Abstract
The pilin of Pseudomonas aeruginosa 1244 is glycosylated with an oligosaccharide that is structurally identical to the O-antigen repeating unit of this organism. Concordantly, the metabolic source of the pilin glycan is the O-antigen biosynthetic pathway. The present study was conducted to investigate glycan substrate recognition in the 1244 pilin glycosylation reaction. Comparative structural analysis of O subunits that had been previously shown to be compatible with the 1244 glycosylation machinery revealed similarities among sugars at the presumed reducing termini of these oligosaccharides. We therefore hypothesized that the glycosylation substrate was within the sugar at the reducing end of the glycan precursor. Since much is known of PA103 O-antigen genetics and because the sugars at the reducing termini of the O7 (strain 1244) and O11 (strain PA103) are identical (β-N-acetyl fucosamine), we utilized PA103 and strains that express lipopolysaccharide (LPS) with a truncated O-antigen subunit to test our hypothesis. LPS from a strain mutated in the wbjE gene produced an incomplete O subunit, consisting only of the monosaccharide at the reducing end (β-d-N-acetyl fucosamine), indicating that this moiety contained substrate recognition elements for WaaL. Expression of pilAO1244 in PA103 wbjE::aacC1, followed by Western blotting of extracts of these cells, indicated that pilin produced has been modified by the addition of material consistent with a single N-acetyl fucosamine. This was confirmed by analyzing endopeptidase-treated pilin by mass spectrometry. These data suggest that the pilin glycosylation substrate recognition features lie within the reducing-end moiety of the O repeat and that structures of the remaining sugars are irrelevant.
Pseudomonas aeruginosa is a gram-negative, opportunistic pathogen that expresses type IV pili (33), fibrous surface appendages that protrude from the poles of the cell (20). Pili contribute to bacterial pathogenicity by initiating the colonization of host tissue through adhesion and mediating motility across surfaces (33). A single pilus fiber is a polymer of a proteinaceous subunit, referred to as pilin (33). Pilin of P. aeruginosa 1244 is modified by glycosylation, a process that requires the presence of the enzyme PilO (6).
Since the discovery of archaeal S-layer glycoproteins (35), numerous accounts of protein glycosylation in prokaryotes have been recorded, especially among surface proteins of pathogens (44, 57). Examples of gram-positive bacteria in which this posttranslational modification has been documented include Streptococcus mutans (9), S. parasanguis (53), Mycobacterium tuberculosis (15), and Bacillus anthracis (55). Among the gram-negative bacteria, much of the research attention has focused on the two glycosylation systems of Campylobacter jejuni (23, 56, 58), in addition to the flagellin glycosylation of P. aeruginosa (3) and Helicobacter pylori (43) and the pilin glycosylation of Neisseria spp. (26, 37, 54) and P. aeruginosa 1244 (6, 7, 51).
P. aeruginosa 1244 pilin contains a single covalently bound glycan (7) that is O linked to the β-carbon of Ser148 (10), the carboxy-terminal residue (8). The pilin glycan is a trisaccharide, structurally identical to the O-antigen repeating unit of the serotype O7 lipopolysaccharide (LPS) of strain 1244 (7), which suggested that the glycan might originate in the same metabolic pathway as O-antigen biosynthesis. Evidence supporting this was provided by the finding that the mutation of genes involved in initial steps of O-antigen biosynthesis (either wbpM or wbpL) abolished pilin glycosylation (14). In addition, expression of heterologous O-antigen gene clusters in P. aeruginosa 1244 allowed for pilin to be decorated with the heterologous saccharide, confirming that pilin glycosylation and O-antigen biosynthesis shared a common metabolic origin (14). Furthermore, the putative oligosaccharyltransferase, PilO, is the only protein required for glycosylation that is not involved in O-antigen or pilin synthesis (14).
P. aeruginosa O-antigen biosynthesis proceeds by the “Wzy-dependent” mechanism (13, 17), during which individual O-antigen repeating units are constructed on the cytoplasmic side of the cell membrane by the sequential addition of nucleotide-activated sugars to the carrier lipid, undecaprenyl phosphate (Und-P) (17, 39). In P. aeruginosa PA103, WbpL catalyzes the transfer of β-d-FucNAc to Und-P (13), followed by the addition of α-l-FucNAc by a previously uncharacterized transferase. Finally, the glucosyltransferase, WbjA, adds β-d-Glc to complete the O11 subunit (12). The putative flippase, Wzx, translocates the undecaprenyl pyrophosphate (Und-PP)-linked O repeat to the periplasmic face (31, 34, 36) of the cell membrane, where O-antigen polymerization (34) is mediated by Wzy (13, 39). O-antigen chain length is regulated by Wzz (41), and the O ligase, WaaL, transfers the entire O-antigen to the core polysaccharide (1, 36). A Wzy mutant cannot synthesize polymerized O-antigen; however, these cells are capable of producing a core that contains a single O-antigen repeating unit, a phenotype referred to as “core + 1” (13). Interestingly, mutation of wbjA yielded cells that produced a core containing an incomplete O subunit and possessed a core + 2/3 phenotype (12). These phenotypes suggest that O-antigen polymerization and complete assembly of the individual O subunit are not necessary for core-O-antigen ligation (12). A similar phenomenon was observed in a mutant of Escherichia coli K-12 that could only synthesize the first sugar of its O subunit (17).
Results from a previous study that investigated pilin specificity in the P. aeruginosa 1244 glycosylation reaction suggested that the positioning of Ser at the pilin C terminus is critical for recognition by the glycosylation machinery (25). Although no other specific recognition features are present, the pilin surface charge must be compatible with the glycosylation apparatus (25). Glycosylation requires an enzyme with a specificity for both the target protein and the glycan source (52, 57). Little is known of glycan features important for recognition in prokaryotic oligosaccharyltransferase-mediated protein glycosylation systems. In the O-linked glycosylation system of P. aeruginosa 1244 pilin, completely assembled individual O-antigen subunits bound to Und-PP (14) are transferred en bloc to pilin (7, 14). A similar model is proposed for C. jejuni N-linked glycosylation (18, 57). As a variety of structurally distinct O subunits can be used as the P. aeruginosa 1244 pilin glycan (14), the apparent saccharide recognition features for catalysis by this system are unclear. However, the 1244 pilin glycosylation machinery does exhibit specificity for the O-antigen subunit, since other Und-PP-linked saccharides, such as peptidoglycan components, are not linked to pilin (7). Elucidating essential structural aspects of the 1244 pilin glycan precursor for glycosylation would contribute to the understanding of an important posttranslational modification.
Results presented here indicate that WbjE catalyzes the addition of the second sugar, α-l-N-acetyl fucosamine, to the O11 subunit of P. aeruginosa PA103. In addition, the present study provides a detailed description of the glycan substrate recognition features of the P. aeruginosa 1244 pilin glycosylation system. These results suggest that the sugar at the reducing end of the O-antigen repeating unit contains the major glycosylation recognition information and that structural aspects of the remaining sugars are unimportant. We present evidence suggesting the carbohydrate moiety at the reducing end of the O repeat also possesses the recognition information necessary for catalysis by WaaL, the O-antigen ligase as was previously observed in E. coli K-12 (17). By examining endopeptidase-treated 1244 pilin produced in trans by O-antigen mutants, we demonstrate a novel approach for the analysis of O subunits. The findings of the present study could have implications for the utilization of the 1244 pilin glycosylation system in vaccine engineering.
MATERIALS AND METHODS
Bacterial strains and media.
P. aeruginosa strains and plasmids used in the present study are listed in Table 1. E. coli DH5α (Invitrogen), SM10 (50), and HB101 (2) were used for genetic manipulations. Luria (L) broth or agar was used for routine growth of bacteria. Cetrimide agar base (Difco) was used for the isolation of P. aeruginosa after triparental matings. For analysis of pilin, overnight cultures were used to inoculate petri plates containing CAYE solid medium, which consisted of 0.75% Casamino Acids, 0.15% yeast extract, and 2% agar (49). The media contained gentamicin (250 μg/ml for P. aeruginosa and 15 μg/ml for E. coli), kanamycin (30 μg/ml for E. coli), carbenicillin (250 μg/ml for P. aeruginosa), ampicillin (50 μg/ml for E. coli), and/or tetracycline (50 μg/ml for P. aeruginosa), as required. When necessary, the medium was supplemented with 5% sucrose or 5 mM IPTG (isopropyl-β-d-thiogalactopyranoside).
TABLE 1.
P. aeruginosa strains and plasmids used in this study
| P. aeruginosa strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| PA103 | Wild type; IATS O11 | 30 |
| 1244N3 | rpoN (Tcr); IATS O7 | 40 |
| PA103 wzy::aacC1 | wzy (Gmr) | 13 |
| PA103 wbjA::aacC1 | wbjA (Gmr) | 12 |
| PA103 wbjE::aacC1 | wbjE (Gmr) | This study |
| Plasmids | ||
| pRK2013 | Kmr; helper plasmid for triparental mating | 42 |
| pLPS2 | Broad-host-range cosmid containing the P. aeruginosa PA103 O11 gene cluster | 22 |
| pEX100T | 5.8-kb gene replacement vector, oriT, sacB+; Apr | 47 |
| pUCGm | Contains a Gmr cassette (aacC1); Apr, Gmr | 46 |
| pEX100T-wbjE::aacC1 | pEX100T with insertion of the Gmr cassette (aacC1) in wbjE | This study |
| pUCP18 | pUC18-derived broad-host-range shuttle vector; Apr/Cbr | 45 |
| pCD207 | pUCP18 with P. aeruginosa PA103 wbjE | This study |
| pMMB66EH | 8.8-kb broad host range expression vector; Apr/Cbr | 21 |
| pPAC46 | pMMB66EH with P. aeruginosa 1244 pilAO | 6 |
| pPAC24 | pMMB66EH with P. aeruginosa 1244 pilA | 6 |
Apr, ampicillin resistance; Cbr, carbenicillin resistance; Tcr, tetracycline resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance.
DNA manipulations.
Plasmid DNA was isolated with Qiaprep Spin Miniprep Kit (QIAGEN Sciences, Valencia, CA) or Wizard Plus Minipreps (Promega Corp., Madison, WI). Restriction endonucleases and modification enzymes (New England Biolabs, Beverly, MA; Boehringer Mannheim Corp., Indianapolis, IN) were used as specified by the manufacturer.
The wbjE sequence was obtained from a PCR fragment amplified from the pLPS2 plasmid. Primers for the amplification of the wbjE gene were 83-30 (5′-GCTGAAACCCCTCGGCTTTAAGG-3′) and 6-95-E (5′-GCATCGCGATTCACCTTCCG-3′) and were purchased from Ransom Hill (Ramona, CA). PCR was performed with the EasyStart PCR Mix-in-a-Tube (Molecular Bio-Products, Inc., San Diego, CA) or with Vent-PCR as described by manufacturer (New England Biolabs, Beverly, MA) and either an Applied Biosystems GeneAmp PCR System 9700 thermocycler or a Perkin-Elmer GeneAmp PCR System 2400 thermocycler. PCR products were purified by agarose gel electrophoresis as previously described (11). After agarose gel electrophoresis, DNA fragments were isolated with a Qiaquick gel extraction kit (QIAGEN) as specified by the manufacturer. Nucleotide sequence determination of both strands of the cloned DNA was carried out by the Biomolecular Research Facility (University of Virginia). Nucleotide sequence fragments were assembled and analyzed with the Gene Construction Kit (SciQuest, Research Triangle, NC) computer program.
In vitro mutagenesis and gene replacement.
A wbjE mutation was made in the chromosome of P. aeruginosa PA103 as follows. A blunt-ended wbjE PCR product, as described above, was cloned into the SmaI site in the pEX100T vector. A nonpolar mutation of wbjE was constructed in vitro by insertion of a gentamicin resistance gene (aacC1), recovered as an 854-bp SalI fragment from pUCGm, into the SalI site in wbjE on pEX100T. This construct (pEX100T-wbjE::aacC1) was introduced into the mobilizing E. coli strain SM10, and SM10/pEX100T-wbjE::aacC1 was conjugated with P. aeruginosa PA103 by pelleting approximately equal numbers of donor (grown overnight at 37°C) and recipient (grown overnight without shaking at 42°C) strains in microcentrifuge tubes and spotting them onto the center of an L agar plate. After incubation for 12 to 18 h at 37°C, the cells were resuspended in 1 ml of L broth and plated on gentamicin-containing cetrimide plates. Colonies arising after 48 h were purified on the same medium and the swabbed onto L agar plates containing both gentamicin and sucrose. Since pEX100T contains the sacB gene, which renders gram-negative bacteria sensitive to sucrose, colonies arising on sucrose- and gentamicin-containing plates carried wbjE::aacC1 on the chromosome and had lost the vector-associated sacB gene. These colonies were tested for sensitivity to carbenicillin to confirm the loss of vector DNA. Gene replacement was confirmed by PCR amplification of chromosomal DNA with primers flanking the aacC1 insertion site in wbjE.
Complementing plasmid.
The same Vent-PCR product that was used for the gene replacement, as described above, was cloned in SmaI-digested plasmid pUCP18. Restriction analysis indicated that the fragment was inserted in the orientation opposite that of the lacZ gene on the plasmid. The recombinant plasmid, referred to as pCD207, was transformed into P. aeruginosa PA103 wbjE::aacC1 by the electroporation protocol of Enderle and Farwell (16). For pilin analysis, triparental mating was used to mobilize pMMB66EH-derived plasmids into P. aeruginosa strains (42).
Isolation of LPS.
P. aeruginosa LPS was isolated by the proteinase K digestion method of Hitchcock and Brown (24), followed by hot-phenol extraction with slight modifications. Cells grown overnight in L broth with appropriate antibiotics were equilibrated to an optical density of 0.5 at a wavelength of 600 nm. This volume of bacterial suspension was then transferred to a microcentrifuge tube and centrifuged at 13,100 × g for 5 min. The supernatant was removed, and the pellet was resuspended in 200 μl of lysis buffer (2% sodium dodecyl sulfate [SDS], 4% β-mercaptoethanol, 10% glycerol, and 0.002% bromophenol blue in 1 M Tris [pH 6.8]), and boiled for 15 min. The lysed cells were treated with RNase and DNase for 30 min at 37°C. The cells were then digested for 3 h at 59°C with 120 μg of proteinase K. The digested lysates were then extracted with an equal volume of 90% phenol for 15 min at 65°C with periodic vortexing. After centrifugation in a microcentrifuge at maximum speed for 10 min, both the aqueous and the phenol layers were transferred to new tubes and extracted with 1 ml of diethyl ether. The aqueous layers from these extractions were removed, pooled, and again extracted with hot phenol. The aqueous layer (blue) was again extracted with 1 ml of ether and centrifuged, and the LPS-containing aqueous layer was diluted 1:3 with fresh lysis buffer and used for SDS-polyacrylamide gel electrophoresis (PAGE) analysis.
SDS-PAGE and Western immunoblotting.
LPS samples were separated by SDS-PAGE as previously described (11), on 12 or 13% acrylamide running gels. LPS was visualized by silver staining with a protocol derived from that of Tsai and Frasch (59). For pilin analysis, cell extracts were separated by SDS-PAGE using 16% Tris-glycine (1244 pilin) or 16% Tricine gels (PA103 pilin) prior to Western blotting (25).
Western immunoblot analyses were performed as previously described (8, 11). The antibody used was anti-1244 pilin monoclonal antibody (MAb) 6.45, which does not recognize PA103 pilin (48). The blot was developed at room temperature with a fluorescein isothiocyanate-conjugated secondary antibody. Fluorescence was detected by using a FluorImager595 (Molecular Dynamics) using the 530 DF30 filter.
Isolation and purification of pili.
Pili from PA103 wbjA::aacC1/pPAC46, PA103 wbjE::aacC1/pPAC46, 1244N3/pPAC46, and 1244N3/pPAC24 were harvested as previously described (49). Briefly, cell cultures were grown in aluminum foil-covered metal pans (68 by 28 by 3 cm) that contained 500 ml of CAYE medium, appropriate antibiotics, and 5 mM IPTG at 37°C for 14 h (1244 N3/pPAC46 and 1244 N3/pPAC24) or 18 h (PA103 wbjA::aacC1/pPAC46 and PA103 wbjE::aacC1/pPAC46). Cells from each pan were suspended in 50 ml of sodium phosphate buffer (49). Cell suspensions were stirred vigorously for 30 min at room temperature. The pili were isolated from the supernatant after centrifugation at 16,000 × g for 30 min at 4°C. Pili were purified by repeated precipitation in the presence of 3% polyethylene glycol 8000 and 0.5 M NaCl (49, 51).
Analytical methods.
GluC (V8 protease; Roche Applied Science) specifically hydrolyzes peptide and ester bonds at the carboxylic side of glutamate residues. GluC endopeptidase digestions of pilin samples were performed as previously described (10). Digested pilin was initially analyzed by SDS-PAGE (17% Tris-glycine gel), followed by Coomassie brilliant blue staining to detect the presence of the ∼5- to 6-kDa fragment (data not shown). The digestion mixture was subjected to two overnight dialyses against 6 liters of deionized water (0.025% sodium azide) at 4°C. Dialyzed samples were analyzed via matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry by Mark Bier of the Mellon Institute Center for Molecular Analysis, Carnegie Mellon University, using a PerSeptive Biosystems Voyager STR with DE and a high m/z detector. O-antigen repeating unit structures (28, 38) were drawn by using ChemDraw 9.0 for comparative analysis.
RESULTS
Structural comparison of O-antigen repeating units.
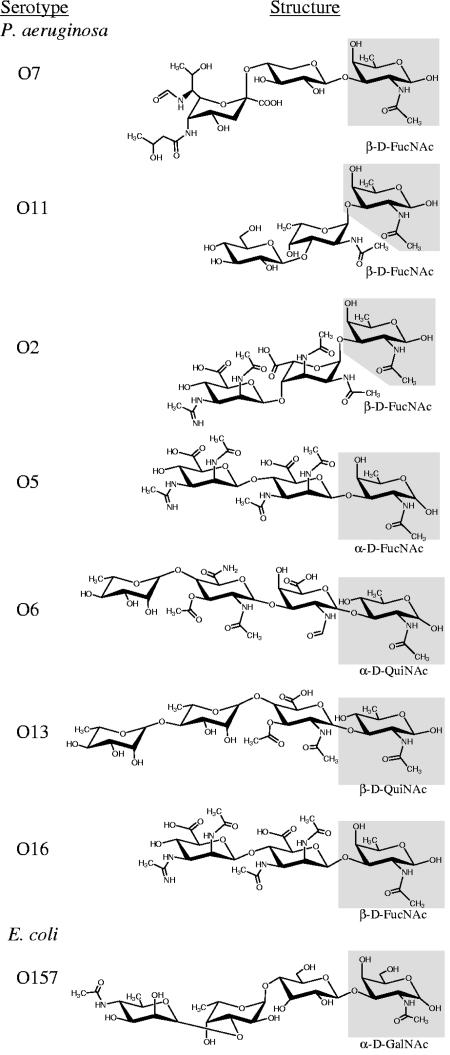
In a previous study that investigated the glycan specificity of PilO, results indicated that nine P. aeruginosa serotypes (seven having known O-antigen structures) and E. coli O157, all possessed an O-subunit that could serve as the 1244 pilin glycan (Fig. 1) (14). This suggested that common structures among these O subunits could be important for recognition by the 1244 glycosylation machinery. Comparative structural analysis of these sugars revealed drastic differences in charge, size, configuration, linkage, and number of sugars (Fig. 1). However, it is evident that the Und-PP/pilin-linked moieties (reducing end) share structural similarities (Fig. 1). Such similarities include a C-1 linkage to the Und-PP carrier, a D configuration, a C-3 linkage to the adjacent sugar, and the presence of a 2-N-acetyl group (Fig. 1). Much of the variance between these O repeats lies within the remaining carbohydrate moieties (Fig. 1). For instance, the serotype O7 repeat of strain 1244 is a trisaccharide in which the sugar in the third position (counting the Und-PP/pilin-linked moiety at the reducing end as the first position), pseudaminic acid, is a 14-carbon sugar in the α configuration. This pseudaminic acid is linked (1→4) to the second sugar, xylose, a β-5-carbon sugar. In contrast, the O6 repeating unit is a tetrasaccharide, in which the second and third moieties are β-sugars (linked 1→3), consisting of seven and nine carbons, respectively, whereas the fourth moiety is in the α configuration and consists of six carbons. Structural comparison of the O repeats from Fig. 1 suggests that the moiety at the reducing terminus may possess the glycan recognition features necessary for catalysis by the pilin glycosylation machinery and that structural aspects of the other saccharides are nonessential.
FIG. 1.
Comparative structural analysis of O-antigen subunits previously shown to be compatible for use in pilin glycosylation (14). Chair conformations of O repeats were drawn by using ChemDraw 9.0 based on structures presented by Knirel and Kochetkov (28) and/or Perry and MacLean (38). Presumably, the sugar at the reducing end (highlighted in gray) is bound to Und-PP prior to pilin glycosylation, O-antigen polymerization, and ligation to the core-lipid A. PilO from strain 1244 (serotype O7) was found to mediate the O linkage between carbon-1 of the moiety at the reducing terminus (β-d-FucNAc) to Ser148 of pilin (7, 10). All O repeats tested for use as the glycan were likely bound to pilin in this fashion.
Characterization of PA103 wbjE::aacC1.
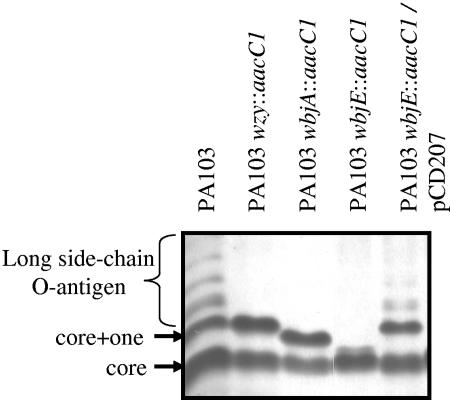
To experimentally determine whether the structural elements of the glycan necessary for glycosylation are within the reducing-end moiety of the O-antigen repeating unit, we required a mutant incapable of complete O-subunit assembly beyond the first sugar, β-N-acetyl fucosamine (Fig. 1). Because little is known of the O-antigen genetics of strain 1244 (serotype O7), we utilized a strain more commonly studied in O-antigen biosynthesis, PA103 (serotype O11), as a genetic background (12, 13). Notably, the first sugar moiety the O7 serotype O subunit is identical to that of the serotype O11 subunit (β-d-FucNAc; Fig. 1). Because wbjE encodes the only remaining hypothetical glycosyltransferase in the O11 biosynthetic gene cluster (12, 13), we hypothesized that WbjE mediated the attachment of α-l-FucNAc to undecaprenyl pyrophosphate-linked β-d-FucNAc during assembly of the O11 O-antigen repeating unit. If this was true, its inactivation would result in an incomplete O-antigen subunit consisting only of β-d-FucNAc. A mutation of wbjE was constructed in vitro by insertion of a gentamicin resistance gene (aacC1). LPS was isolated from PA103, PA103 wzy::aacC1 (13), PA103 wbjA::aacC1 (12), PA103 wbjE::aacC1, and PA103 wbjE::aacC1/pCD207 (Table 1). This LPS was subjected to SDS-PAGE, followed by silver staining, which revealed that PA103 makes a long side chain O-antigen (Fig. 2) that was not detectable in the LPS mutants. O antigen was noted in the PA103 wbjE::aacC1 mutant containing the complementing clone pCD207, indicating that the wbjE mutation was nonpolar (Fig. 2).
FIG. 2.
Analysis of LPS isolated from P. aeruginosa PA103 and derivatives. P. aeruginosa LPS was isolated by the proteinase K digestion method of Hitchcock and Brown (24), followed by hot-phenol extraction with slight modifications. Samples were separated by SDS-PAGE (11), followed silver staining.
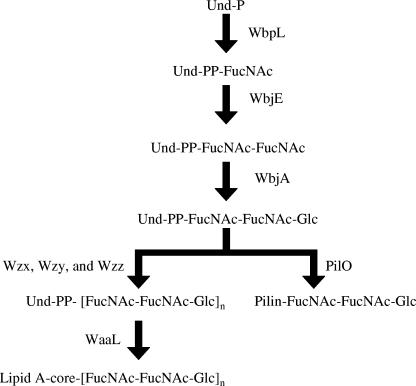
The size of the LPS core was compared in these strains (Fig. 2). LPS from all strains contained a band corresponding to a complete core (Fig. 2). As anticipated, LPS from PA103 and an O-antigen polymerase mutant, PA103 wzy::aacC1, contained core + 1 (13), and PA103 wbjA::aacC1 makes a core and a core + 2/3 (Fig. 2) (12). A small amount of LPS that migrated faster than the core + 2/3 was observed with PA103 wbjE::aacC (Fig. 2). Complementation with the plasmid pCD207 resulted in the expression of a complete core + 1 (Fig. 2). These data suggest a core + 1/3 phenotype for PA103 wbjE::aacC1, indicating that WbjE adds the α-l-FucNAc to undecaprenyl pyrophosphate-bound β-d-FucNAc (Fig. 3) and, as in PA103 wbjA::aacC1, an incomplete O subunit can be transferred to the core (12).
FIG. 3.
Proposed pathway for the assembly of P. aeruginosa serogroup O11 O-antigen and pilin glycosylation. WbpL initiates O-antigen synthesis by the addition of nucleotide-activated N-acetyl fucosamine onto Und-P (41). We propose that WbjE adds the second N-acetyl fucosamine, whereas WbjA adds the terminal glucose to complete the O11 subunit (12). Subsequent O-antigen biosynthesis steps involve the transportation to the periplasm (Wzx), polymerization of O-antigen units (Wzy), chain length determination (Wzz), and linkage of the O antigen to the LPS core (WaaL) (13, 41). We have previously noted that LPS from PA103 wbjA::aacC1 contains a core + 2/3, indicating that the O-antigen subunit need not be complete prior to transfer to the lipid A-core (12). In pilin glycosylation, PilO mediates the transfer of a single, complete O-subunit from Und-PP to pilin (14).
Pilin glycan substrate recognition.
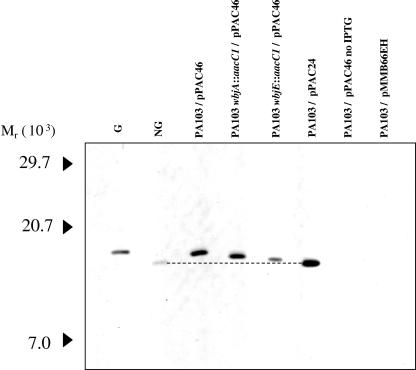
To determine whether the solitary reducing-end sugar in the O-antigen repeat could be transferred to P. aeruginosa 1244 pilin, we expressed pPAC46 (which contains pilAO1244) in PA103 wbjE::aacC1. To further examine the effect of incomplete synthesis of the O-subunit on glycosylation, pPAC46 was also expressed in PA103 wbjA::aacC1. Subsequently, plasmid-encoded pilin was assayed for glycosylation by subjecting extracts of these cells to Western blot analysis using a 1244 pilin-specific MAb as a probe (Fig. 4). Pilin produced by 1244N3/pPAC46 and 1244N3/pPAC24 were used as glycosylated and nonglycosylated standards (6), respectively. This blot revealed that pilin produced by PA103 wbjE::aacC1/pPAC46 had a slightly higher apparent molecular weight than the nonglycosylated pilin standard (Fig. 4). PA103 wbjA::aacC1/pPAC46 synthesized pilin that had a slightly lower apparent molecular weight than both the glycosylated pilin standard and the heterologously glycosylated pilin of PA103/pPAC46 (Fig. 4) (14). However, the apparent molecular weight of PA103wbjA::aacC1/pPAC46 was higher than pilin produced by PA103 wbjE::aacC1/pPAC46 (Fig. 4). This blot suggests that the incomplete O subunits synthesized by the wbjE and wbjA mutants are being utilized for glycosylation, since the difference in the apparent molecular weight of the pilin produced is consistent with the size differences we observed in the LPS (Fig. 2 and 4). It is therefore likely that in PA103 wbjE::aacC1/pPAC46, the pilin glycan is a single β-FucNAc and a disaccharide in PA103 wbjA::aacC1/pPAC46 while, as previously shown, PA103 pPAC46 produces pilin glycosylated with the entire O11 subunit (14) (Fig. 1).
FIG. 4.
Western blot analysis of plasmid-encoded P. aeruginosa 1244 pilin. Cell extracts were prepared from overnight plate cultures in which induction of pilAO1244 (pPAC46) or pilA1244 (pPAC24) was accomplished by the addition of 5 mM IPTG to the medium unless indicated otherwise. The blot was probed with 1244 pilin-specific MAb 6.45. G is glycosylated 1244 pilin, and NG is nonglycosylated 1244 pilin. The dotted line was drawn from the centers of the NG pilin and the PA103/pPAC24 pilin bands (also nonglycosylated) to better illustrate the difference in molecular weight between the pilins shown on this blot.
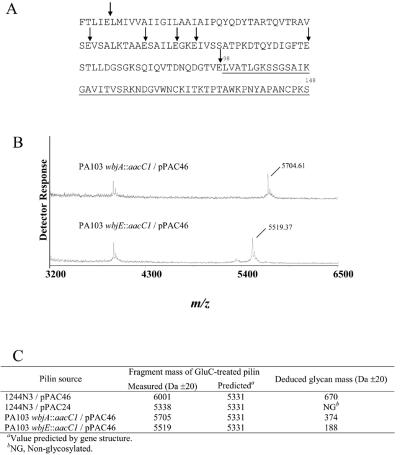
To conclusively determine the glycosylation status of pilin produced by PA103 wbjA::aacC1/pPAC46 and PA103 wbjE::aacC1/pPAC46, purified pilus preparations were treated with the endopeptidase, GluC (10), followed by MALDI-TOF analysis (Fig. 5). The largest pilin fragment produced by GluC digestion (Fig. 5A) consists of the C-terminal 51 residues, which contains the glycosylation site, with a molecular weight predicted from the amino acid sequence to be 5,331 (10). GluC-treated pilins produced by 1244N3/pPAC46 and 1244N3/pPAC24 were used as glycosylated and nonglycosylated controls (6), respectively (spectra not shown; Fig. 5C). The largest fragment mass from the GluC-treated glycosylated pilin control contained covalently bound material with a molecular weight of 670 (Fig. 5C), which is consistent with the value determined for the strain 1244 glycan structure as previously determined (7). The nonglycosylated pilin produced a GluC fragment within the range of the value predicted by the amino acid sequence (Fig. 5C). GluC-treated pilin from PA103 wbjA::aacC1/pPAC46 produced a fragment with a molecular weight of 5,705 (±20) with no signal seen at the mass predicted by the pilin gene, indicating the addition of material with the molecular weight of ∼374 had been covalently linked to the protein (Fig. 5B and C). Endopeptidase treatment of pilin produced by PA103 wbjE::aacC1/pPAC46 produced a fragment with a mass of 5,519 Da (±20), revealing the presence of additional covalently bound material with a molecular weight of ∼188 (Fig. 5B and C). These data support the description of the LPS phenotypes (Fig. 2) and the pilin Western blot analysis (Fig. 4) in reference to the size of the incomplete O-antigen subunits. The additional value of ∼374 for GluC-treated PA103 wbjA::aacC1/pPAC46 pilin is consistent with the molecular weight predicted for the first two carbohydrate moieties of the O11 subunit (12). This suggested that the pilin glycan produced here consists of the disaccharide, α-l-FucNAc-(1→3)-β-d-FucNAc-(1→Ser148). Pilin produced by PA103 wbjE::aacC1/pPAC46 contained a modification with a molecular weight of 188, a value consistent with the first carbohydrate moiety of the O11 subunit, suggesting that this pilin glycan likely consists of β-d-FucNAc-(1→Ser148). The unlabeled peak detected in both spectra (Fig. 5B) was likely generated due to the presence of one of the peptide fragments of the GluC-digested 1244 pilin that did not contain the glycan or was due to a contaminant of the pilus preparation. These results confirm that WbjE adds the second sugar, α-l-N-acetyl fucosamine, to the O11 O-antigen subunit. Furthermore, the first sugar of this subunit, β-d-N-acetyl fucosamine, can be transferred to the core oligosaccharide, as well as pilin, indicating that this sugar contains the substrate recognition features necessary for catalysis by both WaaL and PilO.
FIG. 5.
GluC digestion of 1244 pilin. (A) Amino acid sequence of 1244 pilin in which vertical arrows indicate GluC cleavage sites. The largest GluC fragment (underlined) consists of residues 98 to 148 and is predicted to have a molecular mass of 5,331 Da. This fragment contains the 1244 pilin glycosylation site, Ser148 (10). (B) MALDI-TOF spectra for GluC-treated pilin produced by PA103 wbjA::aacC1/pPAC46 and PA103 wbjE::aacC1/pPAC46 are shown. (C) The deduced glycan mass is listed for each of the pilins subjected to GluC digestion and MALDI-TOF.
DISCUSSION
The results of this study indicated that WbjE catalyzed the addition of the second sugar, α-l-N-acetyl fucosamine, to the O11 subunit (Fig. 3). Because the first sugar of the O11 subunit is identical to the reducing-end moiety of the O7 subunit of strain 1244, and therefore the 1244 pilin glycan (7), pilAO1244 was expressed in the wbjE mutant and pilin was tested for glycosylation to analyze glycan substrate recognition. Pilin produced contained a modification that had a molecular weight consistent with a single β-d-FucNAc. This suggested that the reducing-end moiety of the O-antigen subunit possesses the necessary structures for glycosylation and that the remaining sugars of the glycan precursor are nonessential.
Previous studies have shown that incomplete O subunits can be ligated to the core oligosaccharide (12, 17, 27, 60). The data presented here indicated that a single β-d-FucNAc was transferred to the core oligosaccharide in the wbjE mutant. It is therefore likely that the structural information recognized by WaaL of P. aeruginosa PA103 was also in the reducing sugar of the O repeat. This interpretation is strengthened by previous data indicating that WaaL is capable of ligating a number of different polysaccharides to the core (19), since all of the O repeats tested possessed similar reducing sugars. In addition, gene replacement of glf in an E. coli K-12 O-antigen mutant strain produced an LPS phenotype like that of the PA103 wbjE::aacC1, indicating a similarity between WaaL specificity of both systems (17). These results are consistent with an earlier study showing that Wzx from several different O-antigen systems seemed to have specificity for the sugar at the reducing terminus of Und-PP-linked O subunit (32). Collectively, these data suggest that among related groups of gram-negative bacteria the reducing sugar of the O repeat acts as a universal foundation, in which enzymes involved in the construction of surface antigens are compatible. Therefore, a variety of sugars can be added to this foundation sugar, which is recognized by machinery critical for surface display, such as PilO and WaaL. If this is true, it appears as if evolution has provided an efficient means to ensure maximum surface diversity by providing congruity between products of horizontally transferred genes. For example, if a heterologous O-antigen biosynthetic operon was passed from one bacterium to another, the saccharide produced would likely be compatible with endogenous PilO and/or WaaL and therefore surface expressed, due to the similarities in O-subunit reducing-end sugars. Because waaL and pilO are not located in the same operon as the O-antigen biosynthetic genes (6, 41), there is little likelihood of cohorizontal transfer, underlying the necessity for cohesion with heterologous O antigens. Notably, WaaL mediates the addition of polymerized O antigen and also has specificity for the common antigen, since this enzyme mediates the ligation of both polysaccharides to the core-lipid A (1). However, PilO does not transfer polymerized O antigen or common antigen subunits onto pilin, since each pilin monomer contains a single glycan identical to the O-antigen repeating unit with no evidence of alternate glycoforms (7). Because PilO does not transfer polymerized O antigen to pilin, this suggests that structural attributes of the long side chain O antigen-Und-PP contribute to the loss of affinity for the pilin glycosylation machinery. This may be due to the size of the polymerized O antigen or to an increase of affinity for WaaL. Additional work will be needed for clarification.
To discern structural components of the reducing-end sugar of the O-repeat important for glycan substrate recognition, cloned heterologous O-antigen biosynthetic operons producing structurally distinct reducing end sugars (29) could be expressed in P. aeruginosa 1244, where the pilin produced may be assayed for presence of the heterologous glycan. For example, the O subunit of Burkholderia cepacia serotype B [β-d-Gal-(1→3)-α-d-Fuc] or those of various P. syringae strains (repeats of l- and/or d-Rha in various arrangements) contain sugars at their reducing termini dissimilar from those discussed here (29). The results of such studies may serve to define the glycan specificity of the 1244 pilin glycosylation reaction. Until these experiments are conducted, one cannot dismiss the possibility that the Und-PP carrier alone, or a minimal component of the bound carbohydrate, such as the C-1, or the ether oxygen, contains the substrate recognition properties for the 1244 pilin glycosylation machinery.
A plasmid encoding pilAO1244 was expressed in the wbjE and wbjA mutants and, subsequently, pili produced were extracted and subjected to GluC digestion in which fragments were analyzed by MALDI-TOF. Due to the intrinsic properties of pili, such as extracellular location, fibrous morphology, and solubility in water, pili can be easily purified from cell cultures. In addition, GluC digestion separates pilin into pieces readily detectable by MALDI-TOF analyses, in which the fragment containing the glycan is the largest (10). This methodology not only represents an efficient means of analysis of the pilin glycan molecular weight but also an expedient technique for analysis an organism's O-antigen repeating unit. In this novel approach, pilin is used as an O-subunit “trap.” Therefore, one may analyze the repeating unit apart from the core and lipid A, a previously unattainable endeavor. The sugars of the glycan could be sequenced and further analyzed by using procedures previously described (7). Alternative means to analyze O antigen include acidic and alkaline degradation of extracted LPS, processes that do not disrupt the core-O-polysaccharide bond, and could potentially chemically modify the O antigen (28).
Previously, LPS biosynthesis and eukaryotic protein glycosylation have been hypothesized to have a common evolutionary origin (4, 5) based on similarities between the eukaryotic dolichol and prokaryotic Und-P carrier lipids, the metabolic assembly of oligosaccharides, in addition to sequence similarity between eukaryotic and prokaryotic glycosyltransferases. Feldman et al. suggested that the N-linked glycosylation systems of C. jejuni and eukaryotes are homologous processes (18). Similar to the 1244 pilin glycosylation system, the C. jejuni N-linked glycosylation system has shown the capability to utilize O antigen for protein glycosylation in an engineered E. coli strain. When pglB from C. jejuni was coexpressed with different LPS biosynthesis gene clusters in an E. coli LPS mutant (waaL and the rhamnosyl transferase wbbL), production of recombinant AcrA containing covalently bound, full-length O-antigen occurred (18). If the eukaryotic N-linked glycosylation system had evolved from a similar system to that of C. jejuni, it is plausible that 1244 pilin glycosylation represents an ancestral form. This conclusion is based on the intrinsic utilization of the O subunit as the 1244 pilin glycan and the above-mentioned similarities between the glycosylation systems of C. jejuni and P. aeruginosa 1244.
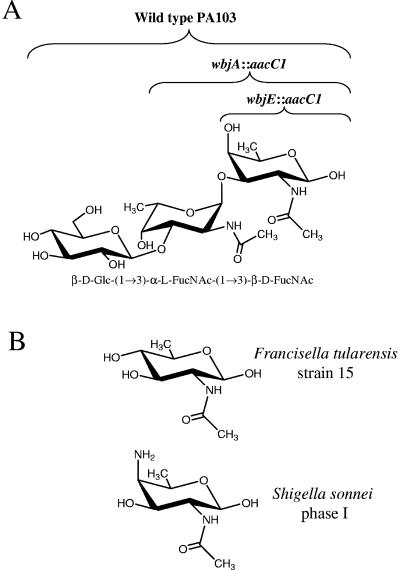
Because O antigen can be covalently linked to protein by both the N-linked glycosylation system of C. jejuni and the P. aeruginosa 1244 pilin glycosylation machinery, these systems could potentially be utilized to produce anti-LPS O-antigen vaccines (10, 18). Previous data have indicated that pure pili from P. aeruginosa 1244 stimulated the production of antibodies in mice against the proteinaceous portion of pilin and the glycan, as well as antibodies against the O antigen of this organism (10). The 1244 glycosylation system may be best applied for vaccine engineering by taking an approach to vaccinate against organisms with few serotypes such as Shigella sonnei, Francisella tularensis, and E. coli serotype O157 (29). As previously mentioned, the E. coli serotype O157 subunit can be utilized as the 1244 pilin glycan, whereas S. sonnei and F. tularensis appear to produce O subunits with reducing-end moieties that are structurally compatible for glycosylation (Fig. 6B). Other than generating a vaccine using pilin as the glycosylated protein, the breadth of protection may be increased by mutation of nonpilin proteins, such as toxoids, to possess the 1244 pilin glycosylation substrate (25). For example, combining expression of the S. sonnei O-antigen gene cluster and mutagenized shiga toxin, allowing compatibility with the 1244 glycosylation machinery, may yield a biologically produced, bivalent antitoxin, anti-LPS vaccine.
FIG. 6.
Saccharide structures. (A) P. aeruginosa serotype O11 repeating unit where brackets above the O subunit indicate saccharide(s) produced by the designated strains. (B) Reducing-end moieties from O subunits of Francisella tularensis strain 15 and Shigella sonnei phase I (29).
Acknowledgments
We thank Paul Kolesar for assistance with the ChemDraw software.
This study was supported by grants from the NIH to P.C. (AI054929) and to J.B.G. (AI35647 and AI37632) and by a grant from the Cystic Fibrosis Foundation to J.B.G. (GOLDBE00P0).
REFERENCES
- 1.Abeyrathne, P., C. Daniels, K. Poon, M. J. Matewish, and J. S. Lam. 2005. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 187:3002-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 68:245-267. [DOI] [PubMed] [Google Scholar]
- 3.Brimer, C. D., and T. C. Montie. 1998. Cloning and comparison of fliC genes and identification of glycosylation in the flagellin of Pseudomonas aeruginosa a-type strains. J. Bacteriol. 180:3209-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bugg, T., and P. Brandish. 1994. From peptidoglycan to glycoproteins: common features of lipid-linked oligosaccharide biosynthesis. FEMS Microbiol. Lett. 119:255-262. [DOI] [PubMed] [Google Scholar]
- 5.Burda, P., and M. Aebi. 1999. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426:239-257. [DOI] [PubMed] [Google Scholar]
- 6.Castric, P. 1995. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology 141:1247-1254. [DOI] [PubMed] [Google Scholar]
- 7.Castric, P., F. J. Cassels, and R. W. Carlson. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 276:26479-26485. [DOI] [PubMed] [Google Scholar]
- 8.Castric, P. A., H. F. Sidberry, and J. C. Sadoff. 1989. Cloning and sequencing of the Pseudomonas aeruginosa 1244 pilin structural gene. Mol. Gen. Genet. 216:75-80. [DOI] [PubMed] [Google Scholar]
- 9.Chia, J., L. Chang, C. Shun, Y. Chang, and J. Chen. 2001. A 60-kilodalton immunodominant glycoprotein is essential for cell wall integrity and the maintenance of cell shape in Streptococcus mutans. Infect. Immun. 69:6987-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comer, J. E., M. A. Marshall, V. J. Blanch, C. D. Deal, and P. Castric. 2002. Identification of the Pseudomonas aeruginosa 1244 pilin glycosylation site. Infect. Immun. 70:2837-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coyne, M., Jr., K. Russell, C. Coyle, and J. B. Goldberg. 1994. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 176:3500-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean, C. R., A. Datta, R. W. Carlson, and J. B. Goldberg. 2002. WbjA adds glucose to complete the O-antigen trisaccharide repeating unit of the lipopolysaccharide of Pseudomonas aeruginosa serogroup O11. J. Bacteriol. 184:323-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, C. R., C. V. Franklund, J. D. Retief, M. J. Coyne, Jr., K. Hatano, D. J. Evans, G. B. Pier, and J. B. Goldberg. 1999. Characterization of the serogroup O11 O-antigen locus of Pseudomonas aeruginosa PA103. J. Bacteriol. 181:4275-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiGiandomenico, A., M. J. Matewish, A. Bisaillon, J. R. Stehle, J. S. Lam, and P. Castric. 2002. Glycosylation of Pseudomonas aeruginosa 1244 pilin: specificity of glycan substrate. Mol. Microbiol. 46:519-530. [DOI] [PubMed] [Google Scholar]
- 15.Dobos, K. M., K. Swiderek, K. H. Khoo, P. J. Brennan, and J. T. Belisle. 1995. Evidence for glycosylation sites on the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. Infect. Immun. 63:2846-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enderle, P., and M. Farwell. 1998. Electroporation of freshly plated Escherichia coli and Pseudomonas aeruginosa cells. BioTechniques 25:954-958. [DOI] [PubMed] [Google Scholar]
- 17.Feldman, M., C. Marolda, M. Monteiro, M. Perry, A. Parodi, and M. Valvano. 1999. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O-antigen assembly is independent of the chemical structure of the O repeat. J. Biol. Chem. 274:35129-35138. [DOI] [PubMed] [Google Scholar]
- 18.Feldman, M., M. Wacker, M. Hernandez, P. Hitchen, C. Marolda, M. Kowarik, H. R. Morris, A. Dell, M. Valvano, and M. Aebi. 2005. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. USA 102:3016-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franco, A., D. Liu, and P. R. Reeves. 1998. The Wzz (Cld) protein in Escherichia coli: amino acid sequence variation determines O-antigen chain length specificity. J. Bacteriol. 180:2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frost, L., and W. Paranchych. 1977. Composition and molecular weight of pili purified from Pseudomonas aeruginosa K. J. Bacteriol. 131:259-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furste, J. P., W. Pansegrau, F. R. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 22.Goldberg, J. B., K. Hatano, G. M. Meluleni, and G. B. Pier. 1992. Cloning and surface expression of Pseudomonas aeruginosa O antigen in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:10716-10720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerry, P., P. Doig, R. A. Alm, D. H. Burr, N. Kinsella, and T. J. Trust. 1996. Identification and characterization of genes required for posttranslational modification of Campylobacter coli VC167 flagellin. Mol. Microbiol. 19:369-378. [DOI] [PubMed] [Google Scholar]
- 24.Hitchcock, P., and T. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horzempa, J., J. E. Comer, S. Davis, and P. Castric. 2006. Glycosylation substrate specificity of Pseudomonas aeruginosa 1244 pilin. J. Biol. Chem. 281:1128-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jennings, M. P., M. Virji, D. Evans, V. Foster, Y. N. Srikhanta, L. Steeghs, P. van der Ley, and E. R. Moxon. 1998. Identification of a novel gene involved in pilin glycosylation in Neisseria meningitidis. Mol. Microbiol. 29:975-984. [DOI] [PubMed] [Google Scholar]
- 27.Klena, J., and C. Schnaitman. 1993. Function of the rfb gene cluster and the rfe gene in the synthesis of O antigen by Shigella dysenteriae 1. Mol. Microbiol. 9:393-402. [DOI] [PubMed] [Google Scholar]
- 28.Knirel, Y. A. 1990. Polysaccharide antigens of Pseudomonas aeruginosa. Crit. Rev. Microbiol. 17:273-304. [DOI] [PubMed] [Google Scholar]
- 29.Knirel, Y. A., and N. K. Kochetkov. 1994. The structure of lipopolysaccharides of gram-negative bacteria. III. The structure of O-antigens: a review. Biochemistry 59:1325-1383. [Google Scholar]
- 30.Liu, P. V. 1973. Exotoxins of Pseudomonas aeruginosa. I. Factors that influence the production of exotoxin A. J. Infect. Dis. 128:506-513. [DOI] [PubMed] [Google Scholar]
- 31.Marino, P., B. McGrath, and M. Osborn. 1991. Energy dependence of O-antigen synthesis in Salmonella typhimurium. J. Bacteriol. 173:3128-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marolda, C., J. Vicarioli, and M. Valvano. 2004. Wzx proteins involved in bioynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 150:4095-4105. [DOI] [PubMed] [Google Scholar]
- 33.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 34.McGrath, B., and M. Osborn. 1991. Location of the terminal steps of O-antigen synthesis in Salmonella typhimurium. J. Bacteriol. 173:649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mescher, M. F., J. L. Strominger, and S. W. Watson. 1974. Protein and carbohydrate composition of the cell envelope of Halobacterium salinarum. J. Bacteriol. 120:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulford, C., and M. Osborn. 1983. An intermediate step in translocation of lipopolysaccharide to the outer membrane of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 80:1159-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parge, H. E., K. T. Forest, M. J. Hickey, D. E. Christensen, E. D. Getzoff, and J. A. Tainer. 1995. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378:32-38. [DOI] [PubMed] [Google Scholar]
- 38.Perry, M., and L. MacLean. 1986. Structure of the O-chain polysaccharide of the phenol-phase soluble lipopolysaccharide of Escherichia coli O157:H7. Biochem. Cell Biol. 64:21-28. [DOI] [PubMed] [Google Scholar]
- 39.Raetz, C., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramphal, R., L. Koo, K. S. Ishimoto, P. A. Totten, J. C. Lara, and S. Lory. 1991. Adhesion of Pseudomonas aeruginosa pilin-deficient mutants to mucin. Infect. Immun. 59:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocchetta, H. L., L. L. Burrows, and J. S. Lam. 1999. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63:523-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruvkun, G., and F. M. Ausubel. 1981. A general method for site-directed mutagenesis in prokaryotes. Nature 289:85-88. [DOI] [PubMed] [Google Scholar]
- 43.Schirm, M., A. Soo, J. Aubry, P. Austin, P. Thibault, and S. M. Logan. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48:1579-1592. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt, M. A., L. Riley, and I. Benz. 2003. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 11:554-561. [DOI] [PubMed] [Google Scholar]
- 45.Schweizer, H. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-112. [DOI] [PubMed] [Google Scholar]
- 46.Schweizer, H. 1993. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 47.Schweizer, H., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 138:15-22. [DOI] [PubMed] [Google Scholar]
- 48.Sidberry, H., B. Kaufman, D. Wright, and J. Sadoff. 1985. Immunoenzymatic analysis by monoclonal antibodies of bacterial lipopolysaccharides after transfer to nitrocellulose. J. Immunol. Methods 76:299-305. [DOI] [PubMed] [Google Scholar]
- 49.Silipigni-Fusco, J. 1987. Studies on the role of somatic pili as virulence and immunity factors in the pathogenicity of Pseudomonas aeruginosa. PhD thesis. University of Pittsburgh, Pittsburgh, Pa.
- 50.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 51.Smedley, J., III, E. Jewell, J. Horzempa, J. Roguskie, J. Syboldt, D. B. Stolz, and P. Castric. 2005. Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. Infect. Immun. 73:7922-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spiro, R. 2002. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12:43R-56R. [DOI] [PubMed] [Google Scholar]
- 53.Stephenson, A., H. Wu, J. Novak, M. Tomana, K. Mintz, and P. Fives-Taylor. 2002. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol. Microbiol. 43:147-157. [DOI] [PubMed] [Google Scholar]
- 54.Stimson, E., M. Virji, K. Makepeace, A. Dell, H. R. Morris, G. Payne, J. R. Saunders, M. P. Jennings, S. Barker, M. Panico, I. Blench, and E. R. Moxon. 1995. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 17:1201-1214. [DOI] [PubMed] [Google Scholar]
- 55.Sylvestre, P., E. Coutre-Tosi, and M. Mock. 2002. A collagen-like surface glycoprotein component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 56.Szymanski, C., S. M. Logan, D. Linton, and B. Wren. 2003. Campylobacter: a tale of two protein glycosylation systems. Trends Microbiol. 11:233-238. [DOI] [PubMed] [Google Scholar]
- 57.Szymanski, C., and B. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. 3:225-237. [DOI] [PubMed] [Google Scholar]
- 58.Szymanski, C., R. Yao, C. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 59.Tsai, C., and C. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, L., J. Radziejewska-Lebrecht, D. Krajewska-Pietrasik, P. Toivanen, and M. Skurnik. 1997. Molecular and chemical characterization of the lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O:8. Mol. Microbiol. 23:63-76. [DOI] [PubMed] [Google Scholar]