppp, the M. tuberculosis PP2C-Family Ser/Thr protein phosphatase. Structure 12:1947-1954. [DOI] [PubMed] [Google Scholar]
- 105.Quirk, P. G., A. A. Guffanti, S. Clejan, J. Cheng, and T. A. Krulwich. 1994. Isolation of Tn917 insertional mutants of Bacillus subtilis that are resistant to the protonophore carbonyl cyanide m-chlorophenylhydrazone. Biochim. Biophys. Acta 1186:27-34. [DOI] [PubMed] [Google Scholar]
- 106.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Robinson, V. L., T. Wu, and A. M. Stock. 2003. Structural analysis of the domain interface in DrrB, a response regulator of the OmpR/PhoB subfamily. J. Bacteriol. 185:4186-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 109.Römling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: The dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 110.Rosario, M. M., K. L. Fredrick, G. W. Ordal, and J. D. Helmann. 1994. Chemotaxis in Bacillus subtilis requires either of two functionally redundant CheW homologs. J. Bacteriol. 176:2736-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y.-W. He, L.-H. Zhang, S. Heeb, M. Cámara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA, 103:6712-6717. [DOI] [PMC free article] [PubMed] [Retracted]
- 112.Ryjenkov, D. A., M. Tarutina, O. M. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Satomura, T., D. Shimura, K. Asai, Y. Sadaie, K. Hirooka, and Y. Fujita. 2005. Enhancement of glutamine utilization in Bacillus subtilis through the GlnK-GlnL two-component regulatory system. J. Bacteriol. 187:4813-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. Ubiquitous protein domain EAL encodes cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sganga, M. W., and C. E. Bauer. 1992. Regulatory factors controlling photosynthetic reaction center and light-harvesting gene expression in Rhodobacter capsulatus. Cell 68:945-954. [DOI] [PubMed] [Google Scholar]
- 116.Shi, L. 2004. Manganese-dependent protein O-phosphatases in prokaryotes and their biological functions. Front. Biosci. 9:1382-1397. [DOI] [PubMed] [Google Scholar]
- 117.Shi, L., M. Potts, and P. J. Kennelly. 1998. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 22:229-253. [DOI] [PubMed] [Google Scholar]
- 118.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shu, C. J., and I. B. Zhulin. 2002. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 27:3-5. [DOI] [PubMed] [Google Scholar]
- 120.Simms, S. A., M. G. Keane, and J. Stock. 1985. Multiple forms of the CheB methylesterase in bacterial chemosensing. J. Biol. Chem. 260:10161-10168. [PubMed] [Google Scholar]
- 121.Slater, H., A. Alvarez-Morales, C. E. Barber, M. J. Daniels, and J. M. Dow. 2000. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38:986-1003. [DOI] [PubMed] [Google Scholar]
- 122.Sonenshein, A. L. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561-566. [DOI] [PubMed] [Google Scholar]
- 123.Stephenson, K., and R. J. Lewis. 2005. Molecular insights into the initiation of sporulation in gram-positive bacteria: new technologies for an old phenomenon. FEMS Microbiol. Rev. 29:281-301. [DOI] [PubMed] [Google Scholar]
- 124.Stock, A., D. E. Koshland, Jr., and J. Stock. 1985. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. Proc. Natl. Acad. Sci. USA 82:7989-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stock, A. M., E. Martinez-Hackert, B. F. Rasmussen, A. H. West, J. B. Stock, D. Ringe, and G. A. Petsko. 1993. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry 32:13375-13380. [DOI] [PubMed] [Google Scholar]
- 126.Stock, A. M., J. M. Mottonen, J. B. Stock, and C. E. Schutt. 1989. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature 337:745-749. [DOI] [PubMed] [Google Scholar]
- 127.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 128.Sumby, P., and M. C. Smith. 2002. Genetics of the phage growth limitation (Pgl) system of Streptomyces coelicolor A3(2). Mol. Microbiol. 44:489-500. [DOI] [PubMed] [Google Scholar]
- 129.Szurmant, H., T. J. Muff, and G. W. Ordal. 2004. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J. Biol. Chem. 279:21787-21792. [DOI] [PubMed] [Google Scholar]
- 130.Szurmant, H., and G. W. Ordal. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tamayo, R., A. D. Tischler, and A. Camilli. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324-33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tanaka, T., T. Fukui, S. Fujiwara, H. Atomi, and T. Imanaka. 2004. Concerted action of diacetylchitobiose deacetylase and exo-β-d-glucosaminidase in a novel chitinolytic pathway in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Biol. Chem. 279:30021-30027. [DOI] [PubMed] [Google Scholar]
- 133.Tarbouriech, N., S. J. Charnock, and G. J. Davies. 2001. Three-dimensional structures of the Mn and Mg dTDP complexes of the family GT-2 glycosyltransferase SpsA: a comparison with related NDP-sugar glycosyltransferases. J. Mol. Biol. 314:655-661. [DOI] [PubMed] [Google Scholar]
- 134.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 11 September 2003, posting date. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [Online.] doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Trach, K. A., J. W. Chapman, P. J. Piggot, and J. A. Hoch. 1985. Deduced product of the stage 0 sporulation gene spo0F shares homology with the Spo0A, OmpR, and SfrA proteins. Proc. Natl. Acad. Sci. USA 82:7260-7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Trempy, J. E., J. E. Kirby, and S. Gottesman. 1994. Alp suppression of Lon: dependence on the slpA gene. J. Bacteriol. 176:2061-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ueda, K., A. Yamashita, J. Ishikawa, M. Shimada, T. O. Watsuji, K. Morimura, H. Ikeda, M. Hattori, and T. Beppu. 2004. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 32:4937-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ulrich, L. E., E. V. Koonin, and I. B. Zhulin. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 13:52-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.van Nimwegen, E. 2003. Scaling laws in the functional content of genomes. Trends Genet. 19:479-484. [DOI] [PubMed] [Google Scholar]
- 143.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 144.Vlamakis, H. C., J. R. Kirby, and D. R. Zusman. 2004. The Che4 pathway of Myxococcus xanthus regulates type IV pilus-mediated motility. Mol. Microbiol. 52:1799-1811. [DOI] [PubMed] [Google Scholar]
- 145.Volkman, B. F., M. J. Nohaile, N. K. Amy, S. Kustu, and D. E. Wemmer. 1995. Three-dimensional solution structure of the N-terminal receiver domain of NtrC. Biochemistry 34:1413-1424. [DOI] [PubMed] [Google Scholar]
- 146.Volz, K. 1993. Structural conservation in the CheY superfamily. Biochemistry 32:11741-11753. [DOI] [PubMed] [Google Scholar]
- 147.Volz, K., and P. Matsumura. 1991. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J. Biol. Chem. 266:15511-15519. [DOI] [PubMed] [Google Scholar]
- 148.Wang, L., Y. P. Sun, W. L. Chen, J. H. Li, and C. C. Zhang. 2002. Genomic analysis of protein kinases, protein phosphatases and two-component regulatory systems of the cyanobacterium Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 217:155-165. [DOI] [PubMed] [Google Scholar]
- 149.Welch, M., K. Oosawa, S. Aizawa, and M. Eisenbach. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. USA 90:8787-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 151.Wheeler, D. L., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, W. Helmberg, D. L. Kenton, O. Khovayko, D. J. Lipman, T. L. Madden, D. R. Maglott, J. Ostell, J. U. Pontius, K. D. Pruitt, G. D. Schuler, L. M. Schriml, E. Sequeira, S. T. Sherry, K. Sirotkin, G. Starchenko, T. O. Suzek, R. Tatusov, T. A. Tatusova, L. Wagner, and E. Yaschenko. 2005. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 33:D39-D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wietzorrek, A., and M. Bibb. 1997. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25:1181-1184. [DOI] [PubMed] [Google Scholar]
- 153.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]
- 154.Yeats, C., S. Bentley, and A. Bateman. 6 February 2003, posting date. New knowledge from old: in silico discovery of novel protein domains in Streptomyces coelicolor. BMC Microbiol. 3:3. [Online.] doi: 10.1186/1471-2180-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yurimoto, H., R. Hirai, N. Matsuno, H. Yasueda, N. Kato, and Y. Sakai. 2005. HxlR, a member of the DUF24 protein family, is a DNA-binding protein that acts as a positive regulator of the formaldehyde-inducible hxlAB operon in Bacillus subtilis. Mol. Microbiol. 57:511-519. [DOI] [PubMed] [Google Scholar]
- 156.Zhang, G., Y. Liu, A. E. Ruoho, and J. H. Hurley. 1997. Structure of the adenylyl cyclase catalytic core. Nature 386:247-253. [DOI] [PubMed] [Google Scholar]
- 157.Zhulin, I. B., A. N. Nikolskaya, and M. Y. Galperin. 2003. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol. 185:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.